Abstract
We report here protein sequences of fossil hominids, from two Neanderthals dating to ≈75,000 years old from Shanidar Cave in Iraq. These sequences, the oldest reported fossil primate protein sequences, are of bone osteocalcin, which was extracted and sequenced by using MALDI-TOF/TOF mass spectrometry. Through a combination of direct sequencing and peptide mass mapping, we determined that Neanderthals have an osteocalcin amino acid sequence that is identical to that of modern humans. We also report complete osteocalcin sequences for chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla gorilla) and a partial sequence for orangutan (Pongo pygmaeus), all of which are previously unreported. We found that the osteocalcin sequences of Neanderthals, modern human, chimpanzee, and orangutan are unusual among mammals in that the ninth amino acid is proline (Pro-9), whereas most species have hydroxyproline (Hyp-9). Posttranslational hydroxylation of Pro-9 in osteocalcin by prolyl-4-hydroxylase requires adequate concentrations of vitamin C (l-ascorbic acid), molecular O2, Fe2+, and 2-oxoglutarate, and also depends on enzyme recognition of the target proline substrate consensus sequence Leu-Gly-Ala-Pro-9-Ala-Pro-Tyr occurring in most mammals. In five species with Pro-9–Val-10, hydroxylation is blocked, whereas in gorilla there is a mixture of Pro-9 and Hyp-9. We suggest that the absence of hydroxylation of Pro-9 in Pan, Pongo, and Homo may reflect response to a selective pressure related to a decline in vitamin C in the diet during omnivorous dietary adaptation, either independently or through the common ancestor of these species.
Keywords: MALDI-TOF/TOF, dietary adaptation, biomolecular preservation, vitamin C, evolution
Biomolecules rarely survive intact in fossil bone and teeth, yet they potentially provide key information on phylogenetic relationships and adaptations. DNA survival in fossil bones and teeth is especially rare in older samples, or in those from hotter climates (1). However, proteins, and therefore phylogenetically informative protein sequences, do persist in much older samples (2, 3). Of the various bone proteins, osteocalcin has been shown to survive particularly well in fossil bone, as demonstrated recently when a complete sequence of fossil Bison priscus osteocalcin was determined with protein extracted from exceptionally well preserved permafrost bones (4).
The protein itself is a small (Mr of 5.9 kDa), negatively charged (pI of 4.0) molecule that is the second most abundant protein in bone. The role of osteocalcin in bone metabolism is uncertain, but it has α-helical domains forming a tightly packed charged molecule that coordinates Ca2+ at the surface of the hydroxyapatite-like lattice of bone mineral crystals (5, 6). In all vertebrate species so far investigated, the osteocalcin protein sequence is highly conserved within the central portion of the molecule, containing the three γ-carboxyglutamic acid (Gla) residues and Ca2+ binding sites (Fig. 1) (6–8), whereas the N terminus displays considerable variation and is the most genetically informative region of the molecule.
Fig. 1.
Osteocalcin amino acid sequences for modern human (9, 10) and Neanderthal (Shanidar 2 Neanderthal). Chimpanzee (Pan troglodytes, Tai Forest, Ivory Coast), gorilla (G. gorilla gorilla, East Africa), and orangutan (Pongo pygmaeus) sequences are also presented, as are selected data for monkey (Macaca fascicularis) (7) and cow (Bos taurus) (11). γ-Carboxyglutamate residues at positions 17, 21, and 24 are indicated by “γ”; hydroxyproline at position 9 is indicated by “O.” Neanderthal residues shaded in gray were unable to be confirmed on CID MS; however, results of peptide mass mapping of tryptic digests are consistent with the first 19 residues being identical to those in the human protein. The new protein sequence data from the primates and Neanderthal were deposited in the Swiss-Prot and TrEMBL database under accession numbers P84348 (Pan troglodytes), P84349 (G. gorilla gorilla), P84350 (Pongo pygmaeus), and P84351 (Neanderthal).
Using a modified version of the methods described in ref. 4, we have attempted to extract osteocalcin from three Neanderthals (specimens 2, 4, and 6) from Shanidar Cave, Iraq, dating to ≈75,000 years old (12), and one Neanderthal [Vi-76/228 (Vi13.1), layer G3] from Vindija Cave, Croatia, dating to ≈40,000 years old (13). These data allowed comparison with both published modern human osteocalcin sequences and those we produced from archaeological, modern humans, and other primates by using both MALDI-TOF/TOF and Edman sequencing.
Methods
Using the method modified from ref. 4, osteocalcin was extracted and purified from bone powder by using Amprep C18 MiniColumns after demineralization (0.5 M sodium EDTA, pH 8.0, 4 h, 25°C). An Applied Biosystems 4700 Proteomics Analyzer was used for MALDI-TOF/TOF analyses. Because the fossil samples are rare and precious specimens, we used minimal sample sizes (≈55 mg of bone); therefore, the results reported here for the Neanderthals are for one extraction per sample. Chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla gorilla) samples were obtained by H.P. from the Swedish Natural History Museum. De novo sequencing of the samples from the modern species was performed via a combination of peptide mass mapping and collision-induced dissociation (CID) product ion MS of selected peptides after tryptic digestion. For Edman sequencing, chimpanzee (Pan troglodytes) and orangutan (Pongo pygmaeus) specimens were obtained by permit from Yerkes Research Center (Emory University, Atlanta). Osteocalcin was extracted and purified from bone powder as described in ref. 7. Tryptic digestion, HPLC purification of peptides, amino acid analysis, and Edman sequencing followed ref. 7, except that 50- to 500-pmol peptide samples were analyzed on an ABI-477A pulsed-liquid phase protein sequencer (Applied Biosystems).
Results
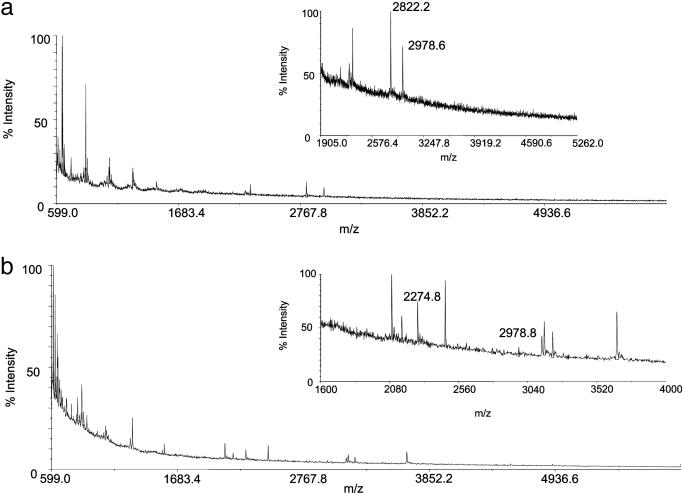
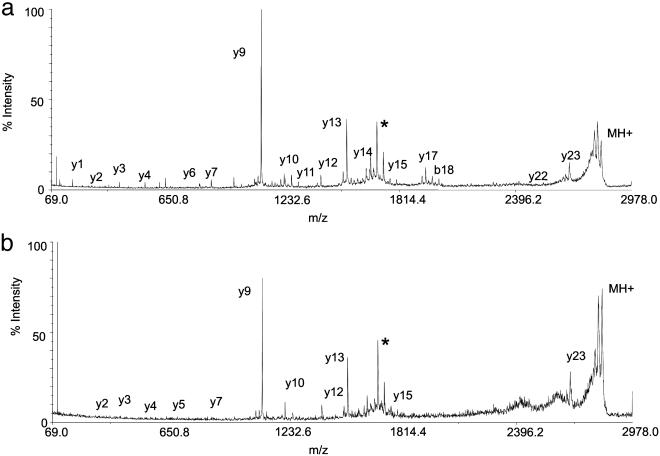
Unlike MS data from the permafrost samples (4), none of the mass spectra of the fossil extracts contained the protonated molecule for osteocalcin. However, after tryptic digestion, masses corresponding to expected tryptic peptides were observed (Fig. 2). Of the Neanderthal samples analyzed, only Shanidar 2 and 6 yielded measurable osteocalcin (Fig. 2). Because of the exceptional limits of detection afforded by the MALDI-TOF/TOF mass spectrometric instrumentation, ion intensities from a tryptic peptide from the Shanidar 2 protein extract were sufficiently high for sequencing after high-energy collision of the ion at m/z 2,822 (Fig. 3). A characteristic pattern of structurally diagnostic fragment ions was observed in product ion spectra from Arg-20–Arg-43 from human (Fig. 3), chimpanzee, and gorilla extracts (data not shown). Submission of the fragment ion data from the Shanidar 2 peptide to a mascot MS/MS ion search of the NCBInr database yielded a match to peptide Arg-20–Arg-43 of human (and other) osteocalcin with an “ions score” of 83 and a probability of <1e–0.5 that this is a random match. For human, chimpanzee, and gorilla samples, CID spectra were obtained from Tyr-1–Arg-19 (human data not shown) and Arg-20–Arg-43 (chimpanzee and gorilla data not shown) peptides to identify the sequences (Figs. 1 and 3, 4, 5, 6).
Fig. 2.
MALDI mass spectra of tryptic digests of bone protein extracts from Neanderthals Shanidar 2 (a) and Shanidar 6 (b). Insets show the expanded regions of the spectra. Only the ion with m/z 2,822.2 from the Shanidar 2 sample was intense enough to perform CID MS, but peaks observed in a MALDI spectrum of a modern human bone protein digest corresponding to osteocalcin tryptic fragments were observed in both Shanidar 2 (m/z 2,822.2, 20–43; m/z 2,978.6, 20–44) and Shanidar 6 (m/z 2,274.8, 1–19; m/z 2,978.8, 20–44) sample spectra.
Fig. 3.
CID product ion spectra of peptide Arg-20–Arg-43 from modern human (a) and Shanidar 2 (b) Neanderthal. Precursor ion, MH+, is represented by the peak at the highest m/z (2,822). Peptide sequence ions produced on cleavage of peptide bonds are labeled by using Roepstorff nomenclature (4). The spectrum from the Neanderthal sample contains a less complete y ion series than that of the modern human, probably as a result of a much smaller quantity of protein surviving in the fossil. The y ion series given by both samples are consistent with the sequence Arg-Glu-Val-Cys-Glu-Leu-Asn-Pro-Asp-Cys-Asp-Glu-Leu-Ala-Asp-His-Ile-Gly-Phe-Gln-Glu-Ala-Tyr-Arg. The peak marked with * is unassigned.
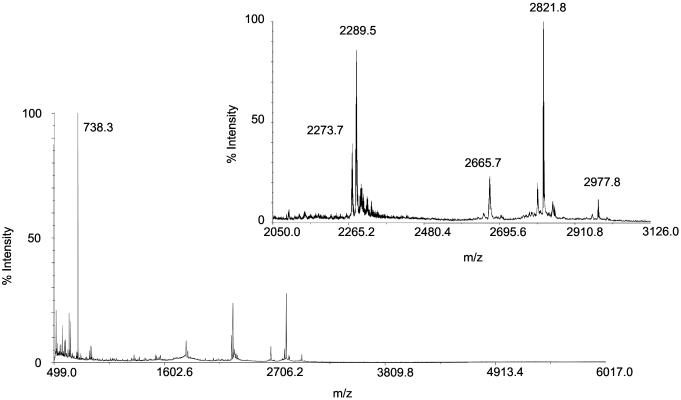
Fig. 4.
MALDI mass spectrum of tryptic digest from gorilla (G. gorilla gorilla) bone protein extract. The Inset shows the expanded region of the spectrum. Tryptic fragments were observed suggesting a mixture of Hyp (m/z 2,289.5) and Pro (m/z 2,273.7) at the ninth position in the 1–19 peptide. Other tryptic peptides observed support the assignment of the proposed sequence (Fig. 1) (m/z 738.3, 44–49; m/z 2,665.7, 21–43; m/z; 2,821.8, 20–43; m/z 2,977.8, 20–44).
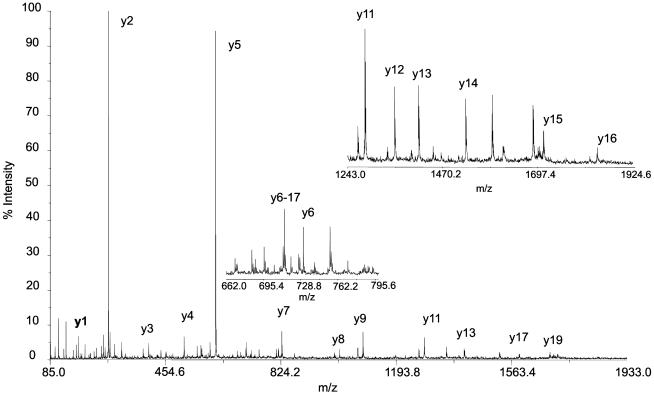
Fig. 5.
CID product ion spectrum of peptide Tyr-1–Arg-19 from chimpanzee (Pan troglodytes). Precursor ion, MH+, is represented by the peak at the highest m/z (2,278.0). Peptide sequence ions produced on cleavage of peptide bonds are labeled by using Roepstorff nomenclature (4); only y ions are labeled. Insets show the expanded region of the spectrum. The sequence given by the y ions series is Tyr-Leu-Tyr-Gln-Trp-Leu-Gly-Ala-Pro-Val-Pro-Tyr-Pro-Asp-Thr-Leu-Glu-Pro-Arg.
Fig. 6.
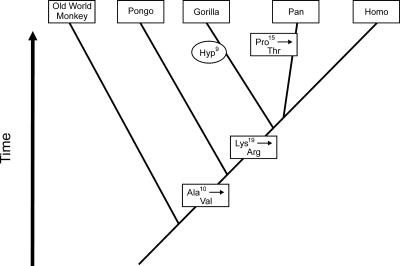
Evolutionary changes in the osteocalcin amino acid sequence in primates. Single-base changes in the codons for Ala-10, Lys-19, and Pro-15 have apparently occurred during the specified intervals, resulting in stable amino acid substitutions in the modern species of the lineages. The presence of Hyp-9 in gorilla may be caused by differences in P4H and cofactor concentrations in this species.
Discussion
The absence of the protonated molecule, and the success of MS after tryptic digestion, suggests that ancient osteocalcin may be partially preserved by condensation into a higher-molecular-weight matrix. The trypsin cleavage product (Arg-20–Arg-43), which spans most of the mineral binding domain (5, 6), was able to be sequenced after CID MS (Figs. 1 and 3). The persistence of the hydrophobic core and calcium binding surface provides direct evidence that adsorption to mineral enhances long-term preservation of ancient proteins. In addition to providing phylogenetic information, MALDI-MS also provides clues as to how the osteocalcin molecule degrades. Osteocalcin peptides extracted from samples from both temperate and permafrost burial environments demonstrate that in fossils with little or no collagen remaining, the first 14 residues of osteocalcin are sometimes missing (C.M.N.-M., unpublished data). These first 14 residues are proline-rich; this finding supports the hypothesis that this region could play a role in binding osteocalcin to collagen (14). Previous attempts to extract collagen from both Shanidar and Vindija Neanderthals proved unsuccessful (M.P.R., unpublished data), and although a peptide with an m/z corresponding to the protonated Tyr-1–Arg-19 fragment observed in spectra of the human sample was detected in the Shanidar 6 sample (but not in Shanidar 2), this was too weak for successful CID MS (Fig. 2). This 1–19 peptide is where most variations between mammal species are observed (8) (Fig. 1), and whereas modern humans and (based on the peptide mass determinations) Neanderthals have identical sequences, gorilla was found to differ by only a single posttranslationally modified amino acid, with a mixture of Hyp and Pro at the ninth position (Figs. 1 and 4).
Chimpanzee shares Pro-9 with orangutan and Homo but has Thr at position 15 (Figs. 1 and 5). Substitution of Thr-15 for Pro-15 is readily explained by a single-base change of C → A at the first position on the 5′ side of the codon for Pro-15 in the chimpanzee osteocalcin gene. This mutation must have occurred in the chimpanzee branch after divergence from the ancestral hominid lineage, because the Pro-15 has been conserved in Homo as well as in primates with earlier divergence: gorilla, orangutan, and Old World monkey, M. fascicularis (Fig. 6). At position 19, Lys-19 occurs in Old World monkey and orangutan but has mutated to Arg-19 in chimpanzee, gorilla, and Homo. The simplest explanation is a single A → G base change in the second position of the Lys codon, yielding the Arg codon, which must have occurred after divergence of orangutan. At position 10, Ala-10 occurs in Old World monkey but changed to Val-10 in orangutan, gorilla, chimpanzee, and Homo. Again, a single-base change in the osteocalcin gene of C → T in the second position of the Ala-10 codon would yield the Val codon (Fig. 6).
The lack of hydroxylation of Pro-9 in chimpanzee, orangutan, and human/Homo osteocalcin is intriguing (i) because it is a posttranslational variation distinct from the evolution of the osteocalcin gene sequence, and (ii) because of the distinctly different properties of the two classes of prolyl-4-hydroxylases (P4Hs) that may target osteocalcin. P4Hs (EC 1.14.11.2) are discriminated by their protein substrate preferences, Km and kinetic properties, and subcellular localization. P4Hs all require molecular O2, l-ascorbic acid, 2-oxoglutarate, and Fe2+ to catalyze the hydroxylation of Pro in peptides (15, 16). Collagen P4Hs (Col-P4Hs) act in the endoplasmic reticulum to modify nascent procollagen chains. The tetrameric enzyme comprises two α subunits (P4HA1, P4HA2, or P4HA3) and two β subunits (P4HB/protein disulfide isomerase). Hydroxyproline accounts for ≈10% of the total amino acid composition of type I collagen and is essential for stability of the collagen triple helix. HIF-1α (hypoxia-inducible factor 1α) P4Hs (HIF-P4Hs) are cytoplasmic and nuclear, targeting Pro-524 and Pro-402 in HIF-1α (17–19). Hydroxyproline at one or both of these sites facilitates binding to the von Hippel–Lindau (VHL) protein that then initiates the ubiquitination and proteasome-mediated degradation of HIF-1α (20). HIF-P4Hs serve as critical sensors for the concentration of dissolved atmospheric molecular O2 because of their poor affinity for O2 (Km = 230–250 μM); in contrast, Col-P4H1 binds O2 strongly (Km = 40 μM) (16). Under hypoxic conditions, HIF-P4Hs are inactive and thus the nonhydroxylated form of HIF-1α accumulates in cells and activates the transcription of genes essential for cell and tissue responses to hypoxia.
The peptide substrate specificities for the HIF-P4Hs (Leu-X-X-Leu-Ala-Pro*-Ala/Tyr) (16, 19) suggest that they may hydroxylate Pro-9 in osteocalcin (Leu-Gly-Ala-Pro*-Ala/Val) (Fig. 1) more efficiently than the Col-P4Hs, which target the Gly-X-Pro*-Gly sequence (15). Both P4H classes are expressed in osteoblasts where osteocalcin and collagen type I are synthesized, but the subcellular localization of osteocalcin hydroxylation has not been established. Interestingly, 14 of 18 vertebrate osteocalcins with a conserved Pro-9 are hydroxylated to Hyp-9, and 13 of these 14 have the sequence Hyp-9–Ala-10 (refs. 4, 7, and 8; P.V.H., unpublished data), whereas gorilla is the lone exception with Hyp-9–Val-10. In all four species where Pro-9 is not hydroxylated, the Pro-9–Val-10 sequence occurs. Thus, hydroxylation of Pro-9 is strongly favored by Ala-10 and resisted by Val-10, pointing to properties of the responsible P4H enzyme. The known sensitivity of proline hydroxylation to concentrations of O2 (for the HIF-P4Hs) and l-ascorbic acid (for all P4Hs) results in partial hydroxylation and Pro/Hyp mixtures at the target sites (16, 21). The Pro-9/Hyp-9 mixture observed in gorilla osteocalcin (Fig. 4) may be explained in this way.
l-gulonolactone oxidase, a microsomal enzyme that catalyzes the terminal step in the biosynthesis of l-ascorbic acid is missing in most primates. Consequently, these organisms are prone to scurvy if the concentration of vitamin C in the diet falls. Paleopathological markers for the diagnosis of scurvy in ancient human skeletons have been described (22), but there is no paleopathological evidence of scurvy (or any other vitamin-specific dietary deficiency) among the Neanderthals or other fossil hominids. A molecule of l-ascorbic acid is consumed for each hydroxylation event (23). With an omnivorous dietary adaptation, especially a shift toward greater carnivory, there are periods when dietary vitamin C would either not be available or present only in reduced amounts. Recent humans, chimpanzees, and orangutans are omnivores; therefore, this difference in the posttranslational hydroxylation of osteocalcin compared to the herbivorous gorilla may relate to increased selective pressure to limit hydroxylation to counteract periods of low dietary vitamin C. Such a dietary adaptation could be inherited from the common chimpanzee/human ancestor or could have arisen independently. Analysis of Hyp in osteocalcin and collagen of other Homo species may resolve this.
The data presented here illustrate the great potential of fossil protein sequences to provide behavioral, phenotypic, and phylogenetic information, especially when compared to related extant species. Now that we have demonstrated that it is possible to extract and sequence intact fossil proteins from fossil hominids, it opens the possibility of sequencing osteocalcin and other proteins from other, earlier, hominid species to better understand the palaeobiological patterning of the hominid line.
Acknowledgments
We thank the late Maja Paunović, Bo Fernholm, and the Swedish Natural History Museum for samples; Jerry Thomas (University of York) for technical assistance; and two referees for their helpful and insightful comments on the paper. This work was supported primarily by U.K. Natural Environment Research Council Environmental Factors in the Chronology of Human Evolution and Dispersal Program Grant NER/T/S/2002/00477 (to M.P.R., C.M.N.-M., and M.J.C.). Additional support was provided by the Max Planck Gesellschaft (C.M.N.-M. and M.P.R.), the Wellcome Trust Bioarchaeology Program (C.M.N.-M. and M.P.R.), the Canadian Natural Science and Engineering Research Council (H.P.), the Canadian Foundation for Innovation (H.P.), the Analytical Chemistry Trust Fund (J.E.T.-O.), the Royal Society of Chemistry Analytical Division (J.E.T.-O.), the British Engineering and Physical Sciences Research Council (J.E.T.-O.), and the National Institutes of Health (P.V.H.).
Author contributions: C.M.N.-M., M.P.R., and E.T. designed research; C.M.N.-M. and P.V.H. performed research; C.M.N.-M. and P.V.H. contributed new reagents/analytic tools; C.M.N.-M., M.P.R., P.V.H., J.E.T.-O., and M.J.C. analyzed data; C.M.N.-M., M.P.R., P.V.H., J.E.T.-O., E.T., P.B.P., I.K., H.P., and M.J.C. wrote the paper; M.P.R. is the grant holder; E.T. contributed Shanidar fossil samples; P.B.P. and I.K. contributed Vindija fossil samples; and H.P. contributed modern primate samples.
Abbreviations: CID, collision-induced dissociation; HIF-1α, hypoxia-inducible factor 1α; P4H, prolyl-4-hydroxylase;
Data deposition: The sequences reported in this paper have been deposited in the Swiss-Prot and TrEMBL database [accession nos. P84348 (Pan troglodytes), P84349 (Gorilla gorilla gorilla), P84350 (Pongo pygmaeus), and P84351 (Neanderthal)].
References
- 1.Smith, C. I., Chamberlain, A. T., Riley, M. S., Stringer, C. & Collins, M. J. (2003) J. Hum. Evol. 45, 203–217. [DOI] [PubMed] [Google Scholar]
- 2.Abelson, P. H. (1954) Carnegie Inst. Washington Year Book 53, 97–101. [Google Scholar]
- 3.Glimcher, M. J., Cohan-Solal, L., Kossiva, D. & de Ricqles, A. (1990) Paleobiology 16, 219–232. [Google Scholar]
- 4.Nielsen-Marsh, C. M., Ostrom, P. H., Gandhi, H., Shapiro, B., Cooper, A., Hauschka, P. V. & Collins, M. J. (2002) Geology 30, 1099–1102. [Google Scholar]
- 5.Hoang, Q. Q., Sicheri, F., Howard, A. J. & Yang, D. S. C. (2003) Nature 425, 977–980. [DOI] [PubMed] [Google Scholar]
- 6.Hauschka, P. V. & Carr, S. A. (1982) Biochemistry 21, 2538–2547. [DOI] [PubMed] [Google Scholar]
- 7.Hauschka, P. V., Carr, S. A. & Biemann, K. (1982) Biochemistry 21, 638–642. [DOI] [PubMed] [Google Scholar]
- 8.Hauschka, P. V., Lian, J. B., Cole, D. E. & Gundberg, C. M. (1989) Physiol. Rev. 69, 990–1047. [DOI] [PubMed] [Google Scholar]
- 9.Poser, J. W., Esch, F. S., Ling, N. C. & Price, P. A. (1980) J. Biol. Chem. 255, 8685–8691. [PubMed] [Google Scholar]
- 10.Celeste, A. J., Rosen, V., Buecker, J. L., Kriz, R., Wang, E. A. & Wozney, J. M. (1986) EMBO J. 5, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price, P. A., Poser, J. W. & Raman, N. (1976) Proc. Natl. Acad. Sci. USA 76, 3374–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinkaus, E. (1983) The Shanidar Neandertals (Academic, New York).
- 13.Wolpoff, M. H., Smith, F. H., Malez, M., Radovèić, J. & Rukavina, D. (1981) Am. J. Phys. Anthropol. 54, 499–545. [Google Scholar]
- 14.Prigodich, R. V. & Vesely, M. R. (1997) Arch. Biochem. Biophys. 345, 339–341. [DOI] [PubMed] [Google Scholar]
- 15.Kukkola, L., Hieta, R., Kivirikko, K. I. & Myllyharju, J. (2003) J. Biol. Chem. 278, 47685–47693. [DOI] [PubMed] [Google Scholar]
- 16.Hirsila, M., Koivunen, P., Gunzler, V., Kivirikko, K. I. & Myllyharju, J. (2003) J. Biol. Chem. 278, 30772–30780. [DOI] [PubMed] [Google Scholar]
- 17.Epstein, A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- 18.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S. & Kaelin, W. G., Jr. (2001) Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- 19.Bruick, R. K. & McKnight, S. L. (2001) Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- 20.Min, J. H., Yang, H., Ivan, M., Gertler, F., Kaelin, W. G., Jr., & Pavletich, N. P. (2002) Science 296, 1886–1889. [DOI] [PubMed] [Google Scholar]
- 21.Gallop, P. M. & Paz, M. A. (1975) Physiol. Rev. 55, 418–487. [DOI] [PubMed] [Google Scholar]
- 22.Ortner, D. J., Butler, W., Cafarella, J. & Milligan, L. (2001) Am. J. Phys. Anthropol. 114, 343–351. [DOI] [PubMed] [Google Scholar]
- 23.Wu, M., Moon, H. S., Pirskanen, A., Myllyharju, J., Kivirikko, K. I. & Begley, T. P. (2000) Bioorg. Med. Chem. Lett. 10, 1511–1514. [DOI] [PubMed] [Google Scholar]