Abstract
In eukaryotes, precursor mRNA (pre-mRNA) splicing removes non-coding intron sequences to produce mature mRNA. This removal is controlled in part by RNA-binding proteins that regulate alternative splicing decisions through interactions with the splicing machinery. RNA binding motif protein 25 (RBM25) is a putative splicing factor strongly conserved across eukaryotic lineages. However, the role of RBM25 in global splicing regulation and its cellular functions are unknown. Here we show that RBM25 is required for the viability of multiple human cell lines, suggesting that it could play a key role in pre-mRNA splicing. Indeed, transcriptome-wide analysis of splicing events demonstrated that RBM25 regulates a large fraction of alternatively spliced exons throughout the human genome. Moreover, proteomic analysis indicated that RBM25 interacts with components of the early spliceosome and regulators of alternative splicing. Previously, we identified an RBM25 species that is mono-methylated at lysine 77 (RBM25K77me1), and here we used quantitative mass spectrometry to show that RBM25K77me1 is abundant in multiple human cell lines. We also identified a region of RBM25 spanning Lys-77 that binds with high affinity to serine- and arginine-rich splicing factor 2 (SRSF2), a crucial protein in exon definition, but only when Lys-77 is unmethylated. Together, our findings uncover a pivotal role for RBM25 as an essential regulator of alternative splicing and reveal a new potential mechanism for regulation of pre-mRNA splicing by lysine methylation of a splicing factor.
Keywords: alternative splicing, protein methylation, protein-protein interaction, proteomics, RNA splicing
Introduction
In eukaryotes, processing of precursor mRNAs (pre-mRNA)5 by the spliceosome to remove non-coding intron sequences and produce mRNA is a fundamental step in gene expression (1). Many transcripts are alternatively spliced, allowing a relatively limited repertoire of genes to encode a wider range of distinct proteins (2, 3). Splicing decisions are governed in part by RNA-binding proteins that promote or suppress alternative splice sites through interactions with splicing machinery (4, 5). Here we focus on the genome-wide role of the putative splicing factor RNA binding motif 25 (RBM25, also called RED120). RBM25 is an RNA-binding protein that is widely conserved across eukaryotic lineages (6). It contains an N-terminal RNA-binding motif domain (RRM), a central glutamate/arginine-rich sequence (ER-rich domain), and a C-terminal PWI domain. In yeast, the RBM25 homolog is a constitutive member of the U1 small nuclear ribonucleoprotein (7), whereas in humans, it co-purifies with intact spliceosomes (8). Human RBM25 is reported to promote apoptosis in HeLa cells by regulating the balance of pro- and anti-apoptotic transcripts of the gene BCL2L1 (9) and to cause truncation of the cardiac voltage-gated Na+ channel encoded by SCN5A during heart failure (10). Collectively, these studies suggest that human RBM25 functions as a splicing factor.
In an earlier study, we observed that RBM25 is mono-methylated at lysine 77 (RBM25K77me1) (11), just N-terminal to the RRM, suggesting that methylation may affect the assembly or function of splicing complexes containing RBM25. Posttranslational modifications of splicing factors, such as phosphorylation and arginine methylation, have been shown to play a central role in regulating pre-mRNA splicing (12–14). Beyond these posttranslational modifications, we and others have shown recently that protein components of the mRNA splicing machinery are frequently methylated on lysine residues, although the functional consequences of these methylation events are not known (11, 15, 16). In other contexts, lysine methylation regulates protein interactions that govern chromatin dynamics, epigenetic programming, and canonical cell signal transduction pathways (17). We have therefore hypothesized that lysine methylation of the splicing machinery represents an important and still unrecognized regulatory process in pre-mRNA splicing.
The goal of this study was to characterize RBM25 as an initial step toward exploring regulatory mechanisms linking lysine methylation of pre-mRNA splicing factors to control of splicing activity. First, we found that RBM25 is required for proliferation in multiple human cell lines, suggesting a central biological role for the protein. High-throughput sequencing revealed that RBM25 promotes inclusion of at least 20% of alternatively spliced cassette exons throughout the human genome. These results argue that RBM25 is a global splicing factor and that its activity is required for fundamental cellular functions. To begin exploring molecular mechanisms of RBM25 activity, we undertook a detailed proteomic analysis of its protein interactions. These experiments showed that RBM25 interacts with core elements of the early spliceosome and with splicing regulators such as serine/arginine-rich splicing factors (SR proteins) and heterogeneous nuclear ribonucleoproteins (hnRNPs). Because lysine methylation often regulates protein interactions, we used quantitative mass spectrometry in combination with peptide interaction experiments to determine whether the sequence around Lys-77 can mediate interactions with RBM25-interacting proteins and to test whether Lys-77 methylation affects these interactions. We found that the splicing factor SRSF2 (also called SC35) binds specifically to the sequence surrounding Lys-77 and that methylation abrogates the interaction. Because SRSF2 and other SR proteins are central regulators of constitutive and alternative splicing (18), we hypothesize that methylation may tune RBM25 activity by affecting its ability to interact directly with SRSF2 during exon definition and pre-mRNA processing.
Results
The spliceosome-associated factor RBM25 is methylated at lysine 77 and required for proliferation in diverse human cell lines
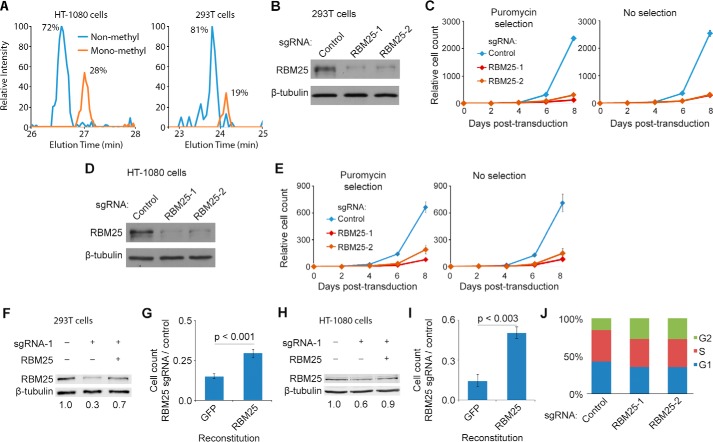
We reported previously that endogenous RBM25 is mono-methylated at Lys-77 (11). To establish the abundance of this modification, we used immunoprecipitation (IP) followed by liquid chromatography and tandem mass spectrometry (LC/MS-MS). We found that the RBM25K77me1 species represents 20–30% of total RBM25 in the 293T and HT1080 cell lines (Fig. 1A). Next, we performed knockdown and reconstitution experiments to test whether RBM25 contributes to the growth or survival of these cells. Depletion of RBM25 using the CRISPR/Cas9 system (19) with two independent small guide RNAs caused a dramatic decrease in proliferation of both the 293T and HT1080 cell lines, whereas reconstitution with ectopic RBM25 restored cell growth (Fig. 1, B–I). Depletion of RBM25 also impaired growth in the PaCa-3 prostate cancer cell line (supplemental Fig. S1). Cell cycle analysis showed that RBM25 depletion in 293T cells causes an accumulation of cells in G2, suggesting impaired ability to undergo mitosis (Fig. 1J), whereas no increase in apoptosis or cell death was observed. Together, these results argue that RBM25 is required for multiple cell types to maintain their proliferative capacity.
Figure 1.
RBM25 is methylated in human cells and required for cell growth in culture. A, RBM25 was immunoprecipitated from HT-1080 and 293T cells, and Lys-77 methylation was measured by in-gel digestion with chymotrypsin followed by LC/MS-MS. Chromatographs show the relative intensity of non-methyl and mono-methylated forms of the peptide containing lysine 77 (amino acids 75–90, KAKENDENC*GPTTTVF, non-methyl and mono-methyl monitored at m/z 604.2807 and 608.9526, respectively). The asterisk indicates carbamidomethylation. B, RBM25 levels in 293T cells were measured by Western blot 4 days after transduction with the CRISPR/Cas9 system. C, growth of transduced 293T cells was tracked for 8 days either with puromycin selection (left panel) or without selection (right panel). Error bars indicate mean ± S.E. (n = 3). D and E, B and C repeated in HT-1080 cells. F, RBM25 knockdown and reconstitution in 293T cells was measured by Western blotting with near-IR fluorescent secondary antibodies. Densitometry indicates RBM25 expression relative to endogenous, normalized to β-tubulin. G, 293T cells expressing exogenous RBM25 or GFP (negative control) were transduced with CRISPR/Cas9 and sgRNA against RBM25 or a non-targeting control. Cells were passaged every 2 days and counted 8 days after transduction (S.E., n = 4, Student's t test). H and I, F and G repeated in HT-1080 cells. J, cell cycle distribution of 293T cells was measured by propidium iodide stain and flow cytometry 4 days after transduction with CRISPR/Cas9. Values are the average of three replicates.
RBM25 is a global splicing regulator that promotes inclusion of target exons
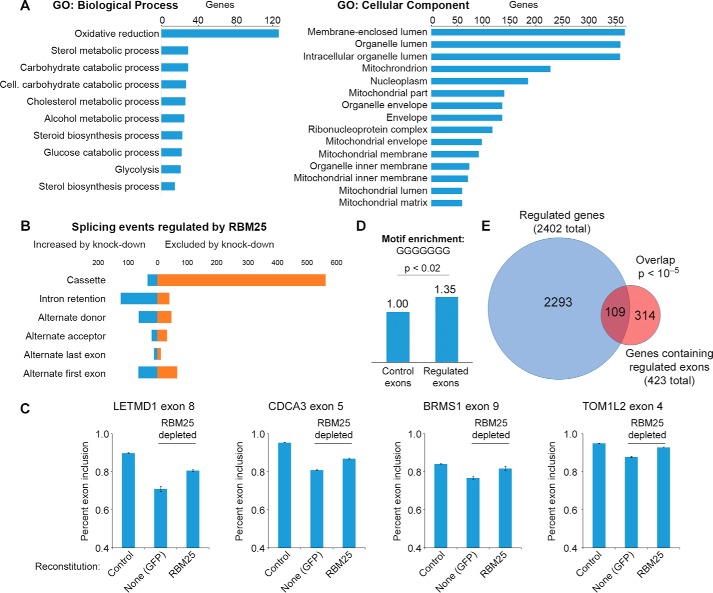
RBM25 has been shown previously to co-purify with the spliceosome and to impact splicing of the gene BCL2L1 (6, 9). However, it is not known whether RBM25 is a general regulator of splicing. To investigate genome-wide functions of RBM25, high-throughput RNA sequencing (RNA-Seq) was used to determine the consequence of RBM25 depletion on transcript levels and splicing in 293T cells. We measured overall gene expression using Kallisto (20) and performed differential expression analysis with Sleuth (21). Setting a 5% threshold for the false discovery rate (FDR), there were 2402 differentially expressed genes of 14,190 genes measured (1423 down-regulated by RBM25 knockdown and 979 up-regulated; supplemental Table S1). Of these, 108 were down-regulated and 86 up-regulated by at least a factor of two. Using the DAVID Bioinformatics Resources (22, 23) to compare gene ontology (GO) terms between regulated genes and all expressed genes, we found that RBM25-regulated genes are enriched for GO terms related to metabolic processes and mitochondrial components (Fig. 2A). The only enriched GO term for molecular function was “cofactor binding.” Based on these results, we conclude that RBM25 impacts gene expression widely throughout the genome and impacts pathways involved in cellular metabolism.
Figure 2.
RBM25 is a global regulator of transcription and alternatively spliced exons. A, RNA-Seq was used to identify genes affected by RBM25 knockdown relative to non-targeting sgRNA. Enriched GO terms were identified using DAVID (Benjamini-Hochberg–corrected p < 0.05). B, splice events regulated by RBM25 knockdown were identified by processing sequencing data with JuncBASE and applying thresholds for 5% FDR and a minimum 10% change in use of the exon or alternative splicing event (e.g. 80% of transcripts to 70% of transcripts). C, endogenous RBM25 was depleted in 293T cells expressing a control vector (GFP) or RBM25, as shown in Fig. 1E. Real-time PCR was used to measure exon inclusion at eight RBM25 target exons identified by RNA-Seq (four shown here and four shown in supplemental Fig. S2). Error bars show S.E., n = 4. D, analysis of motif enrichment (25) was used to compare the frequency of motifs resembling GGGGGGG in exons positively regulated by RBM25 to a background of alternatively spliced exons. E, overlap of differentially expressed genes and genes containing exons positively regulated by RBM25. Statistical significance of the overlap was calculated using Fisher's exact test.
Next, to examine the effect of RBM25 depletion on pre-mRNA splicing at a genomic scale, we used JuncBASE (24) to measure the “percent spliced in” (PSI) of known and novel splicing junctions. Statistically significant differences between wild-type and RBM25-depleted samples were identified by comparing the number of reads supporting inclusion and exclusion for each splice event using Fisher's exact test, filtering to control the FDR at less than 5%, and selecting biologically meaningful differences by requiring the average PSI to change by at least 10% between groups (see “Experimental procedures”). Overall, this analysis identified 1068 splice events regulated by RBM25 (supplemental Tables S2 and S3). Approximately half of these events were classified as increased skipping of exon cassettes upon RBM25 knockdown (Fig. 2B), suggesting that RBM25 is normally a splicing activator. We note that many of the down-regulated exons are not annotated as alternatively spliced, indicating that RBM25 supports inclusion of alternative exons as well as “constitutive” exons that may have relatively weak splicing signals or be alternatively spliced in specific biological contexts. We also observed a modest increase in intron retention, which is a splicing defect that can accompany exon skipping. We next used real-time PCR to test exon skipping in independently prepared 293T cells with RBM25 knockdown and reconstitution. We observed increased skipping at eight of eight exons selected from the genome-wide analysis of RBM25 regulation (Fig. 2 and supplemental Fig. S2). These differences are specifically due to loss of RBM25, as reconstitution of RBM25 in the knockdown cells restored splicing at these eight exons (supplemental Fig. S2). The effect of RBM25 knockdown generalizes to other cell types because the same effects were observed with knockdown and reconstitution in HT-1080 cells (supplemental Fig. S2).
Bind-n-Seq data from the ENCODE Consortium indicates that RBM25 binds to poly(G) sequences (ENCODE experiment ENCSR759QKO). Using analysis of motif enrichment (AME) (25), we found that motifs resembling GGGGGGG are enriched among RBM25-regulated exons relative to randomly selected alternatively spliced exons (Fig. 2D). Specifically, these exons are 35% more likely to contain the motif with up to one mismatch than control exons. These results, along with the Bind-n-Seq data, indicate that RBM25 has considerable flexibility around its optimal target sequence. Using genes containing alternative splicing events as the background set, we did not detect significant enrichment of specific GO terms among RBM25-regulated exons. However, we did observe a significant overlap among genes showing changes both in expression and alternative splicing events upon RBM25 depletion (Fig. 2E).
The set of exons down-regulated by loss of RBM25 represents ∼20% of all alternatively spliced exons that were measured in this experiment. Based on these results, we conclude that RBM25 regulates exon inclusion fidelity to act as a global regulator of pre-mRNA splicing. In addition, RBM25 influences overall transcript levels, likely through its function in regulating exon inclusion, which, in turn, influence a broader gene expression program.
RBM25 interacts with the exon definition splicing machinery
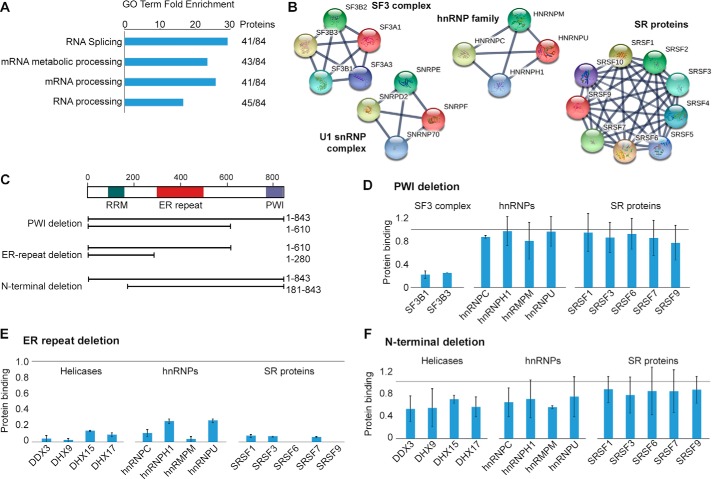
RBM25 is reported to associate with the spliceosome, but its network of protein interactions has not been characterized in detail. Because splicing regulators often act through protein interactions, we set out to determine the RBM25 interactome. To this end, co-IP was performed to enrich interacting proteins of endogenous RBM25 from cell lysate treated with RNase A to disrupt indirect interactions bridged by RNA. We then employed quantitative LC/MS-MS with stable isotopic labeling in cell culture (SILAC) (26) to identify specific binding partners of RBM25. Our analysis identified 84 proteins that are strongly enriched in the RBM25 co-IP relative to the IgG control (supplemental Table S4). Roughly half of these proteins are annotated with GO terms linked to RNA splicing and RNA processing, representing a 20- to 30-fold increase over the background frequency of these terms (Fig. 3A). We visualized the RBM25 interactome using the STRING protein interaction database to locate cliques of interacting proteins (27). In particular, the interactome includes the U1 snRNP complex, responsible for defining the 3′ end of exons; the SF3 complex, which participates in defining the branch point sequence at the 3′ splice site; many SR proteins that participate in defining exons (28); and several hnRNPs (Fig. 3B). These interactions are consistent with a role for RBM25 in establishing, bridging, or stabilizing the splicing commitment complex during exon definition (29). These data point to a model in which RBM25 participates in coordinating assembly of the splicing machinery on its target exons.
Figure 3.
RBM25 interacts with splicing machinery through several protein domains. A, quantitative LC/MS-MS using SILAC was used to compare proteins bound by RBM25 co-IP relative to IgG control. DAVID was used to determine GO terms enriched among RBM25-interacting proteins (selected terms are shown, p < 10−40). B, the STRING protein interaction database was used to visualize the RBM25 interactome. Selected cliques of interacting protein families or complexes are shown. C, RBM25 lacking specific domains were expressed with an N-terminal FLAG tag, and FLAG co-IP with quantitative LC/MS-MS with SILAC was used to measure the effect on protein interactions. D–F, the effect of each domain deletion was measured in duplicate with isotopic labels reversed. Bar plots show the average amount of protein bound by the short form relative to the long form of RBM25 (all normalized to a molar quantity of RBM25), and error bars show high and low values for each pair of measurements. The light gray horizontal line indicates 1:1 binding. Selected protein families are shown.
To further understand how the three domains of RBM25 participate in protein interactions, we systemically deleted each domain and measured changes in protein interactions using the same co-IP strategy and quantitative LC/MS-MS (Fig. 3C). Deletion of the C-terminal PWI domain largely eliminated binding to components of the SF3 complex, but binding to other proteins, including SR proteins and hnRNPs, was unaffected (Fig. 3D and supplemental Table S5). An even shorter fragment containing only the N terminus and RRM (lacking the PWI domain and the ER rich domain) greatly reduced, but did not eliminate, almost all interactions with splicing proteins (Fig. 3E) and was unable to rescue splicing defects results from RBM25 knockdown (supplemental Fig. S3). Deletion of the N-terminal sequence containing Lys-77 and the adjacent RRM modestly weakened many of the protein interactions of RBM25 (Fig. 3F). The ER-rich domain has been reported to be required for RBM25 localization in nuclear speckles (9). Our data indicate that the ER-rich domain is a major determinant of interactions with exon definition factors and components of the early spliceosome. These results suggest that the ER-rich domain is critical for establishing key protein–protein interactions by mediating proper localization of RBM25, directly binding to protein partners, or a combination of the two mechanisms. In contrast, the RRM and PWI domains contribute to but are not are not required for most RBM25 protein interactions, arguing that the RBM25 interactome is established cooperatively through its various domains.
SRSF2 interacts preferentially with a non-methylated RBM25 peptide spanning Lys-77
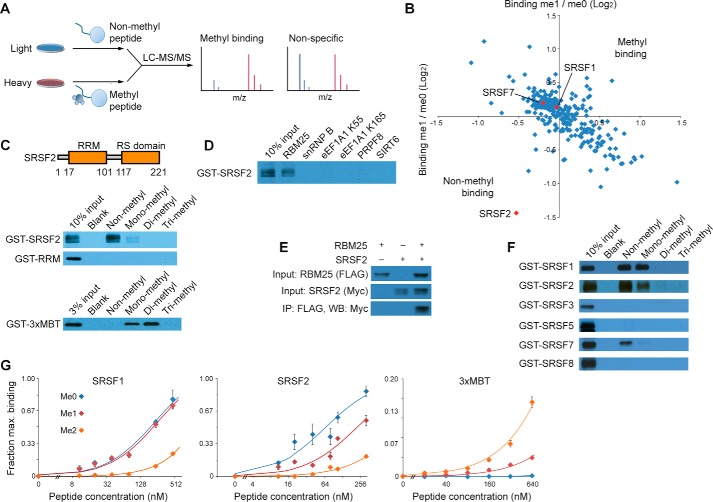
In an effort to characterize the function of RBM25 methylation, we began by investigating whether a known nuclear lysine methyltransferase (KMT) enzyme is responsible for the modification. However, a biochemical activity screen of more than 30 recombinant KMTs did not identify a candidate enzyme that could be physiologically responsible for generating RBM25K77me1 (supplemental Fig. S4). We therefore focused on identifying “reader” proteins able to recognize the methylation state of RBM25. To identify protein interactions that are specifically sensitive to RBM25 methylation, we performed a proteomic screen using biotinylated peptides spanning Lys-77 (amino acids 67–84) and harboring either non-methyl or mono-methyl at Lys-77 (RBM25K77me0 and RBM25K77me1, respectively). SILAC-labeled nuclear extract was incubated with immobilized RBM25K77me0 or RBM25K77me1 peptides, bound proteins were isolated, and quantitative LC/MS-MS was used to identify methyl-sensitive binding proteins (Fig. 4A) (30). The pulldown was performed in duplicate with isotopic labels reversed to control for differences between the light and heavy SILAC-labeled cells. Many RBM25-interacting proteins from earlier co-IP experiments bound to the RBM25 peptide regardless of its methylation state (Fig. 4B). This is consistent with a model where the RBM25 N terminus collaborates with the ER-rich domain to form a network of interactions with the splicing machinery. Although the analysis did not show enrichment of any RBM25K77me1-binding proteins, it did identify the protein SRSF2 as a candidate that binds preferentially to the non-methylated peptide relative to the methylated peptide. We observed the same result in a second independent pair of pulldown and LC/MS-MS experiments (supplemental Table S6).
Figure 4.
SRSF2 binding to the RBM25 Lys-77 peptide is regulated by methylation. A, immobilized non-methyl or methyl peptides centered on RBM25 Lys-77 were used to capture proteins from nuclear extract of 293T cells prepared with SILAC. Bound proteins were identified and quantified using LC/MS-MS. B, relative binding to methyl over non-methyl peptide was measured in two experiments with isotopic labels reversed. Each axis represents one experiment with the ratio of bound protein shown on a log2 scale. Several SR proteins are highlighted in red. C, recombinant SRSF2 or the indicated domain was expressed with N-terminal GST in E. coli and tested for binding to immobilized peptide. Bound proteins were visualized by Western blotting for the GST tag. 3xMBT was used as a positive control for mono- and dimethylated peptides. D, SRSF2 was tested for binding to a panel of non-methylated peptides from proteins involved in transcription, splicing, and translation. E, RBM25 and SRSF2 were expressed by transient transfection in 293T cells with FLAG and Myc tags, respectively. Following FLAG, co-IP bound SRSF2 was measured by Western blotting. F, a panel of SR proteins was tested for binding to immobilized RBM25 peptides. G, SRSF1, SRSF2, and 3xMBT were incubated with varying concentrations of the indicated immobilized RBM25 peptide, and the amount of bound protein was measured by quantitative Western blotting for GST with near-IR fluorescent secondary antibodies. Error bars indicate mean ± S.E. (n = 3).
SRSF2 is the founding member of the SR family of splicing factors that are involved in exon definition and alternative splicing regulation (31, 32). To test whether the interaction with RBM25 peptide is direct, we performed pulldown assays using recombinant SRSF2 and immobilized RBM25 peptide harboring methyl states from 0 to 3 at Lys-77. Consistent with the quantitative proteomic screen, SRSF2 bound to RBM25K77me0 peptide, and the interaction was largely abrogated by mono-methylation at Lys-77 (Fig. 4C). Di- and trimethyl at Lys-77 completely eliminated the interaction; we note that these two methylated species of RBM25 have not yet been identified in cells. SRSF2 contains two domains, an N-terminal RNA-binding RRM and, C-terminal, the arginine/serine-rich (RS) domain. Deletion of the RS domain abolished the interaction with the RBM25 peptide (Fig. 4C). This is consistent with earlier studies indicating that the RS domain is primarily responsible for SRSF2 protein interactions (33); however, we were unable to express the RS domain alone to directly test whether it was sufficient for the interaction. As a positive control, we employed the 3xMBT domain of L3MBTL1, which is a universal binder for mono- and dimethylated peptides (11). This domain bound to RBM25K77me1 and RBM25K77me2, demonstrating the integrity of the methylated peptides (Fig. 4C). Finally, a panel of peptides, all with similar length, and non-methylated lysine at a central position did not interact with SRSF2, suggesting that SRSF2 interacts specifically with the amino acid sequence around RBM25 Lys-77 (Fig. 4D). Taken together, we conclude that SRSF2, in an RS domain–dependent manner, binds specifically to RBM25 peptide and that methylation at Lys-77 blocks this interaction. Consistent with the in vitro data, SRSF2 interacts with RBM25 in co-IP experiments when the two proteins are overexpressed in 293T cells (Fig. 4E).
There are several SR proteins in the human proteome. To determine the specificity of this interaction among the family, we expressed these proteins in vitro and tested their binding to the RBM25 peptides (Fig. 4F). SRSF1 (also called SF2) also binds strongly to the RBM25K77me0 peptide. Unlike SRSF2, this binding is not affected by addition of one methyl group even though it is largely abolished with me2 and me3 peptides. Other SR proteins tested show no more than weak binding to the RBM25 peptide.
Because SRSF2 exhibited low expression and poor stability in solution, we were unable to analyze its binding with isothermal titration calorimetry. As an alternative, we compared the strength of binding to non-methyl and methylated peptides by varying the concentration of immobilized peptide in peptide pulldown assays while maintaining the peptide in large molar excess relative to SRSF2. The fraction of bound protein was measured using quantitative Western blotting with fluorescence near-IR secondary antibodies. Consistent with earlier binding assays, SRSF2 bound most strongly to RBM25K77me0 peptide and much more weakly to me1 peptide, and the binding to me2 peptide was essentially undetectable in this assay (Fig. 4G). In contrast, SRSF1 binds similarly to me0 and me1 peptides. As a positive control, 3xMBT binds me1 and me2 peptides in a manner consistent with its affinity for methylated lysine, as measured by isothermal titration calorimetry (34). Finally, we used the RNA-Seq data (supplemental Table S3) to test whether sequences matching the binding motif of SRSF2 (35) occur in exon sequences regulated by RBM2. Analysis using AME (25) found that high-scoring SRSF2 motifs occur 11% more often among RBM25-regulated exons than among all alternatively spliced exons (1.8 per 100 bases compared with 1.6 using FIMO (36)), representing a small but significant enrichment (p < 0.02). These data suggest a physiological connection between RBM25 and SRSF2 but also that these two proteins are only likely to interact at a subset of their respective splicing targets.
Discussion
Here we have provided evidence that RBM25 is a splicing factor that broadly promotes inclusion of alternative and weak constitutive exons through interactions with the splicing machinery responsible for exon definition. Because alternative splicing is regulated by coordinated assembly of factors bridging the 3′ and 5′ splice sites of exons, we postulated that methylation of RBM25 at Lys-77 may regulate interactions with one or more of these factors (29, 37). Indeed, we found that the region of RBM25 centered on Lys-77 interacts specifically with SRSF2, likely via the SRSF2 RS domain, and Lys-77 methylation of RBM25 abrogates this interaction. To our knowledge, this is the first indication that lysine methylation may regulate protein interactions among splicing factors. These data lead us to hypothesize a model where methylation of RBM25 at Lys-77 tunes RBM25 activity when it is present on the same mRNA or pre-mRNA transcript as SRSF2. Because RBM25 Lys-77 mono-methylation preferentially affects binding to the SRSF2 RS domain but not to other members of the SR protein family, the relative abundance or activity of distinct SR proteins is a mechanism that could lead to different behaviors among RBM25 target exons and between cell types. Our detailed characterization of RBM25 protein interactions and genome-wide splicing function provides a basis for further work identifying pathways or splicing events regulated by methylation. Because we and others have encountered toxicity in cell culture with even small perturbations to RBM25 and SRSF2, we expect that a more promising direction for future work will identify the enzyme or enzymes responsible for RBM25 methylation so that the methylation event can be perturbed directly. Unfortunately, the 30 enzymes we tested do not methylate RBM25; however, we estimate that there are about 90 additional genes encoding candidate KMTs that might methylate RBM25 (38).
The ability of lysine methylation to regulate RBM25 protein interactions suggests a potential paradigm for how this modification might regulate transcription and alternative splicing. Because lysine methylation is disproportionally detected on proteins involved in pre-mRNA splicing and mRNA processing generally (11), we propose that a lysine methylation network may form a regulatory layer that tunes the output of specific splicing events by modulating protein–protein and, possibly, protein–RNA contacts throughout the splicing reaction. Many proteins involved in exon definition, and notably SRSF2 itself, are the sites of frequent mutations, including myelodysplastic syndrome, acute myeloid leukemia, and many forms of neurodegeneration (39, 40). It will be important to see how the splicing factor lysine methylation contributes to the pathways regulated in these diseases and to determine whether methylation-dependent pathways in splicing can reveal new therapeutic strategies to treat these diseases.
Experimental procedures
Plasmids and transfections
CRISPR/Cas9 knockdown used the lentiCRISPRv2 plasmid, which stably expresses both Cas9 and a small guide RNA (sgRNA) as described (Addgene plasmid 52961 (19)). RBM25 guide RNA sequences were as follows: 5′-ACAGTGGGTACTAAGACCTA-3′ (RBM25 sgRNA 1), 5′-GTACCCACTGTGTCTATGGT-3′ (RBM25 sgRNA 2), and 5′-CGGCATACTCACTGCGAGTG-3′ (non-targeting sgRNA). Briefly, lentiCRISPRv2 containing the indicated sgRNA was packaged into lentiviral particles using plasmids pCMV-VSV-G (Addgene plasmid 8454) and pCMV-dR8.2 (Addgene plasmid 8455) and sterile-filtered at 0.22 μm, and target cells were transduced overnight with 4 μg/ml Polybrene followed by a medium change. Selection with puromycin at 2 μg/ml began 2 days after transduction where applicable. RBM25 reconstitution used the PiggyBac transposon system to integrate the indicated gene under control of a Tet-repressed CMV promoter (the vector was a gift from Joanna Wysocka). Cells were co-transfected with RBM25-containing and PiggyBac transposase expression plasmids at a 4:1 ratio and selected using 2 μg/ml puromycin starting 2 days later. Close to endogenous expression occurred without addition of doxycycline. Recombinant protein expression in Escherichia coli used pGEX 6p-1 (GE Healthcare).
Cell culture and lysis
Cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), l-glutamine at 2 mm (Gibco), and penicillin-streptomycin (Gibco). Cell cycle analysis was performed 4 days after transduction with lentiCRISPRv2 using propidium iodide stain (Sigma-Aldrich), flow cytometry on a FACScan analyzer (BD Biosciences), and analysis using FlowJo (FlowJo, LLC). Transfections used Trans-IT 293 or TransIT-LT1 (Mirus Bio) according to the instructions of the manufacturer. For SILAC experiments, cells were grown in light amino acids or heavy amino acids (13C615N2-l-lysine/13C615N4-l-arginine) (Silantes) for at least 6 days as described previously (41). For Western blotting experiments, cells were lysed in radioimmunoprecipitation assay buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, and 0.1% SDS supplemented with PMSF (Sigma-Aldrich) and protease inhibitor mixture (Roche)). For co-immunoprecipitation, cells were lysed in radioimmune precipitation assay buffer without SDS supplemented with 10 μg/ml RNase A and sonicated for 5 min. For proteomic peptide interaction experiments, nuclei were collected following hypotonic lysis, and proteins were extracted by incubation in high-salt buffer as described previously (42), with the modification that 10 μg/ml RNase A was added to the lysis buffers. RNA was extracted using RNeasy Plus Mini Kits (Qiagen).
Antibodies, peptides, and protein sequences
Antibodies used were as follows: RBM25 (Bethyl Labs, A301-068A), FLAG M2 for Western blotting (Sigma-Aldrich, F1804), immobilized FLAG M2 (Sigma-Aldrich, A2220), glutathione S-transferase (custom rabbit polyclonal), and Myc tag (Pierce Fisher, MA1-21316). Peptides used were as follows: RBM25 Lys-77 (amino acids 67–84, NP_067062.1), snRNP B (amino acids 41–59, NP_003082.1), eEF1A1 Lys-55 (amino acids 47–64, NP_001393.1), eEF1A1 Lys-165 (amino acids 157–174, NP_001393.1), PRPF8 (amino acids 1514–1433, NP_006436.3), and SIRT6 (amino acids 337–355, NP_057623.2). Peptides were supplied by ChinaPeptides (Shanghai, China) at >95% purity, verified by HPLC and LC/MS. All peptides had lysine-biotin at their C terminus. Sequences of expressed proteins were as follows: RBM25, NP_067062.1; SRSF1, NP_008855.1; SRSF2, NP_001182356.1; SRSF3, NP_003008.1; SRSF5, NP_001307143.1; SRSF7, NP_001026854.1; and SRSF8, NP_115285.1.
Recombinant protein expression
GST fusion proteins were expressed in BL21 E. coli by overnight culture at 20 °C in LB (10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl) supplemented with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (Sigma) and purified using glutathione-Sepharose 4B (GE Healthcare) as described previously (42). Protein concentrations were measured using the Coomassie Plus assay (Pierce).
In vitro radiolabeling
In vitro radiolabeling to test KMT activity on the RBM25 RRM (amino acids 1–181 with N-terminal GST fusion) was performed as described previously (43).
High-throughput RNA sequencing
The RBM25 depletion experiment consisted of comparison between two biological replicate control samples and two RBM25 depletion samples. Transcriptome library preparation and sequencing were performed by BGI Americas using Illumina HiSeq 4000 with 150 base-paired end reads. After filtering to remove adaptor sequences, data in FASTQ format were used for further analysis.
Gene-level transcriptome analysis
Kallisto version 0.43.0 was used to quantify reads for gene-level expression analysis by aligning reads to GRCh38 transcripts with 20 bootstrap samples (20). Differential expression used Sleuth (21) and was used to perform a pairwise comparison between control sgRNA and RBM25-targeted sgRNA. Significant differences were filtered for false discovery rate using q < 0.05.
Splicing-level transcriptome analysis
Reads from all libraries were processed using the same workflow. Paired-end reads were mapped using HISAT2 v. 2.0.3 with default parameters. Mapped reads were subsequently converted from sequence alignment map format to binary alignment map format using Samtools v. 0.1.19. Binary alignment map files were processed through JuncBASE v1.2 run with default parameters to quantify alternative splicing events (20). Gencode v24 annotations were used to build a reference intron-exon junction database. A series of filtering steps was used to identify splicing events affected by RBM25 depletion or overexpression. First, splice events described as “jcn_only_AD” and “jcn_only AA” were excluded, and events were considered when there were at least 40 reads supporting each inclusion and exclusion across the four samples being compared. For each splicing event, Fisher's exact test was used to compare the number of inclusion and exclusion reads. Each sample from each group was compared with each sample in the other group (four pairwise comparisons). Splicing events were considered different between conditions when they changed in the same direction in all four comparisons with a false discovery rate of 5% using Benjamini-Hochberg correction. Statistically significant differences were considered biological meaningful when the average PSI calculated by JuncBASE changed by at least 10.
Functional analysis of gene-level transcription and RBM25-regulated cassette exons
Gene lists were submitted to the DAVID Bioinformatics Resources 6.7 using genes expressed in the dataset as background (22, 23). The threshold for significant association of gene ontology terms was Benjamini-Hochberg-corrected p < 0.05. Motif analysis used the AME and FIMO tools from the MEME suite (25, 36). Enrichment of the RBM25 motif used GGGGGGG and counted positive hits when they had no more than one mismatch. Enrichment of the SRSF2 motif used the frequency matrix derived from Liu et al. (35) and counted matches when they matched with p < 0.01. A similar degree of SRSF2 motif enrichment was observed with p < 0.001 and p < 0.0001.
Real-time PCR (RT-PCR)
RNA samples were converted to cDNA using the SuperScript III first strand synthesis system (Thermo Fisher). RT-PCR was performed using 25 ng of cDNA with SYBR Green RT-PCR Master Mix with a LightCycler 480 system (Roche). RT-PCR primers are listed in supplemental Table S7.
IP and co-IP with LC/MS-MS
RBM25 methyl site stoichiometry was determined by IP using 1.5 μg of RBM25 antibody immobilized on 20 μl of protein A Dynabeads (Life Technologies). 3 mg of total protein extract were used for each experiment. After washing and boiling in SDS buffer proteins were separated by SDS-PAGE and visualized by silver stain (SilverQuest kit, Thermo Fisher). Bands were cut, destained, and digested in-gel using chymotrypsin (Promega) (42). Recovered peptides were dried and desalted using C18 stage tips (Thermo Fisher). Endogenous RBM25 co-IP used at least 3 mg of protein extract prepared in normal growth medium or light and heavy SILAC medium as indicated. Antibody was cross-linked to beads using disuccinimidyl suberate according to the instructions of the manufacturer (Thermo Fisher), and proteins were eluted using 100 mm glycine (pH 2.0). FLAG co-IP of overexpressed RBM25 used 10 μg of magnetic FLAG M2 beads (Sigma) eluted using 3xFLAG peptide according to the instructions of the manufacturer. Each co-IP experiment was performed in duplicate with isotopic labels reversed. Matched light and heavy SILAC co-IP samples were combined before elution. Proteins were separated briefly by SDS-PAGE and visualized by silver stain, and each lane was processed by in-gel digestion using trypsin (Promega). Peptides were separated by HPLC using an Ekspert nanoLC 420 (AB Sciex) and analyzed with an Orbitrap Elite or Orbitrap Fusion mass spectrometer (Thermo Scientific). Acquisition used data-dependent selection of the top 20 ions on the Orbitrap Elite or dynamic acquisition speed on the Orbitrap Fusion with dynamic exclusion, followed by collision-induced dissociation or higher-energy collisional dissociation and analysis of fragment ions in the ion trap of the Orbitrap Elite or the Orbitrap of the Orbitrap Fusion. Data were analyzed by searching the human UniProt proteome using MaxQuant version 1.3.0.5 (41) with a 1% false discovery rate for proteins and peptides and allowing as variable modifications methionine oxidation and acetylation of protein N termini. Likely interacting proteins were defined by at least four peptides identified in both co-IP replicates and RBM25 over a control ratio of total peptide intensities of less than 0.2 in both replicates. SILAC ratios calculated by MaxQuant were not used for co-IP experiments because they are unstable or not reported when one pair is below the limit of detection. For FLAG co-IP with domain deletions, to avoid overexpression artifacts, we considered only proteins also identified as likely interacting partners. In each case, the ratio of protein bound by the short fragment relative to the long fragment was calculated using total peptide intensities normalized by the amount of RBM25 in each channel (itself calculated by the SILAC ratios of peptides present in both the long and short forms of RBM25).
Peptide pulldowns and LC/MS-MS
RBM25 peptides were immobilized by saturating 10 μl of streptavidin T1 Dynabeads (Life) for at least 1 h while rotating at room temperature in PBS. Beads were washed twice in peptide binding buffer (50 mm Tris (pH 7.5), 225 mm NaCl, and 0.5% Nonidet P-40) and incubated while rotating overnight at 4 °C with 600 μg of nuclear extract from 293T cells prepared in light or heavy SILAC medium. Pulldowns were washed twice in binding buffer and twice in PBS. Bound proteins were prepared for digestion by suspending beads in 50 μl of digest buffer (100 mm Tris (pH 8.0)) supplemented with 2 m urea (Sigma-Aldrich) and 10 mm dithiothreitol, incubation for 20 min with shaking, addition of iodoacetamide (Sigma-Aldrich) to 55 mm final concentration, and shaking in darkness for 10 min. Trypsin (Promega) was added to 12 ng/μl and incubated with shaking for 2 h. Beads were removed, and the digest was placed in darkness overnight. Digestion was stopped by adding 10 μl 10% formic acid (Sigma-Aldrich), and peptides were desalted using C18 stage tips (Thermo Scientific). LC/MS-MS and data analysis were performed as described above.
Recombinant protein peptide pulldowns
RBM25 peptides were immobilized as before on 4 μl of T1 Dynabeads beads using binding buffer (50 mm Tris (pH 7.5), 225 mm NaCl, and 0.15% Nonidet P-40). Beads were washed twice in binding buffer and incubated while rotating at 4 °C for 2 h with 300 ng of recombinant protein in 400 μl of total volume (volume and/or bead and protein concentrations adjusted as needed for binding curves). Pulldowns were washed twice in binding buffer and eluted by addition of SDS buffer. SR proteins for input loading control were stabilized by preincubation for 2 h with non-methylated RBM25 peptide at 10 μg/ml. SDS-PAGE and Western blotting were visualized using secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories) with chemiluminescent reagent (GE Healthcare) or near-IR secondary antibodies (LI-COR Biosciences) with Odyssey Imager (LI-COR Sciences). Western blots for binding curves were performed in triplicate and normalized to loading of input material. Best fit curves were fit by determining maximum binding and an equilibrium constant by minimizing the sum of squared error across all concentrations (because of avidity effects, these should not be taken as a measurement of Kd).
Author contributions
S. M. C. conceived the project, carried out the experiments, analyzed the data, and prepared the manuscript. C. M. S. performed the JuncBASE analysis for alternative splicing. Z. Y. assisted with the experiments. J. E. E. supervised the mass spectrometry. A. N. B. supervised analysis of the RNA-Seq data. O. G. conceived the project, supervised the experiments, and prepared the manuscript. all authors provided input on the final manuscript.
Supplementary Material
This work was supported in part by NCI, National Institutes of Health Grant R01CA172560 (to O. G.). O. G. is a co-founder of EpiCypher, Inc. and Athelas Therapeutics, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S7 and data.
The RNA-Seq data are available at the GEO under accession number GSE89140.
- pre-mRNA
- precursor mRNA
- RRM
- RNA-binding motif domain
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- IP
- immunoprecipitation
- RNA-Seq
- RNA sequencing
- FDR
- false discovery rate
- GO
- gene ontology
- PSI
- percent spliced in
- AME
- analysis of motif enrichment
- SILAC
- stable isotopic labeling in cell culture
- KMT
- lysine methyltransferase
- sgRNA
- small guide RNA
- 3xMBT
- triple malignant brain tumor domain.
References
- 1. Green M. R. (1986) Pre-mRNA splicing. Annu. Rev. Genet. 20, 671–708 [DOI] [PubMed] [Google Scholar]
- 2. McKeown M. (1992) Alternative mRNA splicing. Annu. Rev. Cell Biol. 8, 133–155 [DOI] [PubMed] [Google Scholar]
- 3. Black D. L. (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103, 367–370 [DOI] [PubMed] [Google Scholar]
- 4. Chen M., and Manley J. L. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu X. D., and Ares M. (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fortes P., Longman D., McCracken S., Ip J. Y., Poot R., Mattaj I. W., Cáceres J. F., and Blencowe B. J. (2007) Identification and characterization of RED120: a conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett. 581, 3087–3097 [DOI] [PubMed] [Google Scholar]
- 7. Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H. V., Mann M., Séraphin B., Rosbash M., Lührmann R., and Fabrizio P. (1998) A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4, 374–393 [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Z., Licklider L. J., Gygi S. P., and Reed R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 9. Zhou A., Ou A. C., Cho A., Benz E. J. Jr., and Huang S. C. (2008) Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell Biol. 28, 5924–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao G., Xie A., Huang S. C., Zhou A., Zhang J., Herman A. M., Ghassemzadeh S., Jeong E. M., Kasturirangan S., Raicu M., Sobieski M. A. 2nd, Bhat G., Tatooles A., Benz E. J. Jr., Kamp T. J., and Dudley S. C. Jr. (2011) Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation 124, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore K. E., Carlson S. M., Camp N. D., Cheung P., James R. G., Chua K. F., Wolf-Yadlin A., and Gozani O. (2013) A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol. Cell 50, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stamm S. (2008) Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 13. Friesen W. J., Massenet S., Paushkin S., Wyce A., and Dreyfuss G. (2001) SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 14. Larsen S. C., Sylvestersen K. B., Mund A., Lyon D., Mullari M., Madsen M. V., Daniel J. A., Jensen L. J., and Nielsen M. L. (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 9, rs9. [DOI] [PubMed] [Google Scholar]
- 15. Cao X. J., Arnaudo A. M., and Garcia B. A. (2013) Large-scale global identification of protein lysine methylation in vivo. Epigenetics 8, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., and Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore K. E., and Gozani O. (2014) An unexpected journey: lysine methylation across the proteome. Biochim. Biophys. Acta 1839, 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howard J. M., and Sanford J. R. (2015) The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bray N. L., Pimentel H., Melsted P., and Pachter L. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 [DOI] [PubMed] [Google Scholar]
- 21. Pimentel H., Bray N. L., Puente S., Melsted P., and Pachter L. (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 [DOI] [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 24. Brooks A. N., Yang L., Duff M. O., Hansen K. D., Park J. W., Dudoit S., Brenner S. E., and Graveley B. R. (2011) Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 21, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McLeay R. C., and Bailey T. L. (2010) Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 27. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manley J. L., and Krainer A. R. (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith C. W., and Valcárcel J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388 [DOI] [PubMed] [Google Scholar]
- 30. Vermeulen M., Mulder K. W., Denissov S., Pijnappel W. W., van Schaik F. M., Varier R. A., Baltissen M. P., Stunnenberg H. G., Mann M., and Timmers H. T. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 31. Fu X. D., and Maniatis T. (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437–441 [DOI] [PubMed] [Google Scholar]
- 32. Bradley T., Cook M. E., and Blanchette M. (2015) SR proteins control a complex network of RNA-processing events. RNA 21, 75–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graveley B. R. (2000) Sorting out the complexity of SR protein functions. RNA 6, 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Fischle W., Wang W., Duncan E. M., Liang L., Murakami-Ishibe S., Allis C. D., and Patel D. J. (2007) Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu H. X., Chew S. L., Cartegni L., Zhang M. Q., and Krainer A. R. (2000) Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell Biol. 20, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grant C. E., Bailey T. L., and Noble W. S. (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berget S. M. (1995) Exon recognition in vertebrate splicing. J. Biol. Chem. 270, 2411–2414 [DOI] [PubMed] [Google Scholar]
- 38. Carlson S. M., and Gozani O. (2016) Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med. 6, a026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scotti M. M., and Swanson M. S. (2016) RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., Chalkidis G., Suzuki Y., Shiosaka M., Kawahata R., Yamaguchi T., et al. (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 [DOI] [PubMed] [Google Scholar]
- 41. Ong S. E., and Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 42. Carlson S. M., Moore K. E., Green E. M., Martín G. M., and Gozani O. (2014) Proteome-wide enrichment of proteins modified by lysine methylation. Nat. Protocols 9, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carlson S. M., Moore K. E., Sankaran S. M., Reynoird N., Elias J. E., and Gozani O. (2015) A proteomic strategy identifies lysine methylation of splicing factor snRNP70 by the SETMAR enzyme. J. Biol. Chem. 290, 12040–12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.