Abstract
Thymidylate synthase (TS) is the sole enzyme responsible for de novo biosynthesis of thymidylate (TMP) and is essential for cell proliferation and survival. Inhibition of human TS (hTS) has been extensively investigated for cancer chemotherapy, but several aspects of its activity and regulation are still uncertain. In this study, we performed comprehensive structural and biophysical studies of hTS using crystallography and thermal shift assay and provided the first detailed structural information on the conformational changes induced by ligand binding to the hTS active site. We found that upon binding of the antifolate agents raltitrexed and nolatrexed, the two insert regions in hTS, the functions of which are unclear, undergo positional shifts toward the catalytic center. We investigated the inactive conformation of hTS and found that the two insert regions are also involved in the conformational transition between the active and inactive state of hTS. Moreover, we identified a ligand-binding site in the dimer interface, suggesting that the cavity in the dimer interface could serve as an allosteric site of hTS to regulate the conformational switching between the active and inactive states. On the basis of these findings, we propose a regulatory mechanism of hTS activity that involves allosteric regulation of interactions of hTS with its own mRNA depending on cellular demands for TMP.
Keywords: conformational change, crystallography, DNA synthesis, drug resistance, nucleotide, thymidylate synthase, antifolate
Introduction
Thymidylate synthase (TS)4 (EC 2.1.1.45) catalyzes the reductive methylation of deoxyuridine monophosphate (dUMP) to thymidylate (TMP) using 5,10-methylenetetrahydrofolate as the methyl group donor. This enzyme provides the sole intracellular source of de novo TMP, an essential precursor for DNA synthesis and repair. Inhibition of TS leads to thymidylate depletion and thymineless death characterized with severe DNA damage, chromosome aberrations, and induction of apoptosis (1, 2). hTS therefore serves as a critical point for intervention in cancer chemotherapy. Inhibitors of hTS, classified into fluoropyrimidines (e.g. 5-fluorouracil) and antifolates (e.g. raltitrexed), have been effective therapeutic agents for various cancers such as non-small cell lung cancer and colorectal cancer (3, 4). However, the long-term effectiveness of these drugs is largely limited by the development of drug resistance (3, 5–7). In attempts to overcome resistance problems, researchers continue to search for effective and specific inhibitors of hTS.
Extensive knowledge of structure and properties of the target protein can aid in formulating more efficient strategies for drug development. TS enzymes for Lactobacillus casei (L. casei) and Escherichia coli (E. coli) have been extensively characterized (8–11). As the protein sequence and primary structure of TS from various sources are highly conserved, L. casei and E. coli TS have been the standards for structural studies of difficult-to-crystalize TS enzymes including hTS. However, there are three variable regions in TS: the N-terminal region and two insert regions (region I, residue 117–128; region II, residue 146–153)5 (supplemental Fig. S1). Despite extensive studies on hTS as a chemotherapeutic drug target in the last few decades, functions and implications of the three regions in hTS have received less attention. In recent years, the N terminus of hTS has been demonstrated to be a primary determinant of the intracellular stability of the enzyme (12–14). Still, roles of the two insert regions remain unclear. Previous studies have reported two distinct states of hTS. One is the active state observed in the crystal structures of TS–nucleotide–(anti)folate ternary complexes (15) where the thiol group of catalytic Cys-195 points toward the pyrimidine ring of dUMP. The other one is an inactive state identified in structures from sulfate-containing conditions (16) and structures bound with dimer-interface ligands (17) where Cys-195 is rotated away from dUMP. hTS has been proposed to be involved in translational regulation of its own mRNA (18–20). It was also proposed that the inactive conformation may be involved in this autoregulation of hTS expression and stabilization of the inactive conformation could be a strategy to overcome the drug resistance problem associated with hTS overexpression (16, 21). Considering that the inactive state has been discovered in hTS only, it is possible that the protein state is related to the unique insert regions in hTS.
Based on the study from Lovelace et al. (22) two hTS mutants (M190K and A191K) in the inactive conformation are explored in this work. To investigate the role of the insert regions in hTS and facilitate devising new strategies in hTS inhibitor design, we have performed comprehensive structural and biophysical studies on the active and inactive conformers of hTS. The active conformer has been explored by obtaining the three-dimensional structures of ternary complexes of three different antifolates. Our data suggest that the two insert regions in hTS are involved in conformational changes triggered by antifolate binding. Moreover, these two regions may play critical roles in conformational switching between the active and inactive states of hTS.
Results
Characterization of wild-type and mutant hTS
Both wild-type and mutant (A191K and M190K) hTS were analyzed using size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS). The results indicated that both wild-type and mutant protein existed as dimers (data not shown). The proteins were further analyzed using a thermal shift assay, differential scanning fluorimetry (DSF), showing that they had distinct melting profiles (Fig. 1a). Melting of wild-type hTS featured a biphasic profile. However, the two mutants had a monophasic melting curve and both gave high initial fluorescence, indicating that the mutants folded with a large area of hydrophobic patches exposed to solvent. Interestingly, when the two mutants were titrated with dUMP, the initial fluorescence decreased (Fig. 1), indicating that the mutants were reversed to the active state by dUMP binding as reported previously (16). The dose-response experiments revealed that A191K had higher affinity for dUMP compared with M190K. This was further confirmed with isothermal titration calorimetry (ITC) (supplemental Fig. S2): A191K had a KD of 4.94 μm for dUMP that was similar to wild-type TS (KD of 1.19 μm), whereas M190K showed two-site sequential binding with one site having significantly lower affinity KD1 at 1.32 mm and KD2 at 1.75 μm.
Figure 1.
Characterization of hTS using DSF. a, distinct melting profiles of wild-type and mutant TS proteins. Wild-type TS gives a biphasic melting curve. Mutants M190K and A191K have monophasic melting profiles but give high initial fluorescence. b, dUMP dose-response (DR) data on wild-type TS. Adding dUMP into wild-type TS converts the melting profile to a monophasic curve by stabilizing the first phase. Further titration leads to concentration-dependent stabilization of the monophasic curve. c, dUMP DR data on A191K. d, dUMP DR data on M190K. Addition of dUMP results in reduction of initial fluorescence on both A191K and M190K. The melting profiles of the mutants are gradually transformed to biphasic curves as observed for wild-type TS. A191K is further stabilized at the first phase and exhibits a monophasic profile in the presence of 2 mm dUMP, but M190K fails to achieve that, suggesting M190K has lower affinity for dUMP.
Conformational changes upon dUMP and antifolate binding
We used one ligand-free crystal form of a N-terminal-truncated protein construct (residues 26–313) to study the conformational changes of hTS triggered by ligand binding. The structures of the TS–dUMP binary complex and TS–dUMP–antifolate ternary complexes were obtained by soaking the compounds into the same crystal form to avoid errors resulting from different crystal packing forces.
The apocrystal structure (unsoaked) TS–Pi adopted the active conformation and had a phosphate ion bound in the position corresponding to the phosphate moiety of dUMP (Fig. 2a). The asymmetric unit (ASU) of the crystal form contained six monomers of hTS. Differences in atomic position among the six subunits were small, except for the C-terminal residues. Only in two of the six monomers, the last six residues at the C-terminal loop were clearly defined with electron density. The difference could be the result of distinct crystal packing interactions as the six subunits reside in non-identical crystallographic environment. The structure of TS–dUMP was virtually identical to the structure of TS–Pi, with the root mean square deviation (r.m.s. deviation) of 0.148 Å for Cα atoms (0.241 Å for all atoms).
Figure 2.
Alignment of ligand-free, binary and ternary complex structures. a, overlay of ligand-free structure (blue) and dUMP-bound structure (white). Phosphate ion and dUMP are shown as sticks. b, front view of superposition of protein structures of TS–dUMP (cyan), TS–dUMP–methotrexate (white), TS–dUMP–raltitrexed (yellow), and TS–dUMP–nolatrexed (orange). c, top view of the complex structures. Ligands dUMP and antifolates are shown as stick. d, front view of the folate binding pocket and positional shift of Trp-109.
Three ternary complex structures with known antifolates were determined (Fig. 2b): TS–dUMP–raltitrexed, TS–dUMP–methotrexate, and TS–dUMP–nolatrexed. These are the first structures of hTS in complex with methotrexate and nolatrexed. An interesting finding is that soaking crystals with nolatrexed resulted in changes of crystal unit cell and space group (Table 1). The unit cell and space group conversion reflects the gain of symmetry and corresponds to a switch from six molecules to two molecules per ASU. Nolatrexed has been reported to behave as a noncompetitive inhibitor of hTS with unknown mechanism (23), but the structure we determined showed that nolatrexed bound competitively to the folate-binding site. The three antifolates bound in a similar manner to folate as previously described (24, 25), with the quinazoline ring packed face to face against the pyrimidine and ribose ring of dUMP. The l-glutamate moiety of raltitrexed and methotrexate bound in the entrance channel to the active site. Different from the well determined density map for quinazoline ring, the electron density of l-glutamate moiety in both structures were not clearly defined, implying that the l-glutamate group retained certain flexibility in the binding site.
Table 1.
Data collection and refinement statistics of N-terminal truncated hTS
| Dataset | TS–Pi | TS–dUMP | TS–dUMP-raltitrexed | TS–dUMP–methotrexate | TS–dUMP–nolatrexed | TS–dUMP–F13 |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Wavelength (Å) | 0.7749 | 0.9537 | 1.0130 | 0.9537 | 1.0130 | 1.0000 |
| Space group | P43212 | P43212 | P43212 | P43212 | P41212 | P43212 |
| Cell dimension (Å) | 109.5, 109.5, 317.5 | 109.7, 109.7, 317.7 | 107.8, 107.8, 315.0 | 109.2, 109.2, 317.1 | 108.3, 108.3, 106.0 | 109.4, 109.4, 315.4 |
| Resolution (Å) | 30 – 2.39 | 30 – 2.00 | 30 – 2.79 | 30 – 1.99 | 48.22 – 2.13 | 30 – 2.69 |
| (outer shell) | (2.49 – 2.40) | (2.07 – 2.00) | (2.90 – 2.79) | (2.07 – 1.99) | (2.20 – 2.13) | (2.80 – 2.69) |
| Rmerge (%)a | 10.5 (49.8)b | 8.3 (40.3) | 12.6 (59.6) | 9.3 (44.5) | 17.3 (86.7) | 11.4 (42.9) |
| 〈I〉/σI | 19.7 (4.03) | 14.3 (3.2) | 14.9 (3.2) | 15.4 (3.2) | 15.5 (4.3) | 20.6 (2.4) |
| Completeness (%) | 86.1 (81.6) | 88.2 (95.2) | 93.0 (93.8) | 91.6 (86.8) | 99.5 (94.4) | 98.4 (91.1) |
| Redundancy | 10.1 (10.3) | 4.0 (4.9) | 6.9 (7.6) | 5.4 (5.9) | 15.8 (15.0) | 10.7 (6.9) |
| No. of observed reflections | 672,785 | 463,678 | 303,589 | 642,721 | 560,607 | 569,527 |
| No. of unique reflections | 66,303 | 116,562 | 43,961 | 119,969 | 35,535 | 53,294 |
| Model refinement | ||||||
| Resolution (Å) | 20.35 – 2.39 | 22.57 – 2.00 | 27.18 – 2.79 | 25.18 – 1.99 | 48.22 – 2.13 | 22.18 – 2.69 |
| No. of protein molecules/ASU | 6 | 6 | 6 | 6 | 2 | 6 |
| Rwork/Rfree (%)c | 21.7/25.1 | 18.1/22.4 | 22.2/26.2 | 21.1/24.7 | 15.35/20.65 | 21.3/24.5 |
| Number of atoms | ||||||
| Protein | 13,578 | 13,797 | 13,497 | 13,857 | 4,600 | 13,502 |
| Ligands | 30 (6 phosphate ions) | 120 (6 dUMP) | 312 (6 dUMP and 6 raltitrexed) | 318 (6 dUMP and 6 methotrexate) | 80 (2 dUMP and 2 nolatrexed) | 110 (4 dUMP and 2 F13) |
| Water | 233 | 1624 | 10 | 934 | 267 | 79 |
| Average B factor (Å2) | 37.0 | 26.0 | 50.0 | 28.0 | 22.0 | 40.0 |
| R.m.s. deviations | ||||||
| Bond length (Å) | 0.0145 | 0.0185 | 0.0119 | 0.0161 | 0.0183 | 0.0129 |
| Bond angle (°) | 1.7575 | 1.8848 | 1.5758 | 1.8721 | 1.8739 | 1.6391 |
| Ramachandran plot (%) | ||||||
| Favoured region | 96.3 | 97.2 | 96.0 | 96.9 | 97.4 | 96.8 |
| Allowed region | 3.5 | 2.5 | 3.5 | 3.0 | 2.3 | 2.9 |
| Outlier region | 0.2 | 0.3 | 0.5 | 0.1 | 0.4 | 0.2 |
| PDB entry | 5X5A | 5X5D | 5X5Q | 5X66 | 5X67 | 5X69 |
a Rmerge is defined as Σ|Ih,i − 〈Ih〉|/ΣIh,i, where Ih,i is the intensity for ith observation of a reflection with Miller index h, and 〈Ih〉 is the mean intensity for all measured Ih and Friedel pairs.
b Values in parentheses are for the highest resolution shell.
c Rwork = Σ|Fo − Fc|/Σ|Fo|, where Fo is the observed structure factor amplitude and Fc is the calculated structure factor amplitude. Rfree is the same as Rwork but was calculated using a random set containing 5% of the data that were excluded during refinement.
Overlaying the structures revealed that nolatrexed triggered the largest conformational changes, with methotrexate the smallest. The radius of gyration calculated for the ternary complex dimer was 23.96 Å for nolatrexed, 23.99 Å for raltitrexed, and 24.08 Å for methotrexate, compared with 24.07 Å for the binary TS–dUMP dimer. Compared with the structures of TS–Pi and TS–dUMP, several stretches of the protein adopted slightly different positions in the ternary complexes, including mainly the α-helixes and connecting loops lining the active site (helix AS-α1, AS-α2, insert region I, insert region II, and the C-terminal loop) as shown in Fig. 2, b and c. These segments were shifted in concert toward the active center of TS upon the binding of raltitrexed and nolatrexed, serving to close the active site. The largest shift of C-terminal loop occurred in the structure of TS–dUMP–nolatrexed, where the C-terminal region moved up to 2.2 Å further into the active site accompanied with a shift of the substrate-binding loop (Fig. 2c). In the shifted helixes, residues moved as units to avoid disruption of the favorable hydrogen-bond patterns in these segments as well as to make interactions with the ligands. The only residue that underwent extra rotation of side chain was the fully conserved Trp-109, of which the indole ring rotated into the active site by 6° in raltitrexed structure and 3° in nolatrexed structure to interact with the quinazoline ring (Fig. 2d). Binding of methotrexate was unable to elicit these positional shifts, which was likely because that its quinazoline ring is too far away to make contacts with the lining segments.
Crystal structures of mutant hTS in the inactive conformation
The crystal structures of hTS in the inactive conformation reported so far were obtained from similar conditions containing ammonium sulfate (16, 17, 22, 26). It is reasonable to question the relevance of these structures when the inactive conformers risk to instead be crystallographic artifacts due to the high concentration of sulfate in the crystallization buffer as several sulfate ions are observed in the structures. Structures from crystal conditions free of sulfate and phosphate are therefore important to evaluate the significance of the previously proposed inactive conformation of hTS. Although most crystals of M190K and A191K were in conditions containing various amounts of sulfate ions from our crystallization screening effort, we managed to get crystals of A191K from a sulfate- and phosphate-free condition.
Three mutant structures were used for following analysis: A191K crystallized in a sulfate-free condition as A191K-sf; A191K crystallized in a sulfate-containing condition as A191K-s; and M190K crystallized in a sulfate-containing condition as M190K-s (Table 2). The structures of A191K-sf and A191K-s were virtually identical (r.m.s. deviation at 0.126 Å for Cα atoms and 0.149 Å for all atoms), both adopting an inactive conformation with the catalytic Cys-195 directed away from the active site (Fig. 3a). This confirms that the inactive conformation is not due to high concentrations of sulfate ions in the crystallization solutions. In the structure of A191K-sf, there were two extra tetrahedral densities near the dUMP-binding site, into which we fitted two phosphate ions (Fig. 3b). As illustrated in Fig. 3c, one of the phosphate ions sits close to but different from the position of the phosphate moiety of dUMP. The highly-conserved residues accommodating the phosphate moiety of dUMP (Arg-50, Arg-215, Arg-175′, and Arg-176′) interacted extensively with the two phosphate ions in the inactive conformer as well, but all in a different conformation compared with the active state. Notably, two additional residues, Asn-183 and Arg-185, which were about 18 Å away from dUMP in the active conformer, came in and interacted with the two phosphate ions in the inactive conformer. It was clear from the structure that the two phosphate ions played an essential role in stabilizing the inactive conformer. Because there was no phosphate added during protein purification or crystallization for the A191K-sf structure, the phosphate ions in the structure were likely picked up in the host cell and co-purified with the protein.
Table 2.
Data collection and refinement statistics of mutant TS A191K and M190K
| Dataset | A191K-sf | A191K-s | M190K-s |
|---|---|---|---|
| Crystallization condition | 100 mm HEPES, pH 7.0, 10% PEG 6000 | 100 mm Tris, pH 8.5, 0.2 m lithium sulfate, 1.26 m ammonium sulfate | 100 mm MES, pH 6.5, 1.6 m ammonium sulfate, 10% 1,4-dioxane |
| Data collection | |||
| Wave length (Å) | 0.9537 | 0.9537 | 0.9537 |
| Space group | P3121 | P3121 | P3121 |
| Cell dimension (Å) | 95.6, 95.6, 79.7 | 95.5, 95.5, 81.2 | 96.0, 96.0, 80.9 |
| Resolution (Å) | 31.29 – 2.10 | 29.16 – 2.31 | 27.70 – 2.20 |
| (outer shell) | (2.21 – 2.10) | (2.44 – 2.31) | (2.32 – 2.20) |
| Rmerge (%) | 10.2 (35.7) | 7.0 (29.0) | 6.2 (35.5) |
| 〈I〉/σI | 15.0 (6.4) | 22.1 (7.4) | 26.7 (7.7) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) | 99.9 (100.0) |
| Redundancy | 10.2 (10.5) | 10.6 (10.2) | 11.9 (12.0) |
| No. of observed reflections | 254,681 | 201,463 | 263,957 |
| No. of unique reflections | 24,955 | 19,057 | 22,239 |
| Model refinement | |||
| Resolution (Å) | 30.60 – 2.10 | 29.16 – 2.31 | 26.96 – 2.20 |
| No. of protein molecules/AU | 1 | 1 | 1 |
| Rwork/Rfree (%) | 17.0/19.8 | 18.6/23.5 | 19.7/24.7 |
| Number of atoms | |||
| Protein | 2225 | 2219 | 2215 |
| Ligands | 10 (2 phosphate ions) | 10 (2 sulfate ions) | 10 (2 sulfate ions) |
| Water | 134 | 75 | 140 |
| Average B factor (Å2) | 33.0 | 39.0 | 39.0 |
| R.m.s. deviations | |||
| Bond length (Å) | 0.0206 | 0.0185 | 0.0210 |
| Bond angle (°) | 2.0412 | 1.8497 | 2.0977 |
| Ramachandran plot (%) | |||
| Favored region | 96.3 | 95.2 | 96.4 |
| Allowed region | 3.7 | 4.8 | 3.6 |
| Outlier region | 0 | 0 | 0 |
| PDB entry | 5X4W | 5X4X | 5X4Y |
Figure 3.
Crystal structures of A191K-sf and A191K-s. a, structural alignment of A191K-sf (gray) and A191K-s (blue). Both are in the inactive conformation with the catalytic cysteine Cys-195 siting outside the active site. b, two phosphate ions bind in the structure of A191K-sf with the 2Fo − Fc density map contoured at 1.0 σ level. c, close up view of the binding site for two conserved phosphate/sulfate ions. P2 is near but different from the phosphate moiety of dUMP (orange). The four arginine residues (Arg-50, Arg-215, Arg-175′, and Arg-176′) that coordinate the phosphate moiety of dUMP undergo positional changes to coordinate the two phosphate ions in the inactive conformation. Upon the conformational changes in the inactive conformation, two distal residues (Arg-185 and Asn-183) in the active conformation move close to the active site and interact with the phosphate ions.
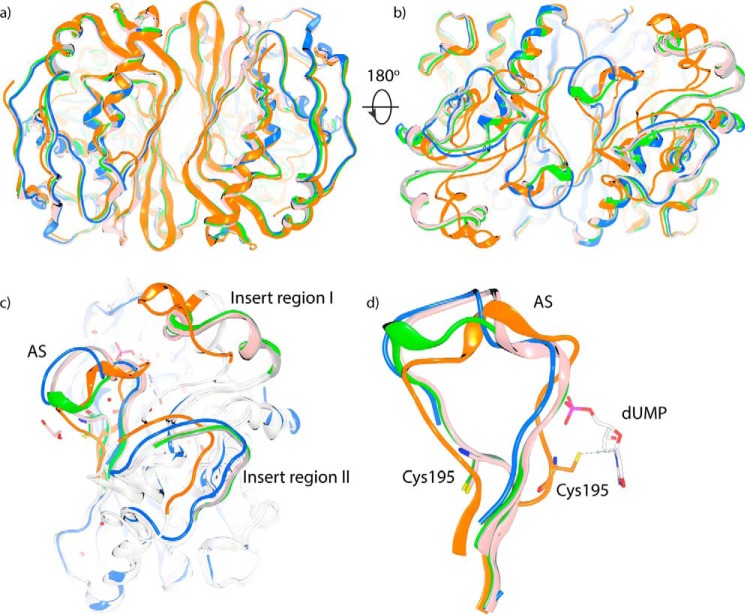
Superposition of the three inactive mutant dimers with inactive wild-type hTS (PDB code 1HW3) and the active dimer of TS–dUMP showed that the dimer interface β-sheet core and the region near the N termini were relatively conserved between the active and inactive states (Fig. 4a). The structural differences were mainly located on the opposite site (Fig. 4b), with the major and most apparent differences in the active site (AS) loop, insert regions I and II as highlighted in Fig. 4c. An interesting finding was that the structures of A191K, M190K, and the inactive wild-type hTS were divergent in these three protein segments, although all adopted an inactive conformation of the active site cysteine. In the comparison shown in Fig. 4d, the AS loop twists the ends of the loop and relocates to the inactive state observed in the structure of the inactive wild-type protein (PDB code 1HW3). In the inactive state of A191K, conformation of the AS loop is further aligned and shifts away from the dimer interface, accompanied with shifting of the external face of insert region II (Fig. 4c). In the inactive conformation of M190K, residues 187–192 in the AS loop reposition to form one additional helical turn. All conformational rearrangements in the AS loop are coupled with adjustment in insert regions I and II by moving away from their original positions that would block the inactive conformation of the AS loop sterically. As a consequence, the inactive dimers of hTS are less compact and the dimer interface becomes more solvent accessible. The exposed hydrophobic dimer interface could explain the high initial fluorescence of DSF melting curves observed in mutant proteins. The relatively lower initial fluorescence in A191K compared with M190K could be correlated to the conformational differences in the accessibility of the dimer interface reflected in the crystal structures of the two mutants.
Figure 4.
Structural comparison of the active and inactive conformations of TS. a, front view of the structural alignment of A191K-sf (gray), A191K-s (pink), M190K (green), TS–dUMP (orange), and the inactive crystal structure of TS (PDB code 1HW3; blue). b, back view of the structural alignment. Significant segmental shifts are observed at this side. c, alignment of the inactive and active conformers with regions undergoing significant conformational changes highlighted. d, structural comparison of the AS loop between the active and inactive states of TS.
Identification of a novel binding site in the dimer interface
In a fragment screening campaign using DSF, we identified fragment F13 as a hit for wild-type hTS. Validation using X-ray crystallography revealed that fragment F13 bound in the dimer interface of the active hTS conformer. As shown in the crystal structure (Fig. 5, a and b), the fragment bound to a fairly polar cleft in the dimer interface formed by Glu-145 (in the insert region II) together with Asn-183, Arg-185, and Asp-186 (in the AS loop) from each hTS monomer. Compared with the structure of TS–dUMP, side chains of residue Arg-185 and Arg-185′ are shifted (3.8 Å for Cζ) to accommodate F13 in the dimer interface (Fig. 5d). Nevertheless, this polar cleft disappeared and was replaced with an exposed and relatively hydrophobic pocket in the inactive conformers due to conformational changes (Fig. 5c). This is the first time that a binding site discovered in the dimer interface of the active hTS conformer though ligand binding has been reported in the inactive conformation (17). No significant effect was observed on hTS enzymatic activity for this fragment at its soluble concentrations (data not shown), which was not uncommon for fragment binders and was often because the affinity and solubility of the fragment were too low to generate a detectable difference in enzymatic assays. However, F13 led to small but significant concentration-dependent stabilization on wild-type hTS in the presence of dUMP using DSF (Fig. 5f). Mutation of Arg-185 to alanine (R185A) did not affect the binding of dUMP but abolished the binding of F13 (Fig. 5, e and f), which was consistent with the structural data that Arg-185 plays a critical role in F13 binding but not in dUMP binding. Moreover, F13 resulted in reduction of the initial fluorescence intensities on both M190K and A191K (Fig. 5, g and h), similar to the effect elicited by the binding of dUMP. The data suggest that ligand binding at this undiscovered binding site could cause conformational switching similarly as dUMP. The binding site in the dimer interface could therefore be a new site for small molecule intervention to modulate hTS activity.
Figure 5.
Binding site identified in the dimer interface. a, crystal structure of F13 binding in the dimer interface of the active hTS conformer. b, electrostatic potential surface for the binding region of F13 in the dimer interface. c, electrostatic potential surface for the region in the inactive conformation corresponding to the binding region of F13. d, superposition of the crystal structure of TS–dUMP (gray) and TS–dUMP–F13 (green) shown in the dimer interface region. Binding of F13 leads to a positional shift of Arg-185 and Arg-185′. The 2Fo − Fc density maps are contoured at the 1.0 σ level around Arg-185 and Arg-185′. e, melting curves of dUMP dose-response on mutant TS R185A. f, dose-response data of F13 on wild-type TS and mutant R185A. Addition of dUMP could drive the protein into the active conformation and also convert the two-phase melting curve into one-phase curve, which facilitates the determination of melting points, therefore the dose-response experiments were carried out in the presence of dUMP. ΔTm values of F13 in the presence of 200 μm dUMP were plotted against the compound concentration from three independent measurements. g, melting curves of the F13 dose-response on A191K. h, melting curves of the F13 dose-response on M190K.
Discussion
The conformational changes of TS induced by folate/antifolate binding have been well described because the first structures of ternary complexes of E. coli TS were determined (9). However, there has been a lack of information on the conformational changes of the hTS protein. In the current study, we have established several structures of the functional complexes of hTS and also compared the crystal structures of unliganded hTS with binary and ternary complexes using the same crystal form via soaking. Overall, the conformational changes taking place in hTS are similar to those observed in E. coli TS: binding of dUMP does not trigger significant protein rearrangement; binding of antifolate induces extensive conformational changes involving mainly the C-terminal loop and protein segments lining the active site moving to close the cavity. Nevertheless, hTS is distinct from E. coli TS in that the two insert regions are also involved in the conformational changes. The two regions undergo positional shifts with a vector toward the active site upon the binding of raltitrexed and nolatrexed. It is notable that these two insert regions are also key players in the conformational switching between the active and inactive states of hTS. In contrast to the closure of the active site in the ternary complexes, they open upon inactivation. It is therefore likely that the two insert regions in hTS function to increase conformational flexibility and cooperativity of the enzyme and thus facilitate to overcome the energy barrier associated with conformational changes.
In this work, we have investigated two inactive hTS mutants (M190K and A191K). DSF data with high initial fluorescence indicates that the mutants have large areas of hydrophobic region exposed to solvent. This turns out to be in agreement with the crystal structures, where the two inactive conformers open insert region II and thus expose the hydrophobic β-sheets in the dimer interface. We have also investigated the binding of dUMP to the two mutants and proved that the inactive conformers could be reversed to the active state by ligand binding in the active site. Together with the previous studies where wild-type protein crystallizes in an inactive conformation (16), the data demonstrate that hTS exists in an equilibrium of active and inactive conformations. There have been several ligands reported in the dimer interface of inactive hTS conformers (17). The discovery of fragment binding in the active dimer interface further supports that the dimer interface of the hTS protein is able to accommodate different types of ligands in the active and inactive states. The cavity in the dimer interface could potentially serve as an allosteric site of the enzyme to regulate the conformational switching of hTS.
The crystal structures obtained in this study provide insights into the mechanism of hTS activation and inactivation. The three different inactive states shown in Fig. 4 possibly represent snapshots of the conformation transition trajectory: the inactive form obtained from wild-type protein as the less ordered intermediate state I; A191K as a more ordered intermediate state II; and M190K as the ultimate stable inactive state. This is supported by our DSF data and ITC data that A191K confronts lower energy barrier to switch between the active and inactive states induced by dUMP binding. However, it is possible that the inactive conformation observed in M190K is not present in the functional cycle of the wild-type protein but a result of a mutation artifact, considering that conformation of the AS loop that contains the mutation site is very different from that in the inactive conformer of wild-type hTS.
In the inactive conformer of A191K-sf, there are two phosphate ions bound in the vicinity of the active site when no additional phosphate ions are added. However, in the active conformation, hTS binds only one phosphate ion in the position corresponding to the phosphate moiety of dUMP at 100 mm phosphate concentration. The difference in anion binding indicates that the conformational switching from the active to inactive state significantly changes the electrostatic properties of the protein. The molecular electrostatic potential shows that the folate-binding region exhibits the largest area of positively charged surface in both the active and inactive conformers (supplemental Fig. S3). This is the region expected to bind to the polyglutamate moiety of activated (anti)folates in cells. Taking the marked changes in propensities for phosphate binding into consideration, it is possible that this positively charged region serves as the folate-binding site in the active state, whereas it is involved in the binding of mRNA in the inactive state of hTS, as proposed in previous studies (18–20). Moreover, the protein segment (residues 107–114) in the folate-binding region is ordered in the active conformation but disordered in the inactive conformers. The disorder/mobility of this segment might correlate with mRNA binding and could be the recognition site of mRNA.
As the sole enzyme involved in de novo synthesis of TMP, hTS is supposed to be tightly regulated to meet the metabolic demands and also to maintain the appropriate nucleotides balance. However, unlike other enzymes involved in nucleotide synthesis that are typically regulated via allosteric interactions and feedback inhibition, regulation of hTS activity has only been reported on its translation and the detailed mechanism for this regulation remains unclear. In this study, we have discovered that the two insert regions in hTS play key roles in the conformational changes of both the active and inactive states. Additionally, a cavity at the dimer interface has been identified, which might function as an allosteric site to regulate the transition between the active and inactive states of hTS. Integrating the experimental data, a regulatory mechanism of hTS activity is proposed: in the resting state of cells, hTS populates the inactive conformation due to an allosteric effector in the dimer interface cavity and binds to its own mRNA thus repressing hTS protein synthesis; when cells start to divide, another allosteric effector binds to the dimer interface to activate hTS and release it from mRNA, leading to an increase in both hTS protein level and enzyme activity to meet the cellular demands. Even though our screening of a metabolic library did not identify any effectors (data not shown), it is possible that the natural effectors are linked to the cell cycle and synthesis of other nucleotides so that the appropriate balance of the four nucleotides required for DNA synthesis is maintained. Further studies are needed to investigate and establish the mechanism.
Experimental procedures
Recombinant protein expression and purification
Recombinant protein expression and purification of hTS protein has been described previously (27). Briefly, gene encoding hTS (GenBankTM accession code: NM_001071.2) was subcloned into pNIC28-Bsa4 vector and expressed in Rosetta BL21(DE3) E. coli (Novagen) in Terrific Broth media. A hTS protein construct with residues 1–25 deleted was subcloned using the same method. All hTS constructs were purified using immobilized metal affinity chromatography followed by gel filtration chromatography on a ÄKTAxpress system (GE Healthcare). Protein was incubated with tobacco etch virus protease in a 1:20 molar ratio (protease:target protein) to remove His6 tag at 4 °C overnight. The final buffer for full-length hTS, M190K, A191K, and R185A was 20 mm HEPES, 300 mm NaCl, 10% (v/v) glycerol, and 1 mm tris(2-carboxyethyl)phosphine, pH 7.5. The buffer for the N-terminal-truncated hTS used for crystallization was 100 mm PO4, 100 mm NaCl, and 2 mm tris(2-carboxyethyl)phosphine, pH 7.4.
Site-directed mutagenesis
The plasmid of full-length hTS was used as a template for site-directed mutagenesis. The mutant constructs were generated by polymerase chain reaction-based methods using KOD XtremeTM Hot Start DNA polymerase (Millipore) according to the manufacturer's instructions. The sequence of forward primer used for the M190K construct was 5′-CCAAGAGATCTTCCTCTGAAGGCGCTGCCT-3′. The sequence of the forward primer used in A191K site-directed mutagenesis was 5′-GAGATCTTCCTCTGATGAAGCTGCCTCCATGCCATG-3′. The sequence of the forward primer used in R185A site-directed mutagenesis was 5′-TGCGCTTGGAATCCAGCGGATCTTCCTCTG-3′. All mutant constructs were verified by DNA sequencing.
SEC-MALS
SEC-MALS was performed using a Superdex-200 15/150 GL column (GE Healthcare) combined with a miniDAWN TREOs light-scattering detector coupled with an Optilab rEX refractive index detector (Wyatt Technology). All experiments were conducted at room temperature at a flow rate of 0.3 ml/min. The injected protein sample was typically 20 μl of 6 mg/ml of protein. Molecular mass calculations were performed using the Astra6.1 software (Wyatt Technology). Input of the refractive increment (dn/dc values) was set at 0.186 ml/g in the molecular mass calculations. The molecular mass was determined across the protein elution peak.
Differential scanning fluorimetry
DSF was performed on the iCycler iQ Real Time PCR Detection System (Bio-Rad), using the 96-well thin-wall PCR plate (Bio-Rad). The experiment was conducted in a buffer of 20 mm HEPES, pH 7.5, and 150 mm NaCl. A total volume of 25 μl of solution containing 0.2 mg/ml of protein, compounds, and 5× Sypro Orange (Invitrogen) diluted from 5000× stock was dispensed into each well of the 96-well plate using a multichannel pipette. The same amount of dimethyl sulfoxide (DMSO) was added instead of compounds in the control wells. The plates were sealed with Microseal B adhesive sealer (Bio-Rad) and heated in iCycler from 25 to 80 °C (56 heating cycles in 28 min). Fluorescent filter was used for Sypro Orange with λexcitation = 492 nm and λemission = 610 nm. The calculation of the midpoint of the curves (Tm) was performed using the software package XLfit (ID Business Solutions).
ITC experiments
Protein was dialyzed with a buffer containing 20 mm HEPES, pH 7.5, and 150 mm NaCl at 4 °C overnight. The protein and compound samples were matched in terms of buffer condition. The protein was loaded into the iTC200 (Microcal) cell. Titrations were initiated with one 0.5-μl injection, followed by 16 to 20 larger volume injections, injected at 180-s intervals, at 25 °C. Stirring speed was 700 rpm. The heat peaks integration and non-linear regression analysis were performed with the Origin software (Microcal) and fitted to a single-site binding model to determine the parameters for stoichiometry N, KD, and ΔH.
Protein crystallization
The ligand-free crystal form of the N-terminal-truncated hTS was obtained in sitting drops of 3 μl comprising an equal volume of protein (about 24 mg/ml) and crystallization buffer at 20 °C. The reservoir contained 500 μl of crystallization buffer composed of 0.1 m sodium calcodylate, pH 6.5, and 25% PEG 4000.
The mutant protein A191K crystallized in two different conditions. Crystals was obtained by mixing protein (31 mg/ml) with crystallization in a 1:1 ratio to set up sitting drops of 2 μl at 20 °C. The crystallization buffer for the crystal form of A191K-sf was 100 mm HEPES, pH 7.0, and 10% PEG 6000. The crystallization buffer for the crystal form of A191K-s was 100 mm Tris, pH 8.5, 0.2 m lithium sulfate, and 1.26 m ammonium sulfate.
The mutant protein M190K crystallized in a buffer containing 100 mm MES, pH 6.5, 1.6 m ammonium sulfate, and 10% 1,4-dioxane at 4 °C. The protein (29 mg/ml) was mixed with crystallization buffer in a 1:3 ratio to set up sitting drops of 2 μl.
Crystals were transferred to the corresponding reservoir solution supplemented with 10% DMSO as cryo-protectant. For soaking with ligands, compounds were prepared at the desired concentrations in cryo-protectant. Crystals were transferred into the soaking buffer and incubated for 10 to 15 min at room temperature, followed by flash freezing in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected on the beamline MX1 and MX2 at the Australian Synchrotron, and on the 13B1 beamline at the National Synchrotron Radiation Research Center, Taiwan. Collected data were indexed and processed with the HKL2000 software (28). The structures were solved by molecular replacement using Phaser (29) with the TS–dUMP–raltitrexed structure (PDB code 1I00) as the search model for wild-type proteins and the inactive conformation of hTS structure (PDB code 1HW3) as the search model for mutant proteins. All the solvent molecules and ligands were removed prior to molecular replacement. Model building and structure refinement were carried out using REFMAC5 (30), respectively. The model was checked and manually improved in Coot (31). Structure validation was carried out using a Ramachandran plot (32). Ligand structures and restraints files were generated using Phenix.eLBOW (33). The statistics of data collection and structure refinement were summarized in Tables 1 and 2. Stereographic pairs of electron densities around the ligands in all structures are shown in supplemental Fig. S4.
Accession codes
The crystal structures of TS–Pi, TS–dUMP, TS–dUMP–raltitrexed, TS–dUMP–methotrexate, TS–dUMP–nolatrexed, and TS–dUMP–F13 have been deposited with the RCSB Protein Data Bank under the accession codes 5X5A, 5X5D, 5X5Q, 5X66, 5X67, and 5X69, respectively. The structures of mutant TS A191K-sf, A191K-s, and M190K-s have been deposited under accession codes 5X4W, 5X4X, and 5X4Y, respectively.
Author contributions
D. C., A. J., A. L., and P. N. conceived the research. D. C. performed the experiments and analyzed data. D. S. did cloning and small-scale protein construct screening. A. J., A. L., and P. N. provided technical advice and contributed to interpretation of data. D. C. wrote the first draft of the manuscript. All co-authors reviewed the results and contributed to the final version of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the Protein Production Platform at Nanyang Technological University for cloning and small-scale protein constructs screening of hTS. We thank the Australian Synchrotron and National Synchrotron Radiation Research Center for support during crystal data collection.
The work was supported by Nanyang Technological University Startup Grant M060080004. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 5X5A, 5X5D, 5X5Q, 5X66, 5X67, 5X69, 5X4W, 5X4X, and 5X4Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The numbering scheme used here is based on hTS.
- TS
- thymidylate synthase
- hTS
- human TS
- TMP
- thymidylate
- dUMP
- deoxyuridine monophosphate
- DSF
- differential scanning fluorimetry
- SEC-MALS
- size exclusion chromatography coupled with multi-angle light scattering
- ITC
- isothermal titration calorimetry
- r.m.s.
- root mean square
- ASU
- asymmetric unit
- DMSO
- dimethyl sulfoxide
- AS
- active site
- PDB
- Protein Data Bank.
References
- 1. Hori T., Ayusawa D., Shimizu K., Koyama H., and Seno T. (1984) Chromosome breakage induced by thymidylate stress in thymidylate synthase-negative mutants of mouse FM3A cells. Cancer Res. 44, 703–709 [PubMed] [Google Scholar]
- 2. Seno T., Ayusawa D., Shimizu K., Koyama H., Takeishi K., and Hori T. (1985) Thymineless death and genetic events in mammalian cells. Basic Life Sci. 31, 241–263 [DOI] [PubMed] [Google Scholar]
- 3. Wilson P. M., Danenberg P. V., Johnston P. G., Lenz H. J., and Ladner R. D. (2014) Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat. Rev. Clin. Oncol. 11, 282–298 [DOI] [PubMed] [Google Scholar]
- 4. Ackland S. P., Clarke S. J., Beale P., and Peters G. J. (2006) Thymidylate synthase inhibitors. Update Cancer Therap. 1, 403–427 [PubMed] [Google Scholar]
- 5. Assaraf Y. (2007) Molecular basis of antifolate resistance. Cancer Metastasis Rev. 26, 153–181 [DOI] [PubMed] [Google Scholar]
- 6. Wang W., Marsh S., Cassidy J., and McLeod H. L. (2001) Pharmacogenomic dissection of resistance to thymidylate synthase inhibitors. Cancer Res. 61, 5505–5510 [PubMed] [Google Scholar]
- 7. Kitchens M. E., Forsthoefel A. M., Barbour K. W., Spencer H. T., and Berger F. G. (1999) Mechanisms of acquired resistance to thymidylate synthase inhibitors: the role of enzyme stability. Mol. Pharmacol. 56, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 8. Kamb A., Finer-Moore J., Calvert A. H., and Stroud R. M. (1992) Structural basis for recognition of polyglutamyl folates by thymidylate synthase. Biochemistry 31, 9883–9890 [DOI] [PubMed] [Google Scholar]
- 9. Matthews D. A., Appelt K., Oatley S. J., and Xuong N. H. (1990) Crystal structure of Escherichia coli thymidylate synthase containing bound 5-fluoro-2′-deoxyuridylate and 10-propargyl-5,8-dideazafolate. J. Mol. Biol. 214, 923–936 [DOI] [PubMed] [Google Scholar]
- 10. Hardy L. W., Finer-Moore J. S., Montfort W. R., Jones M. O., Santi D. V., and Stroud R. M. (1987) Atomic structure of thymidylate synthase: target for rational drug design. Science 235, 448–455 [DOI] [PubMed] [Google Scholar]
- 11. Montfort W. R., Perry K. M., Fauman E. B., Finer-Moore J. S., Maley G. F., Hardy L., Maley F., and Stroud R. M. (1990) Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate. Biochemistry 29, 6964–6977 [DOI] [PubMed] [Google Scholar]
- 12. Peña M. M. O., Melo S. P., Xing Y.-Y., White K., Barbour K. W., and Berger F. G. (2009) The intrinsically disordered N-terminal domain of thymidylate synthase targets the enzyme to the ubiquitin-independent proteasomal degradation pathway. J. Biol. Chem. 284, 31597–31607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peña M. M., O., Xing Y., Y., Koli S., and Berger F., G. (2006) Role of N-terminal residues in the ubiquitin-independent degradation of human thymidylate synthase. Biochem. J. 394, 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang X., Gibson L. M., Bell B. J., Lovelace L. L., Peña M. M., Berger F. G., Berger S. H., and Lebioda L. (2010) Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability. Biochemistry 49, 2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Almog R., Waddling C. A., Maley F., Maley G. F., and Van Roey P. (2001) Crystal structure of a deletion mutant of human thymidylate synthase Δ(7–29) and its ternary complex with Tomudex and dUMP. Protein Sci. 10, 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phan J., Steadman D. J., Koli S., Ding W. C., Minor W., Dunlap R. B., Berger S. H., and Lebioda L. (2001) Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 276, 14170–14177 [DOI] [PubMed] [Google Scholar]
- 17. Cardinale D., Guaitoli G., Tondi D., Luciani R., Henrich S., Salo-Ahen O. M., Ferrari S., Marverti G., Guerrieri D., Ligabue A., Frassineti C., Pozzi C., Mangani S., Fessas D., Guerrini R., et al. (2011) Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 108, E542–E549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brunn N. D., Dibrov S. M., Kao M. B., Ghassemian M., and Hermann T. (2014) Analysis of mRNA recognition by human thymidylate synthase. Biosci. Rep. 34, e00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin X., Mizunuma N., Chen T., Copur S. M., Maley G. F., Liu J., Maley F., and Chu E. (2000) In vitro selection of an RNA sequence that interacts with high affinity with thymidylate synthase. Nucleic Acids Res. 28, 4266–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu E., Koeller D. M., Casey J. L., Drake J. C., Chabner B. A., Elwood P. C., Zinn S., and Allegra C. J. (1991) Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 88, 8977–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berger S. H., Berger F. G., and Lebioda L. (2004) Effects of ligand binding and conformational switching on intracellular stability of human thymidylate synthase. Biochim. Biophys. Acta 1696, 15–22 [DOI] [PubMed] [Google Scholar]
- 22. Lovelace L. L., Johnson S. R., Gibson L. M., Bell B. J., Berger S. H., and Lebioda L. (2009) Variants of human thymidylate synthase with loop 181–197 stabilized in the inactive conformation. Protein Sci. 18, 1628–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Webber S., Bartlett C. A., Boritzki T. J., Hillard J. A., Howland E. F., Johnston A. L., Kosa M., Margosiak S. A., Morse C. A., and Shetty B. V. (1996) AG337, a novel lipophilic thymidylate synthase inhibitor: in vitro and in vivo preclinical studies. Cancer Chemother. Pharmacol. 37, 509–517 [DOI] [PubMed] [Google Scholar]
- 24. Fauman E. B., Rutenber E. E., Maley G. F., Maley F., and Stroud R. M. (1994) Water-mediated substrate/product discrimination: the product complex of thymidylate synthase at 1.83 .ANG. Biochemistry 33, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 25. Fritz T. A., Liu L., Finer-Moore J. S., and Stroud R. M. (2002) Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access. Biochemistry 41, 7021–7029 [DOI] [PubMed] [Google Scholar]
- 26. Lovelace L. L., Minor W., and Lebioda L. (2005) Structure of human thymidylate synthase under low-salt conditions. Acta Crystallogr. D Biol. Crystallogr. 61, 622–627 [DOI] [PubMed] [Google Scholar]
- 27. Almqvist H., Axelsson H., Jafari R., Dan C., Mateus A., Haraldsson M., Larsson A., Martinez Molina D., Artursson P., Lundbäck T., and Nordlund P. (2016) CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nat. Commun. 7, 11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Ramachandran G. N., Ramakrishnan C., and Sasisekharan V. (1963) Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7, 95–99 [DOI] [PubMed] [Google Scholar]
- 33. Moriarty N. W., Grosse-Kunstleve R. W., and Adams P. D. (2009) Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.