Abstract
Signal transducer and activator of transcription 3 (STAT3) is activated by the IL-6 family of cytokines and growth factors. STAT3 requires phosphorylation on Ser-727, in addition to tyrosine phosphorylation on Tyr-705, to be transcriptionally active. In IL-6 signaling, the two major pathways that derive from the YXXQ and the YSTV motifs of gp130 cause Ser-727 phosphorylation. Here, we show that TGF-β-activated kinase 1 (TAK1) interacts with STAT3, that the TAK1-Nemo-like kinase (NLK) pathway is efficiently activated by IL-6 through the YXXQ motif, and that this is the YXXQ-mediated H7-sensitive pathway that leads to STAT3 Ser-727 phosphorylation. Because NLK was recently shown to interact with STAT3, we explored the role of STAT3 in activating this pathway. Depletion of STAT3 diminished the IL-6-induced NLK activation by >80% without inhibiting IL-6-induced TAK1 activation or its nuclear entry. We found that STAT3 functioned as a scaffold for TAK1 and NLK in vivo through a region in its carboxyl terminus. Furthermore, the expression of the STAT3534–770 region in the nuclei of STAT3-knockdown cells enhanced the IL-6-induced NLK activation in a dose-dependent manner but not the TGFβ-induced NLK activation. TGFβ did not cause STAT3 Ser-727 phosphorylation, even when the carboxyl region of STAT3 was expressed in the nuclei. Together, these results indicate that STAT3 enhances the efficiency of its own Ser-727 phosphorylation by acting as a scaffold for the TAK1-NLK kinases, specifically in the YXXQ motif-derived pathway.
Keywords: gp130, YXXQ motif, signaling specificity
The signal transducer and activator of transcription (STAT) family plays pivotal roles in a variety of systems and in development, in response to cytokines and growth factors. STAT family members are activated in the cytoplasm by tyrosine phosphorylation, form dimers, and enter the nucleus, where they act as DNA-binding transcription factors (1). Of the seven known STATs, STAT1, 3, 5A, 5B, and 6 are phosphorylated at one or two serine residues in their carboxyl-terminal transactivation domain (1, 2) and at a critical tyrosine. The serine phosphorylation enhances the transcriptional activity in the case of STAT1 (3), STAT3 (3, 4), and STAT6 (5). STAT3 can be activated by a variety of cytokines, including the IL-6 family, by using gp130 as a common receptor subunit (6), Granulocyte colony-stimulating factor (G-CSF) and erythropoietin, and growth factors, EGF, platelet-derived growth factor, and hepatocyte growth factor, and cytoplasmic tyrosine kinases, including Src and v-Eyk (reviewed in ref. 7). IL-6 uses STAT3 in its major signaling pathway and concomitantly activates the Ras/Raf/ERK and PI3-kinase pathways (7). IL-6, therefore, activates multiple genes, including acute-phase reactants, and the junB, tis11, stat3, c-myc, and c-fos genes, mostly through STAT3 (8–14). In the IL-6 receptor system, the tyrosine-phosphorylated YXXQ motif of gp130 is critical for recruiting STAT3 for subsequent phosphorylation at Tyr-705 by the associated Jak kinases (15). In addition, Abe et al. (4) showed that the YXXQ motif has another important role in activating STAT3, by phosphorylating Ser-727 through an H7-sensitive kinase pathway in response to IL-6, even at low concentrations. At higher concentrations of IL-6, another PD98059-sensitive pathway derived from the YSTV motif of gp130 also participates in STAT3 Ser-727 phosphorylation (4).
TGF-β-activated kinase 1 (TAK1) is a mitogen-activated protein MAP kinase kinase kinase that is activated by the TGFβ family, IL-1β, TNFα, and the Toll-like receptor family (16–19). TAK1 works with TAB1 (17), TAB2 (20), and TAB3 (21–23) to activate downstream kinases, including the IκB kinases, leading to NF-κB activation, and MAP kinase kinase (MKK) 3/6 and MKK4/7, leading, respectively, to p38 and JNK activation (16, 18, 24, 25). Recently, TAK1 was shown to activate Nemo-like kinase (NLK) in TGFβ (26), Wnt5a (27), and Wnt1 (28) signaling. More recently, Ohkawara et al. (29) showed that the TAK1-NLK pathway plays a critical role in TGFβ-induced mesodermal development in Xenopus embryos by phosphorylating STAT3 at Ser-727. However, it remains unclear how TAK1 and NLK select their downstream targets in the different signaling pathways.
In this study, we identified TAK1 in the STAT3 complex by using a combination of tandem affinity purification (TAP) and MS. We then found that TAK1 and its downstream kinase, NLK, were selectively activated by IL-6 through the YXXQ motif of gp130. We showed that TAK1 and NLK are critical components of the YXXQ-derived H7-sensitive kinase pathway leading to STAT3 Ser-727 phosphorylation. We further showed that STAT3 specifically enhances the IL-6-induced TAK1-dependent NLK activation, but not TGFβ-induced NLK activation, by acting as a specific scaffold for the kinases involved in the IL-6 signaling. Thus, the upstream TAK1, which is differentially activated by different signaling pathways, seems to select its specific downstream effector kinases in conjunction with a specific scaffold protein, which is STAT3 in the case of the IL-6 signal.
Methods
Cytokines, Abs, and Reagents. The recombinant human IL-6 and G-CSF were gifts from Ajinomoto (Kawasaki, Japan) and Kirin Brewery (Tokyo), respectively. All other cytokines were from PeproTech (London). Rabbit anti-STAT3 and anti-TAK1 Abs were described (18, 30). Anti-phospho-Ser-727 and phospho-Tyr-705 STAT3 Abs were from Cell Signaling Technology (Beverly, MA). Rabbit anti-GST Ab was from Upstate Biotechnology (Lake Placid, NY). Anti-HA Mo Ab (12CA5) was from Boehringer Mannheim (Mannheim, Germany). All other Abs were from Santa Cruz Biotechnology. H7, 1-(5-iso-quinolinyl-sulfonyl)-2-methylpiperazine, and all other reagents were from Sigma-Aldrich (St. Louis).
Cell Lines. All of the cell lines were grown in DMEM supplemented with 10% FCS and the appropriate antibiotics. HepG2-G108YRHQ and HepG2-G108YSTV were described (4). The HepG2 cell line with knocked-down stat3 mRNA (HepG2-STAT3KD) and its derivative cell line with RNA interference (RNAi)-resistant STAT3 (HepG2-KD-STAT3R) were described (14). For the kinase assays, HepG2 cells were cultured in DMEM containing 0.5% FCS for 36 h before stimulation.
Plasmids. The expression vectors encoding the hemagglutinin (HA)-STAT3 and HA-TAK1 were described (18, 31). The TAP tag was attached to the amino terminus of mouse STAT3 (ref. 32 and modified by K.N.), and the TAP-Stat3 cDNA was inserted into the pEF-BOS expression vector (a gift from S. Nagata, Osaka University, Osaka). pCMV-Myc-NLK was generated by inserting a PCR-generated cDNA encoding human NLK into pCDNAMyc (a gift from K. Iwai, Osaka City University, Osaka). For the lentivirus (LV)-mediated expression system, the Myc-NLK cDNA was subcloned into a pCSII-EF-MCS-IRES2-Venus vector (a gift from H. Miyoshi, RIKEN, Tsukuba, Japan). A cDNA fragment encoding STAT3534–770 was made by PCR and fused to NLS-FLAG at the amino terminus, and the resultant cDNA was subcloned into the pCSII-EF-MCS-IRES2-Venus vector. All constructs were verified by DNA sequencing. pEF-GST and pEF-GST-STAT3533–770 were described (4).
TAP and MS Analysis. TAP was performed as described (32). Briefly, TAP-STAT3 was stably expressed in 293T-G133 cells. Protein complexes interacting with STAT3 were then sequentially affinity-purified by using IgG-Sepharose, Tobacco etch virus protease digestion, and calmodulin beads. The eluates of calmodulin beads were digested with trypsin, and the resultant peptides were subjected to splitless nanoflow liquid chromatography coupled to nanoelectrospray tandem MS (33).
EMSA. 32P-labeled oligonucleotides containing an NF-κB-binding site (5′-AGTGAGGGGACTTTCCGAGTG-3′) from the intercellular adhesion molecule-1 promoter and nuclear extracts from HepG2 cells were used.
LV-Mediated Expression of RNAi. We generated RNAi constructs in the plasmid pU6i-cassette (Genofunction) (34). The target positions of the RNAi against human TAK1 and human NLK were as follows: TAK1, GAGATCGACTACAAGGAGA; NLK, GGCGCACCATCATCAGCAC. To construct the lentiviral RNAi expression system, the cassette containing the U6 promoter and the RNAi was transferred into an LV vector, CS-RfA-EG, with the EF1α promoter-driven GFP gene (a gift from H. Miyoshi). LVs were prepared according to the previously described method (35), except that a three-vector system was used instead of a four-vector system (36).
In Vitro Kinase Assay. The in vitro kinase assays were performed as described (26). For the TAK1 kinase assays, endogenous TAK1 was used, and in some experiments, transiently expressed Flag-tagged TAK1 was used, with recombinant MKK6 from Escherichia coli as a substrate. For the NLK kinase assays, Myc-tagged NLK protein was extracted from various HepG2 cell lines expressing Myc-NLK, and subjected to the in vitro kinase assay to detect autophosphorylation. In some experiments, GST-STAT3534–770 produced in E. coli was used as a substrate.
Results
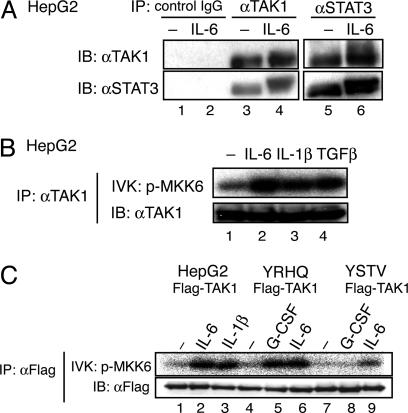
Identification of TAK1 as a STAT3-Interacting Protein That Is Activated by IL-6 Through the YXXQ Motif in gp130. To identify molecules acting with STAT3, we first obtained STAT3-associated complexes by the TAP method (32) from gp130-stimulated 293T-G133 cells (4) stably expressing TAP-tagged STAT3. We analyzed digests of the complexes by using direct nanoflow liquid chromatography-coupled tandem MS (33) and identified multiple peptides. One peptide sequence, QELVAELDQDEK, corresponded to residues 521–532 of human TAK1. We then tested whether endogenous TAK1 and STAT3 formed complexes in HepG2 cells. As shown in Fig. 1A, lanes 3 and 4, anti-TAK1 immunoprecipitates of WCEs from unstimulated and IL-6-stimulated HepG2 cells contained STAT3. The STAT3–TAK1 interaction was reciprocally confirmed by detecting TAK1 in the anti-STAT3 immunoprecipitates (Fig. 1 A, lanes 5 and 6).
Fig. 1.
Endogenous TAK1 binds to STAT3 and is activated through the YXXQ motif in gp130. (A) HepG2 cells were untreated (–) or treated with IL-6 for 15 min. WCE was immunoprecipitated (IP) with control IgG (lane 1–2), anti-TAK1 (lanes 3 and 4), or anti-STAT3 (lanes 5 and 6) Abs. Immunoprecipitates were resolved on SDS/PAGE and immunoblotted (IB) with the indicated Abs. (B) HepG2 cells were untreated (–) or treated with 20 ng/ml IL-6, IL-1β, or TGF-β for 10 min. The proteins immunoprecipitated with anti-TAK1 Ab were divided into two aliquots. One aliquot was subjected to an in vitro kinase assay (IVK) with recombinant MKK6 as the substrate (Upper), and the other aliquot was used in an immunoblot to examine the amount of immunoprecipitated TAK1 (Lower). (C) HepG2, HepG2-G108-YRHQ, or HepG2-G108-YSTV cells were transfected with pEF-Flag-TAK1 plasmid, and untreated (–) or treated with the indicated cytokines at 20 ng/ml for 10 min. The anti-Flag immunoprecipitates of WCEs were subjected to the kinase assay (Upper) and immunoblot analysis with an anti-Flag Ab (Lower) as in B.
We next tested whether IL-6 could activate endogenous TAK1. Anti-TAK1 immunoprecipitates of WCEs from unstimulated or IL-6-stimulated HepG2 cells were subjected to an in vitro kinase assay by using MKK6 as the substrate. As shown in Fig. 1B, IL-6 activated endogenous TAK1 as efficiently as did IL-1β and TGFβ. We next tested whether a tyrosine-phosphorylated motif in gp130 was required for TAK1 activation. As shown in Fig. 1C, stimulation of HepG2-G108YRHQ cells that express a chimeric receptor consisting of G-CSFR-gp130 with the cytoplasmic portion of gp130 truncated to 108 amino acid residues and a YXXQ motif peptide, efficiently increased the TAK1 kinase activity. In contrast, this increase was not seen in HepG2-G108YSTV cells that contain a YSTV motif instead of the YXXQ motif. These results indicated that the YXXQ motif was responsible for the TAK1 activation.
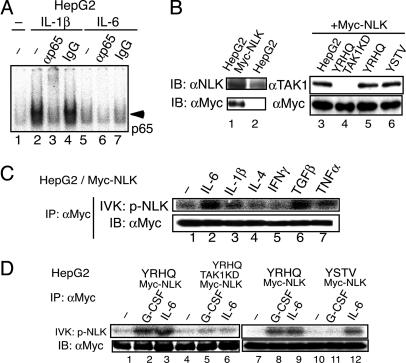
IL-6 Preferentially Activates NLK Via the YXXQ-TAK1-Mediated Pathway. Once we discovered that IL-6 activated TAK1, we investigated which downstream pathways were activated through TAK1 in response to IL-6. First, we examined NFκB activation by using an EMSA. In contrast to the efficient induction of NFκB containing p65 by IL-1β (Fig. 2A, lanes 2 and 3), IL-6 caused only slight NFκB DNA-binding activity in HepG2 cells (Fig. 2 A, lanes 5 and 6). p38 and JNK activation were also tested, with no apparent activation observed in the IL-6-stimulated HepG2 cells (data not shown). We then examined whether IL-6 activated NLK. To efficiently test the NLK kinase activity, we stably expressed Myc-tagged NLK (Myc-NLK) in HepG2 cells, HepG2-G108YRHQ, or HepG2-G108YSTV, and a HepG2 cell line in which TAK1 was knocked down (described later and shown in Fig. 3A), with an LV expression system. The expression levels of Myc-NLK were very similar in these cell lines (Fig. 2B, lanes 3–6), and the level of Myc-NLK was close to that of endogenous NLK in HepG2 cells (Fig. 2B, lanes 1 and 2). The Myc-NLK was recovered with an anti-Myc Ab from HepG2 cells that had either been left unstimulated or stimulated, as indicated, for 10 min. The Myc-NLK was then tested for its autophosphorylation kinase activity (Fig. 2C). IL-6 activated NLK efficiently, as did TGFβ, IL-1β, and TNFα; two other cytokines, IFNγ and IL-4, did not activate NLK. Consistent with the role played by the YXXQ motif in the IL-6-induced TAK1 activation, stimulation of the chimeric GCSF-R-gp130 receptor containing the YXXQ motif, but not the YSTV motif, caused NLK activation at the level similar to that obtained with IL-6 (Fig. 2D, lanes 1–3 and 7–12). Together with the finding that the YXXQ-derived signal-induced and IL-6-induced NLK activation was severely impaired in TAK1-knockdown HepG2 cells (Fig. 2D, lanes 4–6), these results indicated that IL-6 preferentially activates the TAK1-NLK pathway through the YXXQ motif.
Fig. 2.
Preferential activation of NLK through the YXXQ-TAK1 pathway. (A) EMSA for NFκB activity. 32P-labeled oligonucleotides containing an NFκB-binding site and nuclear extracts from HepG2 cells that were untreated (–) or stimulated with IL-1β or IL-6 for 30 min were used. The reaction mixtures were preincubated with no additions (lanes 1, 2, and 5), anti-p65 Ab (lanes 3 and 6), or control rabbit IgG (lanes 4 and 7). (B) HepG2 cells (lanes 1 and 3), HepG2-G108YRHQ (lane 5), HepG2-G108YSTV (lane 6), or HepG2-G108YRHQ with TAK1 KD (lane 4, see also Fig. 3A) cells were infected with LV-Myc-NLK. WCEs from parental (lane 2) and the transfected cells were immunoblotted (IB) with an anti-NLK Ab (Left Upper, lanes 1 and 2), an anti-TAK1 Ab (Right Upper, lanes 3–6) or anti-Myc Ab (Lower). The total amount of endogenous and exogenous NLK in the HepG2-Myc-NLK cells was estimated as ≈2- to 2.2-fold of that of endogenous NLK in the parental cells. (C and D) The various HepG2-Myc-NLK cells shown in B were left unstimulated or stimulated with the indicated cytokines at 20 ng/ml for 10 min. The anti-Myc immunoprecipitates (IP) of WCE were divided into two aliquots. Each aliquot was subjected to the NLK kinase assay in the absence of exogenous substrate (Upper) or immunoblot analysis with the anti-Myc Ab (Lower).
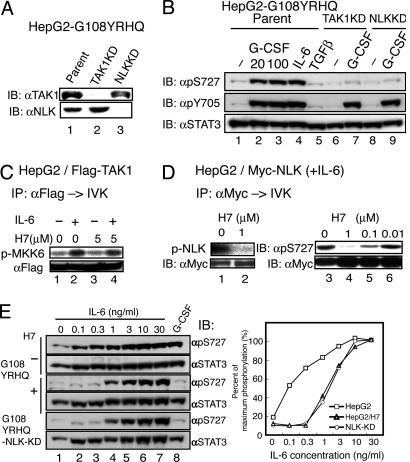
Fig. 3.
TAK1 and NLK are components of the YXXQ-mediated H7-sensitive kinase pathway leading to the phosphorylation of STAT3 Ser-727. (A) HepG2-G108YRHQ cells were infected with LV-U6-small interfering RNA against TAK1 and NLK to deplete TAK1 and NLK (TAK1KD and NLKKD, respectively). The silencing effect was evaluated by immunoblotting with the indicated Ab. (B) HepG2-G108YRHQ cells and the TAK1KD or NLK KD HepG2-G108YRHQ cells were left unstimulated or stimulated with the indicated cytokines at 20 ng/ml (lanes 2, 4, and 5) or at 100 ng/ml (lanes 3, 7, and 9) for 15 min. The WCEs were immunoblotted with the indicated Abs. (C) The Flag-TAK1 immunoprecipitates from unstimulated or IL-6-stimulated HepG2 cells were subjected to the in vitro kinase assay as in Fig. 1B in the absence or presence of 5 μM H7. (D) HepG2-Myc-NLK cells were stimulated with IL-6 at 20 ng/ml for 15 min. The immunoprecipitates of WCEs with anti-Myc Ab were subjected to the in vitro NLK kinase assay as in Fig. 2C (lanes 1 and 2) or the in vitro NLK kinase assay by using GST-STAT3 (534–770) made in E. coli (lanes 3–6) in the presence of the indicated amounts of H7. Phospho-Ser-727 was detected with an antiphospho-Ser-727 Ab (Right Upper). The amount of NLK used for each kinase assay is shown in Lower. (E) HepG2-G108YRHQ cells pretreated with nothing (–) or H7 at 50 μM (+) and HepG2-G108YRHQ-NLK-KD cells were left unstimulated or stimulated with the indicated concentrations of IL-6 (lanes 2–7) or 20 ng/ml G-CSF for 15 min. The WCEs were immunoblotted with the indicated Abs (Left). The intensities of each band were quantified by a densitometer (GS-700, Bio-Rad), and the percentage of STAT3 in the phosphorylated form was calculated (Right). The value in HepG2 cells stimulated with 30 ng/ml IL-6 was defined as 100%, because almost all of the STAT3 was phosphorylated on Ser-727, as judged by its migration pattern on SDS/PAGE (data not shown). Average values of two independent experiments are shown.
The TAK1-NLK Pathway Is the YXXQ-Mediated H7-Sensitive Pathway That Leads to STAT3 Ser-727 Phosphorylation. To test the roles of endogenous TAK1 and NLK in the IL-6- and YXXQ-derived signaling, we knocked down the expression levels of TAK1 or NLK in HepG2-G108YRHQ cells by the RNAi method, using a lentiviral vector system expressing a U6-driven loop-type small interfering RNAs against the tak1 and nlk mRNAs. The LV without an RNAi sequence was used as the control. The small interfering RNAs against the tak1 and nlk mRNAs effectively inhibited the production of their corresponding proteins (Fig. 3A). As shown in Fig. 3B, the YXXQ signal efficiently caused Ser-727 phosphorylation as did IL-6 at 20 ng/ml, and the YXXQ-mediated Ser-727 phosphorylation was severely impaired in both the TAK1- and NLK-depleted cells, whereas the phosphorylation of STAT3 on Tyr-705 was intact in these cells, indicating that the TAK1-NLK pathway is critical for STAT3 Ser-727 phosphorylation by the YXXQ-derived pathway. It was also noted that TGFβ did not cause STAT3 Ser-727 phosphorylation in HepG2 cells despite the strong NLK activation (Fig. 3B, lane 5). TAK1 kinase activity was resistant to H7 at 5 μMin the in vitro kinase assay (Fig. 3C, lanes 3 and 4), whereas the NLK kinase activity was very sensitive to H7 in the autophosphorylation kinase assay (Fig. 3D, lanes 1 and 2), and in an assay using GST-STAT3534–770 made in E. coli as the substrate (Fig. 3D, lanes 3–6) with an of IC50 <0.1 μM. We then tested the role of this pathway in STAT3 Ser-727 phosphorylation at various IL-6 concentrations. We compared the levels of STAT3 Ser-727 phosphorylation in HepG2-G108YRHQ cells and NLK-knockdown-HepG2-G108YRHQ cells in response to increasing doses of IL-6. As shown in Fig. 3E, STAT3 Ser-727 phosphorylation by IL-6 was detected even at 0.1 ng/ml IL-6 and peaked at ≈10 ng/ml in the parental cells. In contrast, in the NLK-KD cells, the Ser-727 phosphorylation responses were suppressed when stimulated with 0.1, 0.3, and 1 ng/ml IL-6, but gradually reached a maximal level similar to that seen in the parental cells at 10 ng/ml. Very similar responses were observed in HepG2 cells pretreated with H7 at 50 μM (Fig. 3E, indicated as H7+) and in the HepG2-G108-TAK1KD cells (data not shown). These results indicated that the TAK1-NLK pathway actually represented the components of the previously characterized YXXQ-derived H7-sensitive kinase pathway that leads to STAT3 Ser-727 phosphorylation.
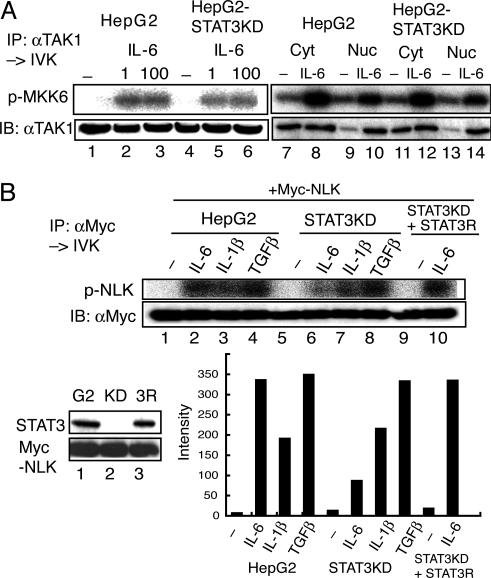
STAT3 Is Required for IL-6-Induced NLK Activation but Not for TAK1 Activation. Because STAT3 interacts with TAK1 even after IL-6 stimulation (Fig. 1 A) and with NLK (29), we explored the role of STAT3 in the YXXQ-derived TAK1-NLK pathway. We used STAT3-knockdown HepG2 cells (HepG2-STAT3KD) and a derivative HepG2-STAT3KD cell line that was reconstituted with short interfering RNA-resistant STAT3 (HepG2-KD-STAT3R) (14). Stimulation with IL-6 at 1 and 100 ng/ml caused TAK1 activation in the HepG2-STAT3KD cells to levels comparable to those obtained in the parental cells (Fig. 4A, lanes 1–6). Interestingly, in response to IL-6, TAK1 translocated into the nucleus (Fig. 6A, which is published as supporting information on the PNAS web site), and the kinase activities in both the cytosolic and nuclear fractions increased even without STAT3 (Fig. 4A, lanes 7–14). These results indicated that the presence of STAT3 did not affect the activation step of TAK1 in response to IL-6.
Fig. 4.
STAT3 is required for IL-6-induced NLK activation but not for TAK1 activation. (A) HepG2 and HepG2-STAT3-knockdown (KD) cells were left unstimulated or stimulated with IL-6 at 1 (lanes 2 and 5), 100 (lanes 3 and 6), or 20 (lanes 8, 10, 12, and 14) ng/ml. Endogenous TAK1 from WCE extracts (lanes 1–6), cytosolic extracts (Cyt, lanes 7, 8, 11, and 12) or nuclear extracts (Nuc, lanes 9, 10, 13, and 14) was immunoprecipitated with an anti-TAK1 Ab, and the kinase activity was measured by using MKK6 as the substrate (Upper). (Lower) Immunoblotting with the anti-TAK1 Ab. (B) HepG2 (G2) cells, HepG2-STAT3KD (KD), and HepG2-STAT3KD reconstituted with RNAi-resistant STAT3 (3R) were infected with LV-Myc-NLK. The expression levels of STAT3 and Myc-NLK are shown in Left Lower. These HepG2 and derivative cells expressing Myc-NLK were left unstimulated or stimulated with the indicated cytokines at 20 ng/ml for 10 min. Kinase assay (Upper) and Western blotting analysis (Lower) of the anti-Myc-immunoprecipitates of WCE are shown. The radioactivity level of each band in the kinase assay was quantified with the BAS 5000 Bioimaging analyzer (Fujix, Tokyo). The activity of NLK in the IL-6-stimulated HepG2-STAT3 KD was estimated to be 17.6 ± 7.6% of that of IL-6-stimulated HepG2 cells (n = 4).
In contrast, the IL-6-induced NLK kinase activity was markedly reduced in the HepG2-STAT3KD cells, to <20% of the activity obtained with the parental cells (Fig. 4B, lanes 2 and 6; see also Fig. 5C; average = 17.6 ± 7.2%, n = 4), and this IL-6-induced NLK activity fully recovered in the HepG2-KD-STAT3R cells (Fig. 4B, lanes 9 and 10), suggesting that STAT3 has a role in the TAK1-dependent NLK activation after the TAK1 activation step of the IL-6 signal. This STAT3-dependency was not observed for the NLK activation by IL-1β or TGFβ (Fig. 4B), indicating a specific role for STAT3 in NLK activation by IL-6 signals.
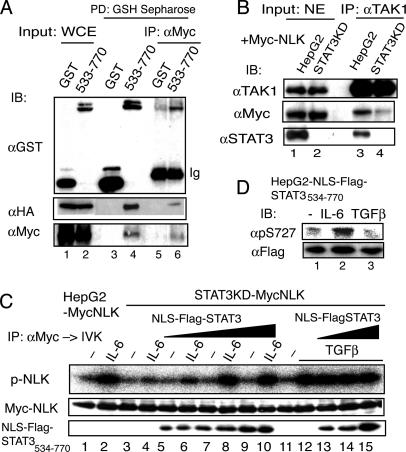
Fig. 5.
The STAT3 molecule acts as a scaffold for TAK1/NLK in the YXXQ-derived pathway. (A) 293T cells were transfected with pCMV-HA-TAK1, pCMV-Myc-NLK, and pEFGST or pEFGST-STAT3533–770 plasmid. The GST-labeled proteins were pulled-down with reduced glutathione (GSH)-Sepharose. The eluates were diluted with 50 mM Hepes buffer (pH 7.8) containing 150 mM NaCl, and subjected to immunoprecipitation using an anti-Myc Ab. The WCEs (lanes 1 and 2), the eluates from the GSH-Sepharose (lanes 3 and 4), and the anti-Myc Ab immunoprecipitates (lanes 5 and 6) were subjected to immunoblotting analysis with the indicated Abs. (B) HepG2-MycNLK and HepG2-STAT3KD-MycNLK were treated with 20 ng/ml IL-6 for 10 min. The nuclear extracts (500 μg each) were immunoprecipitated with an anti-TAK1 Ab. Nuclear extracts (20 μg, lanes 1 and 2) and the immunoprecipitates (lanes 3 and 4) were separated and probed with the indicated Abs. (C) HepG2-STAT3KD-MycNLK cells were infected with three doses of lentiviral preparation LV-EFNLS-FlagSTAT3534–770. HepG2-MycNLK-, HepG2-STAT3KD-MycNLK-, and NLS-FlagSTAT3534–770-expressing cells were stimulated with IL-6 (lanes 2, 4, 6, 8, and 10), or TGF-β (lanes 12–15) at 20 ng/ml, for 10 min or left unstimulated. The anti-Myc-immunoprecipitates (IP) of WCEs were subjected to the in vitro NLK kinase assay (p-NLK) and immunoblot (IB) with the anti-Myc Ab. (Lower) Immunoblots using an anti-STAT3 Ab indicate that the lysate samples contained increasing amounts of NLS-FlagSTAT3534–770 protein in the WCE. (D) HepG2 cells expressing NLS-FlagSTAT3534–770 were left untreated or treated with IL-6 or TGFβ for 15 min. The WCEs were separated on SDS/PAGE and probed with the indicated Abs.
STAT3 Enhances the TAK1-Dependent NLK Activation by Acting as a Scaffold Specifically in the YXXQ-Derived Signal. We previously showed that the carboxyl-terminal region of STAT3 from residues 533 to 770 was enough to be phosphorylated on Ser-727 by the YXXQ-derived pathway (4). Consistently, GST-STAT3533–770 fusion protein interacted with HA-TAK1 or Myc-NLK when coexpressed in 293T cells (data not shown). We then examined the nature of the interactions among STAT3, TAK1, and NLK. Glutathione beads were loaded with WCEs from cells coexpressing GST or GST-STAT3533–770, Myc-NLK, and HA-TAK1. The protein complexes containing GST and GST-STAT3533–770 were then recovered from the beads. As shown in Fig. 5A, TAK1, and NLK, interacted with GST-STAT3533–770 (lane 4), but not with GST (lane 3). The eluates from the beads were then further immunoprecipitated with an anti-Myc Ab to obtain the NLK complexes. Anti-Myc immunoprecipitates of the eluates contained HA-TAK1 and GST-STAT3533–770 in addition to Myc-NLK (lane 6). Control GST did not cause this coimmunoprecipitation (lane 5). Taken together with the fact that TAK1 does not directly interact with NLK (28), these results indicate that STAT3 formed a ternary complex with HA-TAK1 and Myc-NLK through its carboxyl-terminal region.
Next, we further tested whether the interaction of endogenous TAK1 and NLK in the nucleus required STAT3 molecules. The Myc-NLK, which was expressed at a level similar to that of endogenous NLK, was located largely in the nucleus with or without IL-6 stimulation, as was endogenous NLK (Fig. 6 B and C). As shown in Fig. 5B, lanes 1 and 2, Myc-NLK was present at a similar level in the nuclear extracts from IL-6-stimulated HepG2-MycNLK and HepG2-STAT3KD-MycNLK cells. Comparable amounts of TAK1 were present in the nucleus (Fig. 5B, lanes 1 and 2) in response to IL-6, even in the absence of STAT3, and efficiently recovered with an anti-TAK1 Ab (Fig. 5B, lanes 3 and 4). NLK and STAT3 clearly coimmunoprecipitated with TAK1 in the nuclear extracts of IL-6-stimulated HepG2-MycNLK cells (Fig. 5B, lane 3), whereas the amount of NLK that coimmunoprecipitated with TAK1 in the nuclear extracts of HepG2-STAT3KD-MycNLK cells (Fig. 5B, lane 4) was markedly reduced to ≈15% of that in Fig. 5B, lane 3. These results indicate that the nuclear TAK1-NLK interaction in response to IL-6 is largely dependent on STAT3.
To substantiate this TAK1–STAT3–NLK interaction, we stably expressed an NLS-FlagSTAT3534–770 at three different levels in HepG2-STAT3KD-MycNLK cells. These cells were left unstimulated or stimulated with IL-6 or TGFβ for 10 min, and the NLK activities were assayed. As shown in Fig. 5C, expression of the carboxyl terminus of STAT3 in the nucleus enhanced the IL-6-induced NLK activation in a dose-dependent manner. Interestingly, the TGFβ-induced NLK activation was not enhanced by the presence of the carboxyl terminus of STAT3 in the nucleus. These results indicate that STAT3 has an enhancing effect on the TAK1-dependent NLK activation, specifically in IL-6 signaling. We further tested whether NLK that was activated by the TGFβ signal in a STAT3-independent manner could phosphorylate STAT3 on Ser-727 in the nucleus. Very interestingly, in contrast to IL-6, TGFβ did not cause Ser-727 phosphorylation of the NLS-FlagSTAT3534–770 expressed in the nucleus (Fig. 5D), suggesting that the substrate specificity of the NLKs that are activated through different pathways depends on the scaffold proteins used in the specific signaling pathways.
Discussion
This study showed that gp130, a member of the cytokine receptor superfamily, efficiently activates the TAK1 through the YXXQ motif in gp130, and this kinase preferentially activates the downstream kinase NLK, which is responsible for STAT3 Ser-727 phosphorylation via the YXXQ motif-derived signal. Furthermore, this study revealed a novel role for STAT3 in IL-6-induced NLK activation but not in TGFβ-induced NLK activation. STAT3 functions as a scaffold for TAK1 and NLK specifically in IL-6 signaling by binding them at its carboxyl-terminal region. The following observations support the idea that this interaction is physiologically significant: (i) most TAK1-NLK interactions in the nuclear extracts of IL-6-stimulated HepG2 cells depended on the presence of STAT3 (Fig. 5B); (ii) the IL-6-induced NLK activation was greatly decreased in HepG2-STAT3KD cells; (iii) STAT3 and TAK1 were independently activated by IL-6, even at low concentrations, and they then translocated into the nucleus (Figs. 4B and 6A), although most NLK resides in the nucleus before stimulation (ref. 36 and Fig. 6B); and (iv) expression of the carboxyl-terminal portion of STAT3 in the nucleus markedly increased the IL-6-induced NLK activation. In contrast, TGFβ neither used STAT3 to activate NLK (Fig. 4B) nor caused the phosphorylation of STAT3 on Ser-727 (Fig. 3B). This finding could not be explained by the fact that TGFβ did not cause the translocation of STAT3, because TGFβ-induced NLK activation could not be enhanced by the forced expression of STAT3 in the nucleus (Fig. 5C), and NLK activated by TGFβ did not cause Ser-727 phosphorylation of the nuclear STAT3 (Fig. 5D). Thus, NLKs activated through different pathways have different substrate specificities, probably reflecting the nature of the complexes containing the upstream TAK1 kinase and the scaffold proteins involved in the specific activation signals.
We previously showed that the IL-6 signals for STAT3 Ser-727 phosphorylation are mediated by at least two kinase pathways, one is a YSTV-motif-derived PD98059-sensitive pathway, and the other is a YXXQ-motif-derived H7-sensitive kinase pathway (4). The kinase pathway described here is fully consistent with the H7-sensitive kinase pathway, because it has all of the characteristics previously shown for the YXXQ-derived H7-sensitive kinase that leads to STAT3 Ser-727 phosphorylation, including the characteristics indicated in Fig. 3 C–E. This pathway was also active in other cell lines tested, including the NIH 3T3 and HEK293T cell lines (H. Kojima, T.S., and K.N., unpublished data). This pathway seems to be rather specific to the receptor systems that use gp130 or to others containing the YXXQ motif, because neither IFNγ nor IL-4 activated NLK. Other cytokines should be tested to see if they activate this pathway.
To date, only a few downstream targets of NLK have been identified, including TCF/LEF (26), STAT3 (ref. 29 and this study), and c-Myb (28). We do not know what other NLK substrates are in the YXXQ-derived pathway at present. Recently, Kanei-Ishii et al. (28) showed that TAK1, which is activated by the Wnt-1 signal, activates NLK through HIPK2, which functions as an intermediate kinase between TAK1 and NLK. NLK then phosphorylates c-Myb at multiple sites, leading to ubiquitination and proteasome-dependent degradation. In their case, c-Myb interacted with HIPK2 and NLK in the nucleus. It would be intriguing to test whether c-Myb works as a scaffold for NLK activation specifically in Wnt-1 signaling, as STAT3 appears to do in IL-6 signaling. Ohkawara et al. (29) showed that the TAK1-NLK pathway is responsible for the phosphorylation of Xenopus STAT3 on Ser-727 in the marginal zone of the 32-cell embryo, and that NLK interacts with and phosphorylates STAT3 on Ser-727 in vitro. Together with results showing that both STAT3 and TAK1-NLK are required for TGFβ-mediated mesoderm induction, they concluded that the TAK1-NLK-STAT3 cascade participates in the TGFβ-mediated mesoderm induction (29). However, in the present report, TGFβ did not cause STAT3 Ser-727 phosphorylation in HepG2 cells, even when the carboxyl portion of STAT3 was expressed in the nucleus, despite the strong activation of NLK by TGFβ. Clearly, more studies need to be performed to understand how different upstream pathways select the downstream kinases and affect the substrate specificity of those kinases.
Supplementary Material
Acknowledgments
We gratefully acknowledge Akihiro Iwamatsu and his laboratory for advice on protein identification. We are indebted to H. Miyoshi for the lentiviral vector system. We thank Y. Niwa for technical assistance and E. L. Barsoumian for encouraging us. This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan, Nippon Boehringer Ingelheim Co., Ltd., GenoFunction, Inc., and the Osaka Foundation for Promotion of Clinical Immunology.
Author contributions: H. Kojima, K.M., and K.N. designed research; H. Kojima, T.S., S.-I.I., H.Z., S.K., H. Kunimoto, and K.N. performed research; T.I. contributed new reagents/analytic tools; H. Kojima, T.S., K.M., and K.N. analyzed data; and H. Kojima and K.N. wrote the paper.
Abbreviations: STAT, signal transducer and activator of transcription; TAK1, TGF-β-activated kinase 1; NLK, Nemo-like kinase; RNAi, RNA interference; TAP, tandem affinity purification; G-CSF; granulocyte colony-stimulating factor; MAP, mitogen-activated protein; MKK; MAP kinase kinase; HA, hemagglutinin; WCE, whole-cell extract; LV, lentivirus.
References
- 1.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell. Biol. 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 2.Decker, T. & Kovarik, P. (2000) Oncogene 19, 2628–2637. [DOI] [PubMed] [Google Scholar]
- 3.Wen, Z., Zhong, Z. & Darnell, J. E., Jr. (1995) Cell 82, 241–250. [DOI] [PubMed] [Google Scholar]
- 4.Abe, K., Hirai, M., Mizuno, K., Higashi, N., Sekimoto, T., Miki, T., Hirano, T. & Nakajima, K. (2001) Oncogene 20, 3464–3474. [DOI] [PubMed] [Google Scholar]
- 5.Pesu, M., Takaluoma, K., Aittomaki, S., Lagerstedt, A., Saksela, K., Kovanen, P. E. & Silvennoinen, O. (2000) Blood 95, 494–502. [PubMed] [Google Scholar]
- 6.Boulton, T. G., Zhong, Z., Wen, Z., Darnell, J. E., Jr., Stahl, N. & Yancopoulos, G. D. (1995) Proc. Natl. Acad. Sci. USA 92, 6915–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano, T., Ishihara, K. & Hibi, M. (2000) Oncogene 19, 2548–2556. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima, K. & Wall, R. (1991) Mol. Cell. Biol. 11, 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima, K., Kusafuka, T., Takeda, T., Fujitani, Y., Nakae, K. & Hirano, T. (1993) Mol. Cell. Biol. 13, 3027–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichiba, M., Nakajima, K., Yamanaka, Y., Kiuchi, N. & Hirano, T. (1998) J. Biol. Chem. 273, 6132–6138. [DOI] [PubMed] [Google Scholar]
- 11.Kiuchi, N., Nakajima, K., Ichiba, M., Fukada, T., Narimatsu, M., Mizuno, K., Hibi, M. & Hirano, T. (1999) J. Exp. Med. 189, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, E., Lerner, L., Besser, D. & Darnell, J. E., Jr. (2003) J. Biol. Chem. 278, 15794–15799. [DOI] [PubMed] [Google Scholar]
- 13.Higashi, N., Kunimoto, H., Kaneko, S., Sasaki, T., Ishii, M., Kojima, H. & Nakajima, K. (2004) Genes Cells 9, 233–242. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, H., Nakajima, R., Kunimoto, H., Sasaki, T., Kojima, H. & Nakajima, K. (2004) Biochem. Biophys. Res. Commun. 325, 541–548. [DOI] [PubMed] [Google Scholar]
- 15.Stahl, N., Farruggella, T. J., Boulton, T. G., Zhong, Z., Darnell, J. E., Jr. & Yancopoulos, G. D. (1995) Science 267, 1349–1353. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi, K., Shirakabe, K., Shibuya, H., Irie, K., Oishi, I., Ueno, N., Taniguchi, T., Nishida, E. & Matsumoto, K. (1995) Science 270, 2008–2011. [DOI] [PubMed] [Google Scholar]
- 17.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E. & Matsumoto, K. (1996) Science 272, 1179–1182. [DOI] [PubMed] [Google Scholar]
- 18.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z. & Matsumoto, K. (1999) Nature 398, 252–256. [DOI] [PubMed] [Google Scholar]
- 19.Irie, T., Muta, T. & Takeshige, K. (2000) FEBS Letters 467, 160–164. [DOI] [PubMed] [Google Scholar]
- 20.Takaesu, G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J. & Matsumoto, K. (2000) Mol. Cell 5, 649–658. [DOI] [PubMed] [Google Scholar]
- 21.Munoz-Sanjuan, I., Bell, E., Altmann, C. R., Vonica, A. & Brivanlou, A. H. (2002) Development (Cambridge, U.K.) 129, 5529–5540. [DOI] [PubMed] [Google Scholar]
- 22.Ishitani, T., Takaesu, G., Ninomiya-Tsuji, J., Shibuya, H., Gaynor, R. B. & Matsumoto, K. (2003) EMBO J. 22, 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, G., Klika, A., Callahan, M., Faga, B., Danzig, J., Jiang, Z., Li, X., Stark, G. R., Harrington, J. & Sherf, B. (2004) Proc. Natl. Acad. Sci. USA 101, 2028–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriguchi, T., Kuroyanagi, N., Yamaguchi, K., Gotoh, Y., Irie, K., Kano, T., Shirakabe, K., Muro, Y., Shibuya, H., Matsumoto, K., et al. (1996) J. Biol. Chem. 271, 13675–13679. [DOI] [PubMed] [Google Scholar]
- 25.Shirakabe, K., Yamaguchi, K., Shibuya, H., Irie, K., Matsuda, S., Moriguchi, T., Gotoh, Y., Matsumoto, K. & Nishida, E. (1997) J. Biol. Chem. 272, 8141–8144. [DOI] [PubMed] [Google Scholar]
- 26.Ishitani, T., Ninomiya-Tsuji, J., Nagai, S., Nishita, M., Meneghini, M., Barker, N., Waterman, M., Bowerman, B., Clevers, H., Shibuya, H., et al. (1999) Nature 399, 798–802. [DOI] [PubMed] [Google Scholar]
- 27.Ishitani, T., Kishida, S., Hyodo-Miura, J., Ueno, N., Yasuda, J., Waterman, M., Shibuya, H., Moon, R. T., Ninomiya-Tsuji, J. & Matsumoto, K. (2003) Mol. Cell. Biol. 23, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanei-Ishii, C., Ninomiya-Tsuji, J., Tanikawa, J., Nomura, T., Ishitani, T., Kishida, S., Kokura, K., Kurahashi, T., Ichikawa-Iwata, E., Kim, Y., et al. (2004) Genes Dev. 18, 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkawara, B., Shirakabe, K., Hyodo-Miura, J., Matsuo, R., Ueno, N., Matsumoto, K. & Shibuya, H. (2004) Genes Dev. 18, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima, H., Nakajima, K. & Hirano, T. (1996) Oncogene 12, 547–554. [PubMed] [Google Scholar]
- 31.Nakajima, K., Yamanaka, Y., Nakae, K., Kojima, H., Ichiba, M., Kiuchi, N., Kitaoka, T., Fukada, T., Hibi, M. & Hirano, T. (1996) EMBO J. 15, 3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 32.Seraphin, B. (2002) Adv. Protein. Chem. 61, 99–117. [DOI] [PubMed] [Google Scholar]
- 33.Natsume, T., Yamauchi, Y., Nakayama, H., Shinkawa, T., Yanagida, M., Takahashi, N. & Isobe, T. (2002) Anal. Chem. 74, 4725–4733. [DOI] [PubMed] [Google Scholar]
- 34.Miyagishi, M. & Taira, K. (2002) Nat. Biotech. 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 35.Miyoshi, H., Blomer, U., Takahashi, M., Gage, F. H. & Verma, I. M. (1998) J. Virol. 72, 8150–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama, K., Wada, K., Miyoshi, H., Ohashi, K., Tachibana, M., Furuki, R., Mizuguchi, H., Hayakawa, T., Nakajima, A., Kadowaki, T., et al. (2004) FEBS Lett. 560, 178–182. [DOI] [PubMed] [Google Scholar]
- 37.Brott, B. K., Pinsky, B. A. & Erikson, R. L. (1998) Proc. Natl. Acad. Sci. USA 95, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.