Abstract
Purpose
Rapid identification of pathogens causing endophthalmitis may improve treatment outcomes through early administration of species-specific medication. The current study reports a new molecular application of peptide nucleic acid–fluorescence in situ hybridization (PNA-FISH) with Staphylococcus-specific molecular PNA probes for the potential rapid detection of common pathogens causing endophthalmitis.
Methods
An experimental study was designed to evaluate the proof of concept at the microbiology laboratory of the Bascom Palmer Eye Institute. Stored culture-positive staphylococci endophthalmitis isolates obtained from prior vitreous samples (n = 15), along with broth as negative controls (n = 5) were used. Inoculum was prepared to a final concentration of 1 × 105 colony-forming units/mL to ensure that the isolates were viable. Smears of samples were fixed and hybridized using QuickFISH protocol with probes for Staphylococcus.
Results
With PNA-FISH technique, Staphylococcus aureus was identified in 9 of 10 samples and coagulase-negative staphylococci were identified in 10 of 10 samples. Detection time was 20 minutes.
Conclusions
This study serves a proof of concept using a new microbial detection system with FISH probes, and may have the potential for clinical use in the rapid and accurate identification of isolates from patients with endophthalmitis.
Keywords: peptide nucleic acid (PNA), fluorescence in situ hybridization (FISH), endophthalmitis, rapid identification
Molecular techniques are increasingly being used to identify pathogens. Fluorescence in situ hybridization (FISH) is a technique whereby DNA probes labeled with fluorophores are attached to a target DNA for identification. The FISH technique has been used for more than 20 years for the detection of genetic disorders including trisomy. A new molecular application of fluorescence hybridization can be used to rapidly identify microorganisms (Staphylococcus, Pseudomonas, Candida species) in blood cultures from patients with septicemia in the intensive care unit. FISH has been shown to improve patient outcomes in terms of survival, hospital stay, and total cost in this patient population.1
In ophthalmology, endophthalmitis remains an important cause of ocular morbidity. Changes in prevalence of causative organisms and their antimicrobial susceptibilities over time have been reported.2,3 The current methodology of plating and subsequent reading of growth is labor and time intensive. Consequently, there exists a growing need for more rapid and accurate detection of pathogens.4 If peptide nucleic acid (PNA)-FISH is proven useful for quick diagnosis of endophthalmitis, it can further be used for diagnosis of various other ocular infections. The current study evaluated a new molecular application of PNA-FISH with Staphylococcus-specific molecular PNA probes for the rapid detection of common pathogens causing endophthalmitis.
Methods
Vitreous isolates from patients with endophthalmitis obtained during vitreous tap or vitrectomy were acquired from the microbiology laboratory at a university teaching center. The study was approved by the institutional review board at the University of Miami, Miller School of Medicine (Miami, FL, USA). The study was performed in accordance with the ethical standards as laid down in the 1964 Helsinki Declaration. Because this study involved only a retrospective evaluation of microbiological isolates with no clinical patient identifiers, informed consent was not obtained. A total of 20 isolates stored from prior cases of endophthalmitis were tested. Of these, five were identified as Staphylococcus aureus, five were coagulase-negative Staphylococcus, five were mixed samples with both Staphylococcus aureus and coagulase-negative Staphylococcus, and five were negative controls (only broth). Isolates aged less than 30 days, identified initially using conventional microbiology tests, frozen in broth or other growth media and stored at −70°C in skimmed milk were used for this study. Standard inoculum was prepared to a final concentration of 1 × 105 colony-forming units/mL to ensure that the isolates were viable. Blood culture bottles and thioglycollate broth were inoculated and incubated at 35°C for 12 to 18 hours. PNA-FISH technique was carried out per the manufacturer's QuickFISH protocol (AdvanDx, Woburn, MA, USA) as follows.
QuickFISH slides were placed on the incubator/slidestation at 55°C throughout the procedure. From the prepared sample of microorganisms, 10 μL was pipetted onto the manufacturer's slides placed on slidestation, each containing a positive and a negative control sample. Fixation solutions (QuickFIX-1 and QuickFIX-2) were then applied sequentially to each sample, mixed, and samples were allowed to completely dry (1–2 minutes with each fixation solution). PNA probes (yellow and blue) were aliquoted onto a coverslip, and the coverslip was then applied to each sample on the slidestation. Samples were allowed to hybridize for 15 minutes at 55°C. Finally, samples were read qualitatively by two independent observers on a fluorescence microscope (using ×60 magnification) to assess detection of organisms based on shape and color (Fig.). It is important to mention that in most of the test specimens, inoculum used was not intraocular fluid and was artificial medium.
Figure.
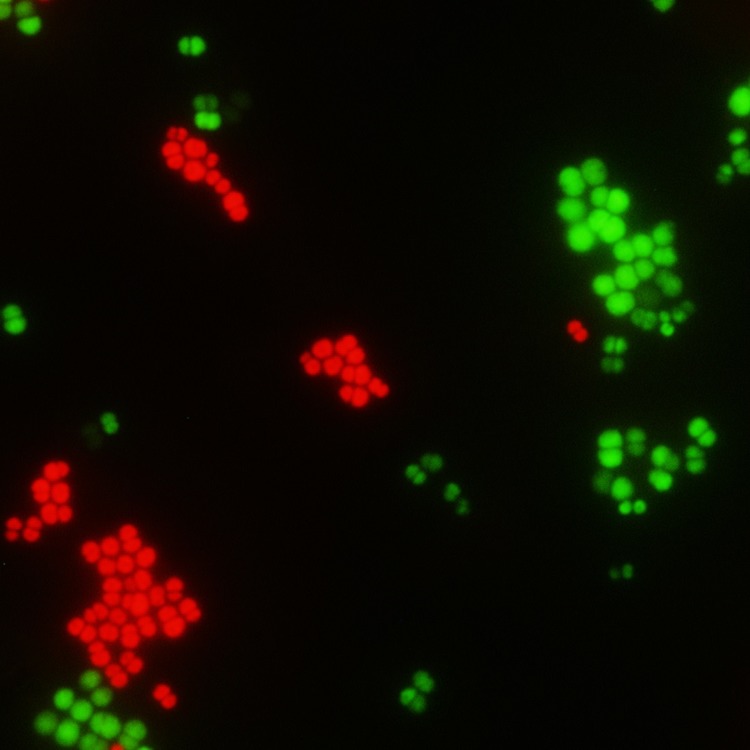
Vitreous sample as seen under fluorescence microscope (×60 magnification), using PNA-FISH/QuickFISH procedure for rapid detection of pathogens. Staphylococcus aureus is depicted as green-colored cocci in groups and coagulase-negative staphylococci as red-colored cocci in groups.
Results
Two independent observers read the samples qualitatively. There were no intraobserver or interobserver disagreements. Using this technique, Staphylococcus aureus was identified in 9 (90%) of 10 samples, whereas coagulase-negative staphylococci was identified in 10 of 10 (100%) samples (Table). No fluorescence was identified in five (100%) of five non-staphylococcal cultures, indicating no false negatives. When calculating the results, the mixed culture was treated as a test for both Staphylococcus aureus and for coagulase-negative staphylococci. Detection time post culture was 20 minutes compared with an average of 1 to 2 days for routine plating methods.
Table.
Comparison of Test Outcomes of PNA-FISH With Staphylococcus-Specific Molecular PNA Probes Versus Culture Growth (Gold Standard) for Rapid Identification of Staphylococcus aureus and Coagulase-Negative Staphylococcus

Discussion
New and emerging molecular technologies for rapid organism recovery and identification directly from patient samples and/or culture media are being used. In this study, PNA-FISH probes were used. PNA probes differ from DNA probes, as the phosphodiester backbone of DNA is replaced by 2-aminoethylglycine linkage. This modification allows for better penetration of the bacterial cell wall and increased rates of sensitivity and specificity. The peptide probes can specifically target the 16S RNA living within bacteria and yeast using species-specific sequences as the target.5
The main advantage primarily is the markedly reduced detection time (approximately 20 minutes) compared with 1 to 2 days with routine culture and plating methods. Additionally, FISH is a low-cost methodology requiring less technical skill as compared with PCR methods.6 Also, PNA probes have high specificity and bind to the target only if there is an exact match in the sequence. PNA-FISH assays help in identifying and diagnosing infectious diseases in a rapid manner within the clinic. PCR is a simple, cheap, easy-to-understand, highly sensitive, and reliable technique to replicate and produce millions to billions of copies of a focused segment of DNA specific for sequencing, cloning, and analysis. One major limitation of PCR is that precise sequence(s) upstream of the target region on each of the two single-stranded templates is required to generate the primers that will allow its selective amplification. Also, errors caused by DNA polymerases and nonspecific binding of primers may result in incorrect interpretation. In the current study, there were no false positives found. Prior studies have shown the ability to detect pathogens in blood cultures as well as in other body fluid samples using a similar method. Poppert et al. showed that FISH could be used to diagnose bacterial meningitis from cerebrospinal fluid samples.6
The current study is limited by small sample size as well as case scenarios with polymicrobial infections. In view of the polymicrobial infections, we are currently working on another laboratory experiment with multiple probes that can detect multiple infections at the same time. Also, it is important to mention that even though PNA-FISH identified almost all the staphylococci organisms tested, the test specimens or inoculum used was not intraocular fluid and artificial. In the form used, PNA-FISH may not be as good or may need to be modified for intraocular fluid samples. Another possible limitation of this method is low sensitivity when the bacterial load is minimal.4 This may not be as much of a concern in endophthalmitis, as bacterial load is generally high on presentation.
In conclusion, current laboratory protocols require labor- and time-intensive methods for the identification of pathogens in endophthalmitis with culture and media plating. The current study is a proof of concept for a microbial detection system using FISH probes. This newer diagnostic system may have clinical benefits for the rapid and accurate identification of isolates from endophthalmitis patients.
Acknowledgments
Supported in part by National Institutes of Health Center Core Grant P30EY014801 (Bethesda, MD, USA), Research to Prevent Blindness Unrestricted Grant (New York, NY, USA), and the Department of Defense (DOD Grant W81XWH-13-1-0048) (Washington, DC, USA).
Disclosure: N. Patel, None; D. Miller, None; N. Relhan, None; H.W. Flynn Jr, None
References
- 1. Forrest GN,, Mehta S,, Weekes E,, Lincalis DP,, Johnson JK,, Venezia RA. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J Antimicrob Chemother. 2006; 58: 154–158. [DOI] [PubMed] [Google Scholar]
- 2. Jones DB. Emerging vancomycin resistance: what are we waiting for? Arch Ophthalmol. 2010; 128: 789–791. [DOI] [PubMed] [Google Scholar]
- 3. Silva RA,, Sridhar J,, Miller D,, Wykoff CC,, Flynn HW., Jr. Exogenous fungal endophthalmitis: an analysis of isolates and susceptibilities to antifungal agents over a 20-year period (1990–2010). Am J Ophthalmol. 2015; 159: 257–264.e1. [DOI] [PubMed] [Google Scholar]
- 4. Schwartz SG,, Flynn HW., Jr. Update on the prevention and treatment of endophthalmitis. Expert Rev Ophthalmol. 2014; 9: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sachse F,, Becker K,, von Eiff C,, Metze D,, Rudack C. Staphylococcus aureus invades the epithelium in nasal polyposis and induces IL-6 in nasal epithelial cells in vitro. Allergy. 2010; 65: 1430–1437. [DOI] [PubMed] [Google Scholar]
- 6. Poppert S,, Essig A,, Stoehr B,, et al. Rapid diagnosis of bacterial meningitis by real-time PCR and fluorescence in situ hybridization. J Clin Microbiol. 2005; 43: 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]


