Abstract
DAF-16/forkhead transcription factor, the downstream target of the insulin-like signaling in Caenorhabditis elegans, is indispensable for both lifespan regulation and stress resistance. Here, we demonstrate that c-Jun N-terminal kinase (JNK) is a positive regulator of DAF-16 in both processes. Our genetic analysis suggests that the JNK pathway acts in parallel with the insulin-like signaling pathway to regulate lifespan and both pathways converge onto DAF-16. We also show that JNK-1 directly interacts with and phosphorylates DAF-16. Moreover, in response to heat stress, JNK-1 promotes the translocation of DAF-16 into the nucleus. Our findings define an interaction between two well conserved proteins, JNK-1 and DAF-16, and provide a mechanism by which JNK regulates longevity and stress resistance.
Keywords: longevity, stress resistance, aging, dauer, insulin-like signaling
The insulin signaling pathway plays a pivotal role in lifespan regulation and stress resistance in many diverse organisms (1). In Caenorhabditis elegans, the phosphatidylinositol 3-kinase signaling cascade downstream of daf-2, an ortholog of the mammalian insulin and insulin-like growth factor-1 (IGF-1) receptor (2), targets DAF-16/forkhead transcription factor (3, 4). DAF-16 then regulates a wide variety of genes involved in longevity, stress responses, metabolism, and development (5, 6). In mammalian systems, other signal transduction pathways including the c-Jun N-terminal kinase (JNK) signaling pathway have been shown to couple to the insulin/IGF-1 pathway (7–9). The JNK family, a subgroup of the mitogen-activated protein kinase superfamily, is part of a signal transduction cascade that is activated by cytokines, including TNF and IL-1, and by exposure to environmental stresses (10). The JNK pathway has been implicated in critical biological processes such as cancer, development, apoptosis, and cell survival (10). In mammalian cell culture, components of the JNK signaling pathway have been shown to interact with the insulin signaling pathway through interaction with either the insulin receptor substrate 1 (7) or protein kinase B (AKT) (8, 9). A recent observation in Drosophila also suggested that the JNK signaling pathway is involved in lifespan regulation and stress resistance, but the mechanism of JNK signaling was not established (11). Therefore, we investigated the role of the JNK signaling pathway in lifespan regulation in C. elegans and its possible connection to the insulin-like signaling pathway.
Experimental Procedures
Strains. All strains were maintained and handled as described (12, 13).
Strain Construction. For jkk-1; lpIn2 and mek-1; lpIn2, jkk-1 and mek-1 males were obtained by heat shock at 30°C for 6 h or spontaneously on the plate and mated to the lpIn2 hermaphrodites. From the mating plate, 20 putative F1 cross progeny were picked to individual plates and allowed to have progeny. From at least two individual F1 plates that segregated both rollers and nonrollers (indicating cross progeny), 20–30 F2 rollers were picked to individual plates and allowed to have progeny. Once the parents had progeny (F3), the F2 parents were tested for hypersensitivity to CuSO4 for mek-1 or the presence of the jkk-1 deletion mutation (5′-AGGAGAAAAGCAAGTTGTCG, 3′-GCAGCAGCTTCTCACAACAC) and jnk-1 overexpression (5′-ACAGTGGAACAGGAGGAGG, 3′-ATGCCTATCTGCCTGAGAGC) by PCR. Then, the plates that segregated 100% rollers (F3) were kept to establish the strain. The crosses were done at 20°C.
For lpIn2; daf-16::gfp and jkk-1; lpIn2; daf-16::gfp, spontaneously obtained daf-16::gfp males were mated to either lpIn2 or jkk-1; lpIn2 hermaphrodites. The crosses were done as described above except that F2 worms were screened for both GFP and the roller phenotype.
For daf-2; lpIn1, daf-2(e1370) males were mated to lpIn1 hermaphrodites. About 20 putative F1 roller cross progeny were transferred to 25°C and allowed to have F2 progeny. From plates segregating rollers and nonrollers, WT and dauers, 15–20 rolling dauers were picked to individual plates and allowed to recover at 15°C. The plates were scored for 100% roller progeny in the F3. The rollers were then retested for dauer formation at 25°C.
Transgenic Worms. jnk-1 genomic DNA including 3 kb of the promoter region, the entire coding region, and 500 bp of 3′ UTR was amplified from N2 genomic DNA by PCR (5′-GCGTCCTCCTGTGCTCACTC, 3′-CCCACGACA ACTGCTACAAC). After gel extraction (Qiagen, Valencia, CA), the 9.3-kb fragment was injected at 50 ng/μl into the gonad of N2 worms along with pRF4 [rol-6(su1066) plasmid] as a coinjection marker (100 ng/μl) (14) to generate stable extrachromosomal transgenic lines. Two independent integrated lines (lpIn1 and lpIn2) were generated from one extrachromosomal line (lpEx1). Several extrachromosomal and integrated lines using at least two different markers were established and showed similar results (data not shown).
Lifespan Analysis. Lifespan assays were performed at 20°C. Adult hermaphrodites from each strain were transferred to fresh nematode growth medium (NGM) plates at 20°C and allowed to undergo one full generation to ensure the worms were well fed and had not gone through dauer. L4s or young adults were then transferred to new NGM plates containing 0.1 mg/ml 5′-flurodeoxyuridine (FUDR) to prevent the growth of progeny (15). Animals were tapped every 2–3 days and scored as dead when they did not respond to the platinum wire pick. We determined survival from the point when the worms were transferred to the FUDR plate, and therefore lifespan is defined as the days the worms survived (day 1). All of the lifespan assays were repeated at least three times. For lifespan analysis on RNA interference (RNAi) plates, a single colony from each RNAi clone was grown in LB broth containing 50 μg/ml ampicillin and 12.5 μg/ml tetracycline to an OD of 0.5–1.0. The bacteria were then seeded onto an NGM plate containing 1 mM isopropyl-thiogalactoside, 50 μg/ml ampicillin, and 0.1 mg/ml of FUDR. Seeded plates were dried overnight at room temperature and then stored at 4°C for subsequent use.
Plasmid Construction and Transfection. Full-length daf-16 a1 and jnk-1α cDNAs were amplified by RT-PCR (Invitrogen) by using total RNA isolated from N2 worms (Ambion, Austin, TX). The cDNAs were cloned into the mammalian expression vector with either Flag tag (p3XFLAG-myc-CMV-26, Sigma) or Xpress tag (pcDNA3.1B, Invitrogen) to obtain Flag-tagged DAF-16 and Xpress-tagged JNK-1. Plasmids were transfected into COS-7 cells and harvested after 48 h. For determining whether DAF-16 could serve as a substrate for JNK-1, N- and C-terminal portions of DAF-16 were amplified by PCR using N2 total RNA and cloned into pET-24b vector (Novagen) between HindIII and XhoI. The His-tagged DAF-16 fusion protein was expressed in Escherichia coli (BL-21) and purified under native condition with Ni-NTA His-Bind resin (Novagen).
Antibody Production. A C-terminal portion of JNK-1α (amino acids 228–451) was cloned into a His-tagged expression vector (pET-24b, Novagen), expressed in bacteria [BL21(DE3)], and purified by using Ni-NTA His-bind resin (Novagen). Polyclonal antiserum was raised against the recombinant protein in rabbit (Capralogics, Hardwick, MA).
Immunoblotting, Immunoprecipitation, and Kinase Assay. For phospho-JNK immunoblotting, worms were grown on 10-cm NGM plates and ground with stainless-steel Dounce homogenizer in lysis buffer (20 mM Tris, pH 7.4/137 mM NaCl/2 mM EDTA/10% glycerol/1% Triton X-100/25 mM β-glycerophosphate/1 mM NaVO3/1 mM PMSF/10 μg/ml leupeptin/10 μg/ml aprotinin). Proteins were separated by SDS/PAGE and immunoblotted with phospho-JNK antibody (Cell Signaling Technology, Beverly, MA) or JNK-1 antibody (raised against C. elegans JNK-1). For immunoprecipitation, COS-7 cells were lysed in the same lysis buffer, and after centrifugation at 14,000 × g for 10 min, the supernatant was precleared with 50 μl of protein G-Sepharose bead (Amersham Pharmacia). The supernatant was then incubated with anti-Xpress antibody (Sigma) along with fresh beads overnight at 4°C. After several washes, lysates were boiled with sample buffer. Proteins were separated by SDS/PAGE and immunoblotted with anti-Flag antibody (Invitrogen). For kinase assay, protein G-Sepharose (Pharmacia LKB) beads were incubated for 3–4 h at 4°C with anti-Xpress antibody, washed twice with the lysis buffer as described above, and then incubated with lystates of COS-7 cells transfected with Xpress-JNK-1 overnight at 4°C. Complexes were washed three times with the lysis buffer and once with kinase buffer (25 mM Hepes, pH 7.4/25 mM β-glycerophosphate/25 mM MgCl2/0.1 mM NaVO3/2 mM DTT). The kinase activity of JNK-1 was measured by adding 20 μl of kinase buffer containing 50 μM [γ-32P]ATP (10 Ci/mmol) and 1 μg of the N- or C-terminal portion of DAF-16 or GST-c-Jun (amino acids 1–79) and incubating at 30°C for 30 min. The reactions were terminated by boiling in sample buffer. Proteins were resolved by SDS/PAGE and analyzed by autoradiography.
Heat Resistance Assay and DAF-16 Translocation Assay. To measure heat stress resistance, 50 young adult worms were picked onto NGM plates and kept at 35°C. Animals were tapped every hour and scored as dead when they did not respond to the platinum wire pick. Assays were repeated at least three times. For DAF-16 translocation assay, ≈10–15 L4s were placed on NGM plates at 35°C. After 30 min, worms were immediately mounted onto the slide with 0.1% of sodium azide in S-basal buffer. We visualised the nuclear translocation of DAF-16 with a fluorescence microscope (Zeiss) equipped with a Hamamatsu (Middlesex, NJ) digital camera and analyzed it by openlab software. We scored each animal as having cytosolic localization, nuclear localization when localization is observed throughout the entire body from head to tail, or intermediate localization when there is a visible nuclear localization but one not as complete as nuclear. The number of worms with each level of nuclear translocation was counted. The translocation assay was repeated at least five times by two different individuals.
PCR Primers Used for Cloning. The primers used were: Flag-DAF-16, 5′-GAAGATCTGGAGATGCTGGTAGATCAGGG, 3′-CGGGGTACCTTACAAATCAAAATGAATAT; Xpress-JNK-1, 5′-CGGGATCCGGAGGAACGATTATCCACAAC, 3′-GCTCTAGATCAGGAATAAATGTCATGGG; Xpress-JNK-1 (APF), 5′-GAGGCATTCATGATGGCTCCTTTCGTTGTGACAAGATAC, 3′-GTATCTTGTCACAACGAAAGGAGCCATCATGAATGCCTC; His-DAF-16 (N-terminal fragment), 5′-CCCAAGCTTGGCCTATACGGGAGCAATGAGC, 3′-CCGCTCGAGCGGACGGA A AGATGATGGA ACG; and His-DAF-16 (C-terminal fragment), 5′-CCCA AGCT TGGCGGAGCCA AGA AGAGGATA, 3′-CCGCTCGAGCGCAATTGGAAGAGCCGATGAA.
Results and Discussion
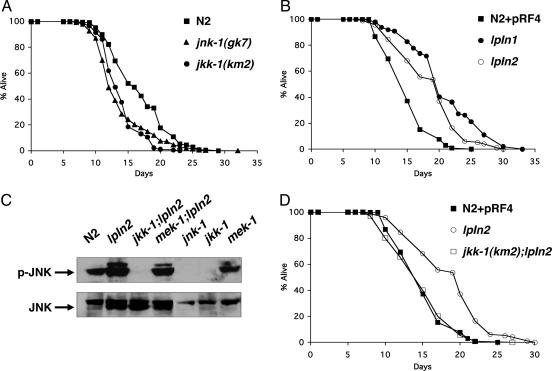
Several orthologs of JNK pathway components have been characterized in C. elegans including: a mammalian JNK ortholog, jnk-1, two MKK4/7 (JNK kinase) orthologs, jkk-1 and mek-1, and a JIP3 (scaffold protein) ortholog, unc-16 (16–19). We started by examining the lifespan of loss-of-function mutants of jnk-1 and jkk-1. Both mutants show a statistically significant decrease in lifespan [WT, 16.8 ± 0.2 days; jnk-1(gk7), 13.8 ± 0.2 days (P < 0.0001); and jkk-1(km2), 13.9 ± 0.2 days (P < 0.0001); Fig. 1A]. However, mutation in the two other components of the C. elegans JNK pathway, mek-1(ks54) and unc-16(e109), show little effect on lifespan (Fig. 5, which is published as supporting information on the PNAS web site). These data indicate that part of the JNK signaling pathway is required to maintain normal lifespan in C. elegans.
Fig. 1.
JNK pathway regulates lifespan dependent on its phosphorylation that requires an upstream kinase, jkk-1. (A) Lifespan analysis of jnk-1(gk7) and jkk-1(km2). All lifespan data are presented as mean lifespan ± standard error: N2, 16.8 ± 0.2 (386 animals); jnk-1(gk7), 13.8 ± 0.2 (333 animals); jkk-1(km2), 13.9 ± 0.2 (189 animals). (B) Lifespan analysis of jnk-1 overexpression lines (lpIn1 and lpn2): N2 + pRF4, 15.2 ± 0.3 (105 animals); lpIn1, 20.9 ± 0.6 (99 animals); lpIn2, 18.8 ± 0.5 (97 animals). (C) Immunoblotting with phospho-JNK antibody (Upper). The same blot was reprobed with JNK antibody (Lower). (D) Lifespan analysis of jkk-1;lpIn2:N2 + pRF4, 15.2 ± 0.3 (105 animals); lpIn2, 18.8 ± 0.5 (97 animals), jkk-1; lpIn2, 15.0 ± 0.3 (130 animals). The lifespan curves were plotted from the pooled data of individual experiments.
We next asked whether the lifespan regulation by jnk-1 is dose-dependent. We amplified 9.3 kb of jnk-1 genomic DNA including 3 kb of the promoter region, the entire coding region, and 500 bp of the 3′ UTR by PCR. The entire PCR fragment was injected into the gonads of WT worms along with the coinjection marker rol-6(su1066) (14) to create jnk-1 overexpression transgenic worms. From one of several extrachromosomal lines (lpEx1), we derived two independent integrated lines (lpIn1 and lpIn2). Both lpIn1 and lpIn2 exhibit similar increases in expression of the jnk-1 transcript, as determined by RT-PCR (Fig. 6, which is published as supporting information on the PNAS web site). lpIn1 and lpIn2 extend lifespan by 40% compared with the control, implying that jnk-1 is a positive regulator of lifespan {N2 + pRF4 [rol-6(su1066) plasmid] control, 15.2 ± 0.3 days; lpIn1, 20.9 ± 0.6 days (P < 0.0001); and lpIn2, 18.8 ± 0.5 (P < 0.0001); Fig. 1B}.
Dual phosphorylation of JNK on conserved Thr and Tyr by an upstream kinase is critical for its function (10). Therefore, we investigated whether the phosphorylation of JNK is required for lifespan regulation. We crossed jnk-1 transgenic worms with loss-of-function mutants of upstream kinases jkk-1(km2) and mek-1(ks54) and analyzed phosphorylation status of JNK-1 by using a phospho-specific antibody that binds activated JNK. We detect more phosphorylated JNK-1 in the jnk-1 overexpression strain (lpIn2; Fig. 1C). However, it is not detectable in either jnk-1(gk7) or jkk-1(km2) mutants. In addition, mutation in jkk-1 completely suppresses the phosphorylation of JNK-1 in jnk-1 overexpression worms (jkk-1; lpIn2; Fig. 1C). In accordance with this result, lifespan extension by jnk-1 overexpression is also suppressed by mutation in jkk-1 (lpIn2, 18.8 ± 0.5 days; jkk-1; lpIn2, 14.9 ± 0.4 days; N2 + pRF4 control, 15.2 ± 0.3 days; Fig. 1D). However, mutation in mek-1(ks54) alone or in combination with jnk-1 overexpression (lpIn2) does not affect JNK-1 phosphorylation (Fig. 1C). These results show that JKK-1 is the upstream kinase of JNK-1 and phosphorylation of JNK-1 is required for lifespan extension.
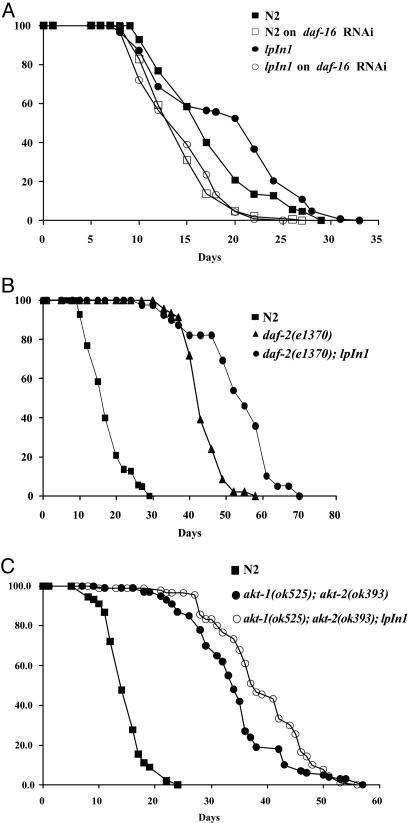
In C. elegans, daf-16 plays a central role in lifespan regulation such that a mutation in daf-16 suppresses the lifespan extension of daf-2, age-1, or other long-lived mutants (20–23). Therefore, we asked whether lifespan extension by jnk-1 overexpression also depends on daf-16. We performed lifespan analysis of jnk-1 overexpression worms on daf-16 RNAi. In agreement with previous work (24), daf-16 RNAi shortens lifespan (WT on control RNAi, 17.6 ± 0.4 days; on daf-16 RNAi, 14.4 ± 0.4 days; Fig. 2A). Lifespan extension by jnk-1 overexpression is completely suppressed by daf-16 RNAi (lpIn1 on control RNAi, 19.1 ± 0.6 days; on daf-16 RNAi, 14.7 ± 0.4 days; Fig. 2 A). This finding suggests that daf-16 is required for lifespan extension by jnk-1 overexpression.
Fig. 2.
jnk-1 regulates lifespan in parallel to the insulin-like pathway, but both converge onto daf-16. (A) Lifespan analysis of jnk-1 overexpression strain (lpIn1) on daf-16 RNAi. All lifespan data are presented as mean lifespan ± standard error: N2 on control RNAi, 17.6 ± 0.5 (125 animals); N2 on daf-16 RNAi, 14.6 ± 0.3 (145 animals); lpIn1 on control RNAi, 19.1 ± 0.6 (147 animals; lpIn1 on daf-16 RNAi, 14.5 ± 0.3 (136 animals). (B) Lifespan analysis of daf-2(e1370);lpIn1: N2, 17.6 ± 0.5 (125 animals); daf-2(e1370), 44.0 ± 0.7 (46 animals); daf-2(e1370);lpIn1, 53.3 ± 1.7 (39 animals). In A and B, the lifespan curves were plotted from the pooled data of the individual experiments. (C) Lifespan analysis of akt-1(ok525);akt-2(ok393) and akt-1(ok525);akt-2(ok393);lpIn1: WT (N2), 14.9 ± 0.4 (90 animals); akt-1(ok525);akt-2(ok393), 34.2 ± 0.8 (100 animals); akt-1(ok525);akt-2(ok393);lpIn1, 38.8 ± 0.9 (90 animals). The representative lifespan curve is shown here, which has been repeated with similar results.
If jnk-1 exerts its effect through the insulin-like pathway, jnk-1 overexpression would not have an additional effect on lifespan in the background of mutations in the insulin-like pathway. However, if combining a hypomorphic mutation in the insulin-like pathway with jnk-1 overexpression produces a further increase in lifespan, then this result would be consistent with the possibility that jnk-1 acts in a parallel pathway. We crossed a jnk-1 overexpression line (lpIn1) with daf-2(e1370) and found that the combination significantly extends lifespan beyond daf-2(e1370)[daf-2, 44.0 ± 0.7 days; daf-2; lpIn1, 53.3 ± 1.7 days (P < 0.0001); Fig. 2B]. Further downstream of daf-2 in the insulin-like signaling pathway are the kinases AKT-1 and AKT-2 (25). Because experiments in mammalian system have shown an interaction between the AKT and JNK pathways (8, 9), we examined the lifespan of lpIn1 in combination with akt-1/akt-2. We generated a double mutant strain containing null mutations (26) in both akt-1(ok525) and akt-2(ok393). However, 100% of the akt-1(ok525); akt-2(ok393) double mutants arrest at dauer larval stage at all temperatures. To circumvent this problem, we grew akt-1(ok525); akt-2(ok393) double mutants and the akt-1(ok525); akt-2(ok393); lpIn1 strain on daf-16 RNAi to bypass the dauer larval stage and then tested lifespan on regular plates. akt-1(ok525); akt-2(ok393); lpIn1 strain shows a lifespan extension beyond the akt-1(ok525); akt-2(ok393) double mutant alone [WT (N2), 14.9 ± 0.4 days; akt-1(ok525); akt-2(ok393), 34.2 ± 0.8 days; akt-1(ok525); akt-2(ok393); lpIn1, 38.8 ± 0.9 days; Fig. 2C]. Similar results were obtained with akt-1/2 RNAi (Fig. 7, which is published as supporting information on the PNAS web site). In addition, lifespan extension by either daf-2 mutation alone or in combination with jnk-1 overexpression (lpIn1) is completely suppressed by daf-16 (refs. 1, 3, and 4 and data not shown). These results suggest that jnk-1 regulates lifespan in parallel to the insulin-like pathway but both converge onto daf-16. Alternatively, it is also possible that jnk-1 acts in a linear pathway by interacting with other upstream members of the insulin-like pathway.
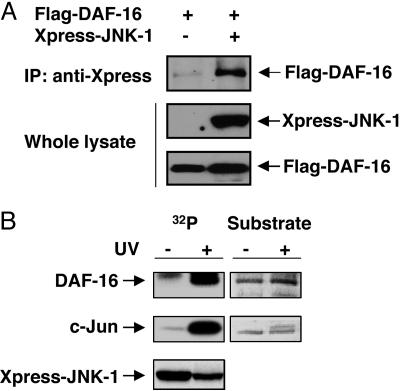
Based on our genetic studies, we next examined whether JNK-1 interacts physically with DAF-16. COS-7 cells were transfected with plasmids encoding either Flag-tagged DAF-16 alone or in combination with Xpress-tagged JNK-1. After coimmunoprecipitation with anti-Xpress antibody, we find that JNK-1 binds to DAF-16 (Fig. 3A). We then examined whether DAF-16 could serve as a substrate for JNK-1. COS-7 cells were transfected with plasmids encoding Xpress-tagged JNK-1 that was then immunoprecipitated from the cell lysate with anti-Xpress antibody and incubated with bacterially expressed N- or C-terminal portions of DAF-16 as a substrate in an in vitro kinase assay. Upon activation of JNK-1 by UV, JNK-1 phosphorylates DAF-16 as well as the mammalian c-Jun protein used as a positive control (Fig. 3B). The phosphorylation was observed only with the N-terminal (amino acids 83–307) fragment of DAF-16, but not with the C-terminal (amino acids 255–470) fragment (data not shown), implying that the JNK-1 phosphorylation site resides in the N-terminal region of DAF-16. Previously, JNK-1 was shown to be activated by dual phosphorylation on Thr-276 and Tyr-278 (17, 19). Therefore, we created kinase-dead JNK-1 construct to determine specificity by replacing TPY residues with APF and performed the in vitro kinase assay by using the N-terminal portion of DAF-16 as a substrate. The kinase-dead JNK-1 (APF) fails to phosphorylate DAF-16. This finding confirms the specificity of JNK-1 phosphorylation of DAF-16 as a substrate (Fig. 8, which is published as supporting information on the PNAS web site). These results demonstrate that JNK-1 directly interacts with and phosphorylates DAF-16 as a separate input from AKT-1/2 and therefore supports a parallel pathway model suggested by our genetic data.
Fig. 3.
JNK-1 interacts with and phosphorylates DAF-16. (A) Coimmunoprecipitation (IP) assay. After transfection with either Flag-DAF-16 alone or in combination with Xpress-JNK-1, cell lysates were immunoprecipitated with anti-Xpress antibody and immunoblotted with anti-Flag antibody. (B) In vitro kinase assay. Cells were transfected with Xpress-JNK-1 and activated by UV, followed by immunoprecipitation with anti-Xpress antibody and incubation with His-DAF-16 (N-terminal fragment) in kinase buffer. (Right) Loading of the substrates [His-DAF-16 (N-terminal fragment) and c-Jun] stained with Coomassie blue is shown. In both A and B, the expression of protein was confirmed by immunoblotting with anti-Flag or anti-Xpress antibody.
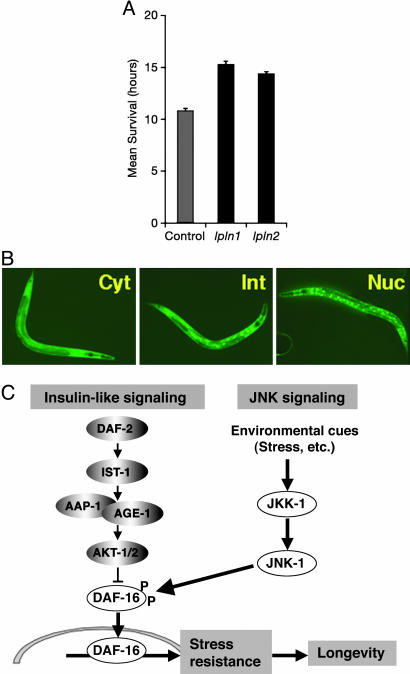
JNK is a stress-responsive gene in diverse organisms (10, 11). Therefore, we examined whether overexpression of jnk-1 confers stress resistance. We monitored survival of young adult worms at 35°C. Mutation in jnk-1(gk7) results in stress sensitivity when compared with WT (Fig. 9, which is published as supporting information on the PNAS web site). jnk-1 transgenic lines show significantly increased resistance to heat stress [mean survival time: N2 + pRF4 control, 10.8 ± 0.2 h; lpIn1, 15.3 ± 0.3 h; lpIn2, 14.4 ± 0.2 h (P < 0.0001); Fig. 4A]. In addition, jnk-1 transgenic lines significantly increase resistance to oxidative stress (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
JNK-1 promotes the translocation of DAF-16 into the nucleus in response to heat stress. (A) Heat stress assay of jnk-1 overexpression strain (lpIn1 and lpIn2). Mean survival time ± standard error was: N2 + pRF4, 10.8 ± 0.2 (138 animals); lpIn1, 15.3 ± 0.3 (147 animals); lpIn2, 14.4 ± 0.2 (249 animals). (B) DAF-16 translocation assay. After heat shock at 35°C for 30 min, the nuclear translocation of DAF-16 was measured, and the number of worms in each category was counted (see Table 1) (28). Cyt, cytosolic; Int, intermediate; Nuc, nuclear. (C) JNK signaling pathway presumably serves as a molecular sensor for various stresses. Upon detecting environmental cues, JNK-1 transmits the signal by phosphorylating and modulating the nuclear translocation of DAF-16. Once DAF-16 enters the nucleus, it enhances the expression of numerous target genes to prevent damage from any harmful stresses. This would then confer increased stress resistance and help to maintain normal life in C. elegans.
In C. elegans, mutation in daf-2 or age-1 confers stress resistance and this resistance depends on daf-16 (27–29). We also found that the stress resistance observed in the jnk-1 overexpression strains also depended on daf-16 (Fig. 11, which is published as supporting information on the PNAS web site). Additionally, in response to stress, DAF-16 translocates from the cytoplasm to the nucleus (30). Therefore, we crossed jnk-1 overexpression lines to a strain containing a daf-16::gfp reporter construct to visualize the nuclear translocation in vivo. We compared daf-16::gfp alone with lpIn2; daf-16::gfp or jkk-1; lpIn2; daf-16::gfp after 30 min of heat shock. The extent of nuclear translocation was categorized as cytosolic, nuclear, or intermediate (Fig. 4B and Table 1) (28). The number of worms with nuclear localization signal is increased in the jnk-1 overexpression (lpIn2; daf-16::gfp) strain, whereas that with cytosolic localization is decreased compared with the control (daf-16::gfp). However, mutation of jkk-1 suppresses the enhancement in nuclear localization in jnk-1 overexpression worms [daf-16::gfp (cytosolic), 16.7 ± 1.4%; lpIn2; daf-16::gfp (cytosolic), 4.3 ± 0.9%; jkk-1; lpIn2; daf-16::gfp (cytosolic), 21.0 ± 2.4%; daf-16::gfp (nuclear), 13.6 ± 1.8%; lpIn2; daf-16::gfp (nuclear), 30.4 ± 2.1%; jkk-1; lpIn2; daf-16::gfp (nuclear), 19.4 ± 2.5%; Table 1]. Together with biochemical evidence, our genetic analysis suggests that JNK-1 interacts directly with DAF-16 and modulates its nuclear translocation in response to stress. Previous studies have also shown that daf-18, a phosphatase in the insulin-like signaling pathway, is also required for daf-16 translocation in response to stress (31). Our data would suggest that this is a unique input into daf-16 for regulation of lifespan and stress.
Table 1. DAF-16 translocation assay.
| % of total worms tested
|
No. of worms tested
|
|||
|---|---|---|---|---|
| Strain | Cytosolic Intermediate Nuclear | |||
| daf-16::gfp | 16.7 | 69.7 | 13.6 | 66 |
| lpln2; daf-16::gfp | 4.3 | 65.2 | 30.4 | 69 |
| jkk-1; lpln2; daf-16::gfp | 21.0 | 59.7 | 19.4 | 62 |
Conditions were 30 min at 35°C.
The JNK signaling pathway serves as a molecular sensor for various stresses. Upon detecting environmental cues, JNK-1 might deliver the signal to DAF-16 by direct interaction and modulation of its nuclear translocation. Once DAF-16 enters the nucleus, it enhances the expression of numerous target genes to prevent damage from any harmful stresses, which would then confer increased stress resistance and help to maintain normal life in C. elegans (Fig. 4C). In addition, it has recently been shown that lifespan extension by Drosophila JNK also depends on D-forkhead (H. Jasper and D. Bohmann, personal communication). In summary, we present results that uncover a role of JNK as a lifespan regulator in C. elegans. We demonstrate a direct interaction between JNK-1 and DAF-16 that modulates the nuclear translocation of DAF-16. These findings provide important clues on underlying mechanism by which an animal can respond and adapt to environmental fluctuations to preserve homeostasis.
Supplementary Material
Acknowledgments
We thank Y. Wang and M. Grabowski for helpful discussions, suggestions, encouragement, and critical reading of the manuscript; S. Lee, P. Hu, and G. Ruvkun (all Harvard Medical School/Massachusetts General Hospital, Boston) for the daf-16, akt-1, and akt-2 RNAi constructs; T. Johnson (University of Colorado, Boulder) for daf-16::gfp strain; and Theresa Stiernagle and the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis), which is funded by the National Institutes of Health National Center for Research Resources, for many C. elegans strains used in this study. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant 5 P30 DK32520, a Career Award in the Biomedical Sciences from the Burroughs Well-come Fund, and an American Federation of Aging Research grant (to H.A.T.).
Author contributions: S.W.O., A.M., R.J.D., and H.A.T. designed research; S.W.O., A.M., N.S., and F.J. performed research; S.W.O., and H.A.T. contributed new reagents/analytic tools; S.W.O., A.M., N.S., F.J., R.J.D., and H.A.T. analyzed data; and S.W.O., A.M., R.J.D., and H.A.T. wrote the paper.
Abbreviations: AKT, protein kinase B; JNK, c-Jun N-terminal kinase; NGM, nematode growth medium; RNAi, RNA interference.
References
- 1.Barbieri, M., Bonafe, M., Franceschi, C. & Paolisso, G. (2003) Am. J. Physiol. 285, E1064–E1071. [DOI] [PubMed] [Google Scholar]
- 2.Kimura, K. D., Tissenbaum, H. A., Liu, Y. & Ruvkun, G. (1997) Science 277, 942–946. [DOI] [PubMed] [Google Scholar]
- 3.Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A. & Ruvkun, G. (1997) Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- 4.Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. (1997) Science 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
- 5.Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., Li, H. & Kenyon, C. (2003) Nature 424, 277–283. [DOI] [PubMed] [Google Scholar]
- 6.Lee, S. S., Kennedy, S., Tolonen, A. C. & Ruvkun, G. (2003) Science 300, 644–647. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre, V., Uchida, T., Yenush, L., Davis, R. & White, M. F. (2000) J. Biol. Chem. 275, 9047–9054. [DOI] [PubMed] [Google Scholar]
- 8.Kim, A. H., Sasaki, T. & Chao, M. V. (2003) J. Biol. Chem. 278, 29830–29836. [DOI] [PubMed] [Google Scholar]
- 9.Kim, A. H., Yano, H., Cho, H., Meyer, D., Monks, B., Margolis, B., Birnbaum, M. J. & Chao, M. V. (2002) Neuron 35, 697–709. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R. J. (2000) Cell 103, 239–252. [DOI] [PubMed] [Google Scholar]
- 11.Wang, M. C., Bohmann, D. & Jasper, H. (2003) Dev. Cell 5, 811–816. [DOI] [PubMed] [Google Scholar]
- 12.Brenner, S. (1974) Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulston, J. & Hodgkin, J. (1988) in The Nematode Caenorhabditis elegans, ed. Wood, W. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 587–602.
- 14.Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosono, R., Mitsui, Y., Sato, Y., Aizawa, S. & Miwa, J. (1982) Exp. Gerontol. 17, 163–172. [DOI] [PubMed] [Google Scholar]
- 16.Byrd, D. T., Kawasaki, M., Walcoff, M., Hisamoto, N., Matsumoto, K. & Jin, Y. (2001) Neuron 32, 787–800. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki, M., Hisamoto, N., Iino, Y., Yamamoto, M., Ninomiya-Tsuji, J. & Matsumoto, K. (1999) EMBO J. 18, 3604–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga, M., Zwaal, R., Guan, K. L., Avery, L. & Ohshima, Y. (2000) EMBO J. 19, 5148–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villanueva, A., Lozano, J., Morales, A., Lin, X., Deng, X., Hengartner, M. O. & Kolesnick, R. N. (2001) EMBO J. 20, 5114–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson, D. W. & Padgett, R. W. (2003) Genes Dev. 17, 813–818. [DOI] [PubMed] [Google Scholar]
- 21.Hekimi, S., Benard, C., Branicky, R., Burgess, J., Hihi, A. K. & Rea, S. (2001) Mech. Ageing Dev. 122, 571–594. [DOI] [PubMed] [Google Scholar]
- 22.Braeckman, B. P., Houthoofd, K. & Vanfleteren, J. R. (2001) Mech. Ageing Dev. 122, 673–693. [DOI] [PubMed] [Google Scholar]
- 23.Kenyon, C. (2001) Cell 105, 165–168. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. S., Lee, R. Y., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nat. Genet. 33, 40–48. [DOI] [PubMed] [Google Scholar]
- 25.Paradis, S. & Ruvkun, G. (1998) Genes Dev. 12, 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertweck, M., Gobel, C. & Baumeister, R. (2004) Dev. Cell 6, 577–588. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, T. E. (2002) Sci. Aging Knowledge Environ. 2002, re4. [DOI] [PubMed] [Google Scholar]
- 28.Lithgow, G. J. & Walker, G. A. (2002) Mech. Ageing Dev. 123, 765–771. [DOI] [PubMed] [Google Scholar]
- 29.Munoz, M. J. (2003) Mech. Ageing Dev. 124, 43–48. [DOI] [PubMed] [Google Scholar]
- 30.Henderson, S. T. & Johnson, T. E. (2001) Curr. Biol. 11, 1975–1980. [DOI] [PubMed] [Google Scholar]
- 31.Lin, K., Hsin, H., Libina, N. & Kenyon, C. (2001) Nat. Genet. 28, 139–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.