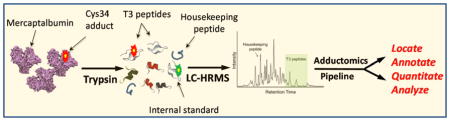
Abstract
An important but understudied class of human exposures is comprised of reactive electrophiles that cannot be measured in vivo because they are short lived. An avenue for assessing these meaningful exposures focuses on adducts from reactions with nucleophilic loci of blood proteins, particularly Cys34 of human serum albumin, which is the dominant scavenger of reactive electrophiles in serum. We developed an untargeted analytical scheme and bioinformatics pipeline for detecting, quantitating and annotating Cys34 adducts in tryptic digests of human serum/plasma. The pipeline interrogates tandem mass spectra to find signatures of the Cys34-containing peptide, obtains accurate masses of putative adducts, quantitates adduct levels relative to a ‘housekeeping peptide’, and annotates modifications based on a combination of retention time, accurate mass, elemental composition and database searches. We used the adductomics pipeline to characterize 43 adduct features in archived plasma from healthy human subjects and found several that were highly associated with smoking status, race and other covariates. Since smoking is a strong risk factor for cancer and cardiovascular disease, our ability to discover adducts that distinguish smokers from nonsmokers with untargeted adductomics indicates that the pipeline is suitable for use in epidemiologic studies. In fact, adduct features were both positively and negatively associated with smoking, indicating that some adducts arise from reactions between Cys34 and constituents of cigarette smoke (e.g. ethylene oxide and acrylonitrile) while others (Cys34 oxidation products and disulfides) appear to reflect alterations in the serum redox state that resulted in reduced adduct levels in smokers.
Graphical Abstract
Biologically-active chemicals enter the body from inhalation and ingestion of xenobiotic substances and are generated endogenously by human and microbial metabolism, oxidative stress, lipid peroxidation, and other processes. Evidence that the risks of acquiring chronic diseases are driven largely by diverse exposures rather than heritable genetic factors 1 has generated interest in the ‘blood exposome’ for studies of disease etiology 2. By comparing exposomes in blood from diseased and healthy subjects, discriminating features can be identified and then targeted for follow-up 3.
An understudied class of exposures includes reactive electrophiles that are generated from metabolic processes. These molecules are potentially toxic because they react with nucleophilic sites in DNA and proteins to produce mutations and post-translational modifications, respectively 4. Although reactive electrophiles cannot generally be measured in vivo, their systemic concentrations have been estimated from targeted studies of their adducts with blood proteins, notably Cys34 of human serum albumin (HSA) and N-terminal Val of hemoglobin (Hb) 5. In fact, Cys34, the only free thiol in HSA, is a nucleophilic hotspot that accounts for about 80% of the antioxidant capacity of serum 6. Oxidation of Cys34-SH to its sulfenic acid (Cys34-SOH) can motivate further oxidization to the corresponding sulfinic acid (Cys34-SO2H) and sulfonic acid (Cys34-SO3H) as well as reactions with low-molecular-weight thiols (primarily cysteine) that produce mixed Cys34 disulfides 7. The relative amounts of oxidized and cysteinylated Cys34 have been related to the redox state of the serum and to global oxidative stress 8.
Although targeted assays can relate Cys34 adducts to specific exposures 5, they are not well suited for discovering unknown causes of human diseases arising from exposures to electrophilic species. With this in mind, we developed an untargeted assay for Cys34 modifications that we refer to as ‘Cys34 adductomics’ 9. The original assay was based on triple-quadrupole mass spectrometry (TQMS) with selected reaction monitoring (SRM) of 77 mass bins between 9 and 351 Da. An untargeted TQMS-SRM approach has also been reported for N-terminal Val adducts of Hb 10.
The Cys34 residue is located on the third largest tryptic peptide of HSA (designated T3) with sequence ALVLIAFAQYLQQC34PFEDHVK and a mass of 2432 Da. Our TQMS method for Cys34 adductomics 9 consisted of steps to 1) purify and quantify HSA, 2) remove mercaptalbumin, 3) reduce HSA disulfides with tris(2-carboxyethyl)phosphine (TCEP) and digest the protein with trypsin, 4) enrich T3-related peptides via off-line HPLC, and 5) infuse the peptides into a TQMS for SRM. Although the original assay demonstrated proof of principle 9, it had low throughput and precluded detection of mixed Cys34 disulfides because of treatment with TCEP prior to digestion. Also, use of TQMS with direct infusion did not provide accurate masses, retention times, and MS2 spectra that facilitate annotation.
Here we present an analytical scheme for Cys34 adductomics that employs LC-HRMS and an adductomics pipeline that acquires and processes data. We validated the methodology with archived plasma from healthy subjects - stratified by smoking status, race, and gender - and detected 43 adducts. Levels of particular adducts were associated with smoking status as well as with race, gender, body mass index (BMI), and self-reported consumption of animal and vegetable fats. These results indicate that the Cys34-adductomics pipeline is suitable for applications in human populations.
MATERIALS AND METHODS
Chemicals and Reagents
Triethylammonium bicarbonate buffer (TEAB), ethylenediamine-tetraacetic acid (EDTA, anhydrous), ammonium sulfate, porcine trypsin, HSA (lyophilized powder, 97–99%), acetonitrile (Ultra CHROMASOLV®, LC-MS grade), acetic acid (LCMS grade), L-glutathione (reduced), L-cysteine, L-homocysteine, iodine, iodoacetamide (IAA), ammonium sulfate, dimethyl sulfoxide (DMSO), hydrogen peroxide (30% aqueous solution) were from Sigma-Aldrich (St. Louis, MO). Methanol (Optima®, LCMS grade), tris(2-carboxyethyl)phosphine (TCEP), formic acid (Optima®, LCMS Grade), were from Fisher Scientific (Pittsburgh, PA). Purified water (18.2 mΩ cm resistivity at 25°C) was prepared with a PureLab purification system (Elga LabWater, Woodridge, IL). The following chemicals were custom synthesized: T3 peptide with sequence ALVLIAFAQYLQQCPFEDHVK (>97%, Biomatik, Wilmington, DE), isotopically modified T3 with sequence AL-[15N, 13C-Val]-LIAFAQYLQQCPFEDH-[15N, 13C-Val]-K (>95%, BioMer Technology, Pleasanton, CA).
Synthesis of Reference Standards
A mixture of oxidized adducts of the T3 peptide [(+O, −2H), +O2 and +O3] and the internal standard (isotopic T3 adducted with iodoacetamide) were prepared as reported previously 11. Standards of disulfides of the T3 peptide with cysteine, homocysteine and glutathione were synthesized as described by Kirihara et al. 12. Briefly, an aqueous solution of the T3 peptide (prepared from a 5 mM stock solution in DMSO) was mixed with each thiol, iodine, and 30% H2O2 in a 1:5:0.01:1 molar ratio. After incubating at room temperature with constant agitation for 3 h, the T3 peptides containing the respective Cys34 disulfides were purified by solid phase extraction using C8 cartridges (Sep-Pak®, 100mg sorbent, 37–55 μm particle size, Waters, Milford, MA). The yields and MS characteristics of all synthesized products were confirmed by LC-HRMS/MS.
Plasma Samples
A set of 34 pooled plasma samples was prepared from 158 young healthy subjects that had been collected with informed consent from a previous study 13 and stored at −80°C for approximately 13 y. Samples of plasma were pooled by combining aliquots from four to six randomly-selected subjects stratified by smoking status (smoker/nonsmoker), race (black/white), and gender. (Pooling of these archived specimens was required to ensure anonymity of subjects). Demographic characteristics and average daily consumption of 131 food items had been obtained from each subject at the time of phlebotomy, and these permitted consumption of animal and vegetable fats to be estimated 13. The following covariates were derived from these data for statistical analyses: smoking status (smoke), race, gender, BMI (bmi) (kg/m2), and consumption of animal fat (afat) and vegetable fat (vfat) (g/d) as described previously 13. (Because samples had been pooled, mean values of bmi, afat and vfat for each pooled specimen were used). Summary statistics of bmi, afat and vfat were previously reported for lipidomics of the same pooled specimens 14 (Table S1). Validation samples were processed randomly in nine daily batches of eight specimens (in duplicate).
Sample Processing
To isolate HSA, five μL of each pooled plasma specimen was added to 60 μL of 50% methanol and incubated at room temperature for 15 min with constant agitation. The samples were centrifuged at 10,000×g for 15 min to remove precipitates and immediately diluted with four volumes of digestion buffer [50 mM TEAB buffer containing 1 mM EDTA (pH 8.0)]. Purified HSA (~ 0.1 mg total protein as determined by spectrophometry at 260/280 nm) in 140 μL of digestion buffer containing 10% methanol was transferred to a 150-μL capacity digestion vessel (MT-96, Pressure Biosciences Inc., South Easton, MA) and one μL of 10 mg/mL trypsin was added (~1:10 molar ratio of trypsin: protein). Proteolytic digestion was performed at 37°C using a pressurized system (NEP2320, Pressure Biosciences Inc., South Easton, MA) that cycled between ambient pressure (15 sec) and 138 mPa (45 sec) for 30 min. After digestion, three μL of 10% formic acid was added and samples were immediately centrifuged for 20 s to precipitate trypsin and protein aggregates. A 40-μL aliquot of each digest was diluted to a final volume of 120 μL with an aqueous solution containing 2% acetonitrile and 0.2% formic acid, and then transferred to a silanized 0.3-ml glass vial (National Scientific, Rockwood, TN) to which 1 μL of the isotopically-labeled internal standard (iAA-T3, 20 pmol/μL) 9 was added.
LC-HRMS Analysis
Freshly-digested samples were analyzed with a LTQ Orbitrap XL HRMS coupled with a Dionex Ultimate® 3000 nanoflow LC system via a Flex Ion nano-electrospray-ionization source (Thermo Fisher Scientific, Waltham, MA), operated in positive-ion mode with a spray voltage of 2.0 kV, a capillary temperature of 200°C and voltage of 35 V. One μL of each digest (~ 10 pmol) was injected using the UltiMate® 3000 Rapid Separation Autosampler with sample vials maintained at 4°C. The peptides were separated on a Dionex PepSwift monolithic nanoflow column (100-μm i.d. × 25 cm) (Thermo Scientific, Sunnyvale, CA, USA), operated at room temperature with a flow rate of 750 nL/min. Mobile phase A was water/0.1% formic acid (v/v) and mobile phase B was acetonitrile/0.1% formic acid (v/v). The gradient involved isocratic flow at 2% B for 5 min, a linear gradient from 2% to 45% B over 30 min, rapid increase to 98% B to wash the column (3 min), and reset to initial conditions over 5 min. The needle was rinsed with 30% methanol after each injection and the column was washed after every fifth injection with oneμL of a solution containing 80% acetonitrile, 10% acetic acid, 5% dimethyl sulfoxide and 5% water. With these conditions the carryover of hydrophobic T3 peptides between injections was less than 3%.
Mass spectra were acquired over the range m/z = 300 to 1200 using the Orbitrap mass analyzer, in profile format, with a resolution setting of 6×104 at m/z = 400. In data-dependent mode, the six most intense ions exceeding 10,000 counts were selected from each full-scan mass spectrum for tandem mass (MS2) analysis using CID. MS2 spectra were acquired in centroid format, using the linear ion trap, with the following parameters: precursor ion mass range m/z = 750 to 1000, isolation width 3 m/z units, normalized collision energy 28%, activation time 30 ms, activation Q 0.25, and default charge state 3+. To avoid the occurrence of redundant MS2 measurements, real-time dynamic exclusion was enabled using the following parameters: repeat count 2, repeat duration 10 s, exclusion list size 500, exclusion duration 90 s, and exclusion precursor ion width ± 20 ppm. Charge-state screening and monoisotopic-precursor-ion selection were enabled. The lock-mass option was enabled to provide a real-time internal mass calibration using a reference list of 8 identified background ions 15. External mass calibration was performed prior to analysis using the standard Thermo LTQ calibration mixture. MS1 and MS2 spectra were acquired and processed using Xcalibur software (version 2.0.7) and Chromeleon Xpress (v. 6.80) (Thermo Fisher Scientific).
Using a Housekeeping Peptide for Quantitation of Adducts
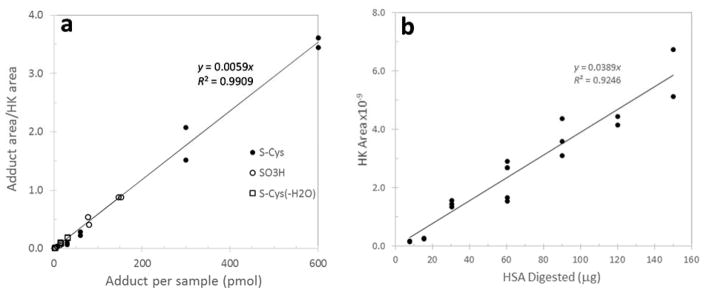
To adjust each sample for the amount of digested HSA, we used the HSA tryptic peptide adjacent to T3 (LVNEVTEFAK, # 41-50, doubly charged precursor ion at m/z=575.3111) as a ‘housekeeping peptide’ (HK). The peak-area ratio (PAR) of each modified T3 peptide relative to HK was used as a linear measure of the adduct concentration. This HK peptide was chosen because it is chemically neutral and unlikely to be a target for PTMs; it is adjacent to T3 and should have similar tryptic digestion efficiency; it has a distinct elution time and symmetrical peak shape; and it ionizes well as a doubly charged species. The linearity of the relationship between the PAR and the adduct concentration between 0.01 and 5 μM was determined with synthetic standards of the Cys34 sulfonic acid and two Cys34 disulfides. Peak-area ratios increased linearly with adduct concentrations over the full range of tested concentrations and did not differ across analytes (Figure 1a). A calibration curve relating the peak area of HK to a given amount of digested HSA (0.01 mg to 0.15 mg in 0.12 ml of digestion buffer) was generated (Figure 1b) to estimate approximate adduct concentrations as pmol adduct/mg of HSA. Although we are unaware of any previous use of a housekeeping peptide for quantitation, this approach is similar to that of a ‘signature peptide’ described by Zhang et al. for quantitation of serum proteins 16.
Figure 1.
Calibration of adduct concentrations relative to a ‘housekeeping peptide’ (HK). To adjust each sample for the amount of HSA in a digest, the HSA tryptic peptide adjacent to T3 (LVNEVTEFAK) is used as the HK. The ratio of peak areas of each modified T3 peptide to that of HK is a linear measure of the adduct concentration. a) The log-log plot of peak area ratios of three synthetic T3 adducts (analyzed in duplicate) at concentrations between 0.01 and 5 μM. b) Calibration curve relating the peak area of HK to the amount of HSA in a digest. This facilitates approximation of the adduct concentration with units of pmol adduct/mg HSA.
MS2 Signatures of Modifications to the T3 Peptide
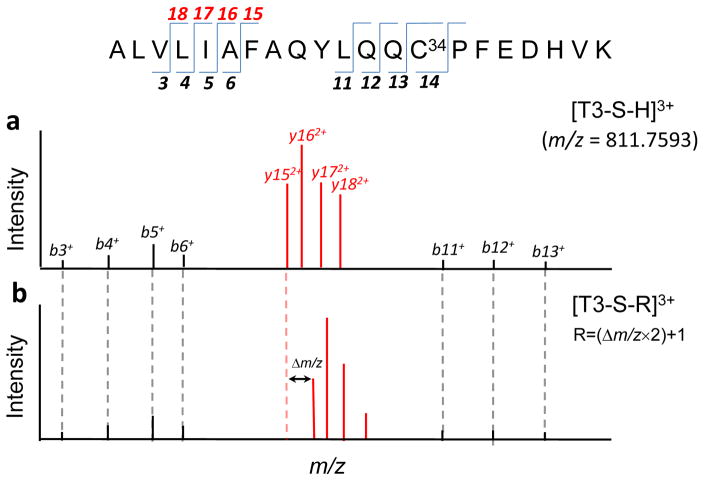
To set the stage for the adductomics pipeline, we illustrate fragmentation of the T3 peptide and its Cys34 modifications produced by CID. Figure 2 is a schematic that displays fingerprints of MS2 fragment ions resulting from precursor ions representing the triply-charged T3 peptide ([T3-SH] 3+ m/z = 811.75936, Figure 2a) and an adducted T3 peptide ([T3-S-R]3+, Figure 2b). All b+-series ions prior to b14 (i.e., b3+ – b6+ and b11+ – b13+) are the same in both spectra, confirming that no other amino acid has been modified at positions prior to Cys34. Note that the most abundant ions in MS2 spectra tend to be y2+-series ions (y152+ – y182+). By comparing Figures 2a and 2b, these prominent y2+ ions from fragmentation of T3-SH and T3-S-R are offset by the addition Δm/z to Cys34, with the added mass R = (Δm/z×2)+MH.
Figure 2.
Schematic showing tandem mass spectra of unmodified and modified T3 peptides. a) Spectrum of the precursor ion for the unmodified T3 peptide [T3-S-H]3+. b) Spectrum of the precursor ion of a T3 peptide modified at Cys34 [T3-S-R]3+ to produce an adduct of added mass R. Note that the indicated pairs of b+-series are the same in both spectra while those of the y2+-series ions are offset by a mass represented by Δm/z = R/2 − MH. [The indicated signature ions were extracted from actual spectra obtained for the T3 peptide (a) and the T3 peptide modified with homocysteine (b)].
The Adductomics Pipeline
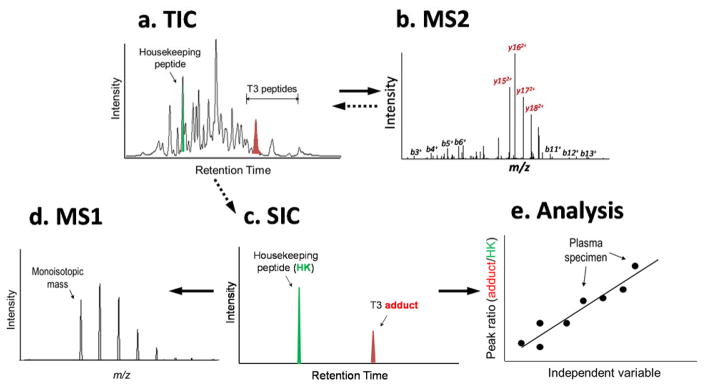
The adductomics pipeline for T3-peptide adducts is illustrated in Figure 3, with each step detailed below. The input data file is embodied by the total ion chromatogram (TIC) generated by LC-HRMS (Figure 3a). Putative T3 adducts are located from MS2 signatures as described above (Figure 3b). Then, selected ion chromatograms (SICs) for each precursor ion and the HK peptide were extracted (Figure 3c) to obtain monoisotopic masses (MIMs) (Figure 3d) and the corresponding added masses of putative adducts. The PARs, estimated from the peak areas of SICs for the T3 peptide and the HK peptide, were used for quantitation and statistical analysis (Figure 1 and 3e).
Figure 3.
Scheme for the adductomics pipeline. a) Total ion chromatograms (TICs) are monitored over the retention-time range of interest for T3 peptides and the housekeeping peptide. b) Tandem mass (MS2) spectra are compiled for all triply charged precursor ions and interrogated for b+- and y2+-series ions as signatures of the T3 peptide and its modifications. c) The selected ion chromatograms (SICs) of each precursor ion and the housekeeping peptide are extracted from the TIC. d) Each SIC is used to obtain an accurate (monoisotopic) mass (MIM) for a given adduct. e) The ratio of peak areas (T3-adduct/housekeeping peptide) is used to adjust adduct intensities for the amount of HSA in each digest and for statistical analysis.
Locating Putative Adducts
The raw data from data-dependent analyses were acquired using Xcalibur software and converted to MZXML format using ProteoWizard Tools 3.0.4624 (http://proteowizard.sourceforge.net) without any filters. All MS2 spectra were processed using in-house software written in R. Spectra displaying at least five of the seven requisite b+-series ions (b3+-b6+ & b11+-b13+) with signal-to-noise (S/N) ≥ 5, at least four of the six y2+-series ions (y142+-y182+) with relative abundance ≥ 20%, as well as cursory visual inspection were designated as putative T3 modifications. To determine whether Cys34 was the site of modification, the following manual checks were followed:
If either the b*14 fragment ion (m/z = 1562.8 Da + added mass) or both the y*82+ ion (m/z = 487.72 + added mass) and y72+ (m/z = 436.2, corresponding to Pro35) fragment ions were detected, then modification at Cys34 was confirmed;
if the b14+ fragment ion (m/z =1562.8, corresponding to unmodified Cys34) was detected, then Cys34 was regarded as the likely site of modification, recognizing that losses of labile adduct moieties can also occur during fragmentation; and
if y72+ was not detected but both y*72+ and y*82+ were observed, then Cys34 was ruled out as the site of modification.
Peak Picking and Quantitation of Putative Adducts
Peak picking and quantitation used the MIMs of the putative T3 adducts within 4 ppm and retention times ± 0.5 min (after adjustment with the internal standard) using Thermo Fisher software (Xcalibur Processing Method,v. 2.0.7). The SIC for each extracted monoisotopic mass with the closest retention time was integrated with the following parameters: retention time (RT) window 15–120 s (depending on co-eluting peaks), RT view width 5 min, genesis peak integration valley detection 15 s, and tailing factor 9.0 (S/N = 3). All peaks having at least three scans were automatically integrated and those with one or two scans were manually integrated. Peaks with ambiguous isotope distributions were excluded. (A representative SIC is shown in Figures S2a–e for each of the 43 putative T3 adducts detected in plasma). The mass added (Δm/z) to the T3 thiolate ion (T3-Cys34-S−) was calculated by subtracting the MIM of the triply charged precursor ion from the corresponding m/z of the modified T3 peptide, multiplying the difference by three and adding the MIM of one hydrogen atom. Peaks representing the SIC for the internal standard were used to monitor retention times and mass accuracies of putative T3 adducts.
Annotation of Putative Adducts
Elemental compositions of masses added to the T3 peptide were obtained with the Molecular Weight Calculator (v.6.49 www.alchemistmatt.com), Elemental Composition Finder from the Xcalibur Qual Browser (version 2.0.7, Thermo Fisher Scientific), and Molecular Formula Finder available in ChemCalc (http://www.chemcalc.org/mf_finder). In silico CID fragmentation of peptides was performed using Protein Prospector (http://prospector.ucsf.edu/prospector/mshome) and Molecular Weight Calculator. The following online libraries were used for database searches by elemental composition: the National Institute of Standards and Technology (NIST, http://www.nist.gov), Open Chemistry Database from NCBI (PubChem, https://pubchem.ncbi.nlm.nih.gov), and the Human Metabolome Database (HMDB, http://www.hmdb.ca/metabolites). Untargeted database searches of MS2 spectra for cysteine modifications employed UNIMOD (www.unimod.org) and DeltaMass (http://www.abrf.org/index.cfm/dm.home). The mass accuracies of identified adducts were calculated based on observed and theoretically calculated monoisotopic masses. The theoretical monoisotopic mass of each triply-charged T3 peptide was visualized by simulating the isotope distribution of a given empirical formula using Xcalibur software. Identities of some adducts were verified by comparing observed monoisotopic masses and retention times to those of their synthetic analogs. To provide additional evidence concerning putative annotations of mixed Cys34 disulfides, we compared mass spectra obtained from protein digests with and without addition of TCEP to reduce all disulfide bonds. Five validation samples that were most heavily populated with T3 adducts were treated with the standard protocol except that TCEP was added before digestion to a final concentration of two mM.
Statistical Analyses
Statistical analyses were performed using SAS software for Windows (v. 9.7, SAS Institute, Cary, NC). First, PARs of the 43 adduct features with sufficient data were multiplied by 1000 (for scaling), log-transformed and adjusted for batch effects with a mixed-effects model, which included batch as a fixed effect and each specimen (analyzed in duplicate) as a random effect. The resulting best linear unbiased predictor for each random effect was added to the respective intercept term to provide a normalized predictor of the natural logarithm of the PAR (×1000). The within-specimen variance component (the error variance, ) was used to estimate the CV for each adduct feature as (Table S3). Six adduct features were excluded from statistical analysis because their estimated intraclass correlation coefficients were zero, indicating that adduct levels did not vary across plasma specimens. Three adduct features were sodiated or potassium products; PARs for these metal adducts were added to those of the parent adducts for statistical purposes. The log-scale predictors of adduct features were returned to natural scale by exponentiation and values below the limit of quantitation (LOQ) were imputed a PAR of as estimated from the seven least-abundant adduct features. Wilcoxon rank-sum exact tests were performed to determine whether each PAR differed between smokers and nonsmokers, males and females, or black and white subjects, using a Bonferroni-adjusted P-value of 0.05/34 = 0.00147 to determine significance. Finally, multivariable regression analyses were performed with the predicted PAR (×1000, in natural scale) for each adduct as dependent variable and race, gender, smoke, bmi, and consumption of animal fat (afat) and vegetable fat (vfat) as independent variables. Backward selection was used with an alpha level for entry of 0.05 and for deletion of 0.10.
RESULTS AND DISCUSSION
Isolation of HSA from Plasma
In our previous Cys34-adductomics assay, we had used ammonium sulfate to precipitate the non-HSA serum proteins followed by buffer exchange 9. Because HSA has the lowest isoelectric point of all major plasma proteins 17, we tested different concentrations of methanol and acetonitrile that would maintain HSA in solution while precipitating other proteins. Using 50% methanol as the extraction solvent, HSA represented 72 – 75% of the protein content in the supernatant as determined by quantitative densitometry of proteins following SDS-PAGE (data not shown). Although this purity is less than that from treatment with saturated ammonium sulfate in our TQMS assay 9 (84%), use of methanol eliminated the need for buffer exchange prior to digestion and also enhanced tryptic digestion (discussed below). Treatment with 50% methanol also effectively precipitated residual Hb.
Tryptic Digestion with Pressure Cycling
Following a conventional proteomics approach, our initial scheme incorporated TCEP to reduce disulfides and denature proteins prior to tryptic digestion 9. However, to include mixed Cys34 disulfides in the assay and to prevent unwanted reactions involving TCEP 18,19, we investigated tryptic digestion without prior reduction of disulfide bonds. In fact, we observed that digestion of HSA with pressure cycling proceeded favorably without reduction of disulfide bonds, as reported previously 20, and was enhanced with use of methanol to purify HSA. Treatment of HSA with 50% methanol – as used in the purification step – reportedly unfolds the ternary structure while maintaining the protein in solution 21. We also observed that the activity of trypsin was slightly higher in the presence of 10–20% methanol and thus adjusted the final concentration to 10% methanol prior to digestion.
Sample Throughput
By eliminating buffer exchanges from the original assay as well as steps to enrich Cys34 prior to digestion and purification of T3 peptides by off-line HPLC 9, sample processing was reduced from about 21 h to one h per batch of 12 plasma samples. The rate-limiting step is the LC-HRMS analysis that requires about 40 min per assay. Thus, we estimate throughput at about 12 duplicate plasma sample (24 assays) per day using a single LC-HRMS platform.
Annotations of Modified T3 Peptides
Table 1 lists 43 T3 peptides and their modifications (designated A1 to A43) that were quantified in plasma samples. (Representative MS2 spectra for the 43 putative T3 adducts listed in Table 1 are shown in Figures S1–A1 to S1–A43). Annotations based on accurate masses, MS2 spectra, and citations to references from literature and database searches are given in Table S2. Three adducts were traced to sodium and potassium products, i.e. A19 = A12+Na, A35 = A29+Na, and A37 = A29+K. Two T3 peptides had MS2 spectra that were identical to those of the unmodified T3 peptide, namely A7 that had the same retention time as the synthetic peptide (28.90 min) and an early-eluting peptide, A6 (27.89 min), which is apparently a labile T3 adduct that disaggregated during electrospray ionization. Adduct A8 was confirmed to be the T3 dimer (30.37 min, 6+ charge state). Three adducts represent Cys34 oxidation products that we had characterized previously 11 [i.e. a Cys-Gln crosslink (A9), the sulfinic acid (A12+ A19), and the sulfonic acid (A15)]. Another three adducts showed mass losses consistent with Cys34 conversion to Gly (A1), dehydroalanine (A2), and oxoalanine/formylglycine (A3). A third set of adducts was derived from products of Cys34 reactions involving addition of cyanide (A11), ethylene oxide (A14) [S-(2-hydroxyethyl)Cys34], acrylonitrile (A18) [S-(2-cyanoyethyl )Cys34], methyl isocyanate (A20) [S-(methylcarbamoyl)Cys34], and methylvinylketone (A21) [S-(3-oxobutyl)Cys34]. The largest class of adducts was comprised of Cys34 disulfides, stemming from reactions of the sulfenic acid with low-molecular-weight thiols in serum, namely, cysteine (Cys) [A29+A35+A37, A24 (following dehydration) & A30 (following deamination)], homocysteine (hCys) [A32, A33 & A28 (following dehydration], cysteinyl glycine (Cys-Gly) [A40 & A38 (following dehydration)], γ-glutamylcysteine (A42) (γ-Gln-Cys), glutathione (A43), N-acetylcysteine (A39), mercaptoacetamide (A22), and mercaptoacetic acid (A23). Upon reanalysis of several plasma specimens that had been treated with TCEP to reduce disulfides prior to tryptic digestion, none of these putative Cys34 disulfides were detected, providing further evidence that they were disulfides (Table 1) (data not shown). Three adducts (A25, A27 and A34) were confirmed as Cys34 modifications with mass accuracies within 1 ppm of theoretical empirical formulas, but were not found in databases. Moreover, A27 and A34 were not detected in TCEP-reduced samples and thus are likely to be disulfides. Scrutiny of the MS2 spectra pinpointed several modifications of the T3 peptide that occurred at a site(s) other than Cys34, notably the methylation product (A10) and several unknown adducts (A4, A5, A13, A16, A17, A36 & A41).
Table 1.
T3 peptides detected in validation samples of plasma from smoking and non-smoking subjects.
| Adduct | Retention time (min) | PAR a (×1000) | Conc.* (pmol/mg HSA) | m/z, 3+ Observed | Mass (Da) added to T3 (Cys-S−) | Adduct composition | m/z,3+ (Theoretical) | ΔMass (ppm) | Annotation |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 27.74 | 0.2950 | 1.31 | 796.43069 | −45.9899 | -CH2S | 796.43012 | −0.7214 | Cys34→Gly |
| A2 f | 28.90 | 0.0512 | 0.227 | 800.43140 | −33.9878 | -SH2 | 800.43012 | −1.5991 | Cys34→Dehydroalanine |
| A3 | 27.55 | 0.0533 | 0.236 | 805.76389 | −17.9903 | -SH2, +O | 805.76175 | −2.6503 | Cys34→Oxoalanine |
| A4 f | 28.01 | 0.8701 | 3.85 | 808.73050 | −9.0905 | Not Cys34 adduct f | |||
| A5 f | 28.38 | 1.289 | 5.71 | 810.45390 | −3.9203 | Not Cys34 adduct | |||
| A6 | 26.56 | 3.042 | 13.5 | 811.76052 | −0.0004 | 811.75936 | −1.4279 | T3 Labile adduct | |
| A7 | 27.90 | 47.71 | 211 | 811.76066 | 0.0000 | 811.75936 | −1.5975 | Unmodified T3 e | |
| A8 f | 30.37 | 1.597 | 7.07 | 811.42490 | 2431.2483 | +C114H172N27O30S | 811.42341d | −1.8979 | T3 Dimer e |
| A9 | 27.32 | 2.605 | 11.5 | 816.41996 | 13.9779 | -H2, +O | 816.41911 | −1.0420 | Cys34-Gln crosslink e |
| A10 | 28.93 | 0.3161 | 1.40 | 816.43214 | 15.0224 | +CH3 | 816.43124 | −1.1083 | Methylation (not Cys34) |
| A11 f | 28.46 | 0.3014 | 1.33 | 820.09202 | 26.0020 | +CN | 820.09111 | −1.1099 | Cyanide adduct |
| A12 b | 27.35 | 2.250 | 9.96 | 822.42369 | 32.9970 | +HO2 | 822.42263 | −1.2847 | Cys34 Sulfinic acid e |
| A13 | 27.17 | 0.0223 | 0.099 | 823.39710 | 35.9173 | Not Cys34 adduct | |||
| A14 c | 27.89 | 0.5273 | 2.33 | 826.43575 | 45.0332 | +C2H5O | 826.43476 | −1.1983 | Ethylene oxide adduct |
| A15 | 27.69 | 0.5681 | 2.51 | 827.75501 | 48.9910 | +HO3 | 827.75427 | −0.8948 | Cys34 Sulfonic acid e |
| A16 | 27.37 | 0.2276 | 1.01 | 829.12640 | 53.1052 | Not Cys34 adduct | |||
| A17 | 28.68 | 0.1978 | 0.876 | 829.39790 | 53.9197 | Not Cys34 adduct | |||
| A18 c | 28.92 | 0.2980 | 1.32 | 829.43689 | 54.0366 | +C3H4N | 829.43487 | −2.4350 | Acrylonitrile adduct |
| A19 | 27.25 | 0.1337 | 0.592 | 829.75070 | 54.9781 | A12+Na | 829.74995 | −0.9062 | Na adduct of A12 |
| A20 | 27.48 | 3.379 | 15.0 | 830.76845 | 58.0313 | +C2H4NO | 830.76651 | −2.3300 | Methylisocyanate adduct |
| A21 | 28.93 | 0.4216 | 1.87 | 835.10757 | 71.0487 | +C4H7O | 835.10664 | −1.1098 | Methylvinylketone adduct |
| A22 | 27.77 | 0.7292 | 3.23 | 841.42510 | 90.0013 | +C2H4NOS | 841.42387 | −1.4574 | S-Mercaptoacetamide |
| A23 f | 28.33 | 0.6193 | 2.74 | 841.75308 | 90.9852 | +C2H3O2S | 841.75187 | −1.4333 | S-Mercaptoacetic acid |
| A24 f | 27.37 | 2.456 | 10.9 | 845.42509 | 102.0012 | +C3H4NOS | 845.42387 | −1.4400 | S-Cys (-H2O) |
| A25 | 29.39 | 0.1792 | 0.793 | 847.10889 | 107.0526 | +C7H7O | 847.10664 | −2.6554 | Unknown |
| A26 f | 27.36 | 0.0586 | 0.259 | 847.76685 | 109.0265 | Unknown | |||
| A27 f | 28.39 | 0.2133 | 0.944 | 849.06977 | 112.9353 | +HO3S2 | 849.06898 | −0.9327 | Unknown |
| A28 f | 28.35 | 0.2295 | 1.016 | 850.09820 | 116.0206 | +C4H6NOS | 850.09575 | −2.8820 | S-hCys (-H2O) |
| A29 b | 26.70 | 557.6 | 2470 | 851.42890 | 120.0127 | +C3H6NO2S | 851.42739 | −1.7684 | S-Cys e |
| A30 f | 27.77 | 4.051 | 17.9 | 851.75702 | 120.9970 | +C3H5O3S | 851.75539 | −1.9167 | S-Cys (NH2→OH) |
| A31 f | 27.76 | 0.2925 | 1.30 | 853.78457 | 127.0797 | Unknown | |||
| A32 f | 26.72 | 4.039 | 17.9 | 856.10035 | 134.0270 | +C4H8NO2S | 856.09927 | −1.2613 | S-hCys e |
| A33 f | 27.10 | 15.62 | 69.1 | 856.10011 | 134.0263 | +C4H8NO2S | 856.09927 | −0.9797 | S-hCys e |
| A34 f | 27.73 | 0.5691 | 2.52 | 857.09988 | 137.0256 | +C4H9O3S | 857.09916 | −0.8370 | Unknown |
| A35 | 26.45 | 1.5866 | 7.02 | 858.75406 | 141.9881 | A29+Na | 858.7547 | 0.7487 | Na adduct of A29 |
| A36 | 26.53 | 2.5561 | 11.3 | 862.09027 | 151.9968 | Not Cys34 adduct | |||
| A37 f | 26.60 | 0.4479 | 1.98 | 864.07784 | 157.9595 | A29+K | 864.07933 | 1.7 | K adduct of A29 |
| A38 f | 27.21 | 0.5339 | 2.36 | 864.43218 | 159.0225 | +C5H7N2O2S | 864.43102 | −1.3386 | S-Cys-Gly (-H2O) |
| A39 f | 27.68 | 0.1491 | 0.660 | 865.43144 | 162.0203 | +C5H8NO3S | 865.43091 | −0.6125 | S-(N-acetyl)Cys |
| A40 f | 26.20 | 49.0529 | 217 | 870.43575 | 177.0332 | +C5H9N2O3S | 870.43454 | −1.3875 | S-Cys-Gly e |
| A41 f | 26.74 | 0.1424 | 0.630 | 872.73090 | 183.9187 | Not Cys34 adduct | |||
| A42 f | 26.71 | 1.9810 | 8.77 | 894.44253 | 249.0536 | +C8H13N2O5S | 894.44159 | −1.0491 | S-γ-Gln-Cys |
| A43 f | 26.38 | 2.1374 | 9.46 | 913.44937 | 306.0741 | +C10H16N3O6S | 913.44874 | −0.6853 | S-Glutathione e |
Approximate concentration
Peak-area ratio (adduct-peak area/housekeeping-peptide-peak area)
Adduct level significantly lower in smoking subjects after Bonferroni adjustment (P-value < 0.00147)
Adduct level significantly higher in smoking subjects after Bonferroni adjustment (P-value < 0.00147)
6+ Charge state
Annotation confirmed with synthetic standard
Adduct disappeared when digested in the presence of TCEP
Dynamic Range and Precision
Estimated PARs covered a 25,000-fold range, from 2.23×10−5 to 0.557 (approximately 0.099 to 2,470 pmol adduct/mg HSA, Table 1). Covariance parameters for linear mixed models that were used for batch adjustment are shown in Table S3. CVs, derived from within-specimen variance components, represent technical variation of the assay that ranged from 0.082 to 1.35 for the 43 features (median = 0.335).
Associations with Smoking and other Covariates
Of the 34 adduct features tested with Wilcoxon rank-sum tests, five had PARs that differed between smokers and nonsmokers, using a Bonferroni-adjusted P-value of 0.00147 (Table S4). Levels of the ethylene oxide (A14) and acrylonitrile (A18) adducts were both higher in smoking subjects (P-value < 0.0001). Ethylene oxide and acrylonitrile are suspected human carcinogens that have previously been associated with Hb adducts in targeted and untargeted analyses of smokers’ blood 10,22. On the other hand, adducts of the Cys34 sulfinic acid (A12+A19) (P-value = 0.00058) and the S-Cys adduct (A29+A35+A37) (P-value = 0.00120) were significantly lower in smokers. When Wilcoxon tests were repeated to investigate race and gender, no significant differences in adduct levels were observed after Bonferroni adjustment (Table S4).
Multivariable regression models were constructed for each adduct feature using the following predictor variables: smoke, race, gender, bmi, afat and vfat. A total of 40 covariate effects (P-value < 0.10) were detected with directions indicated by arrows in Table 2. The amounts of variation explained by the models (Adj. R2) ranged from 13.7% to 89.2% (median = 27.0%). Seven of the models pointed to associations that were significant after Bonferroni adjustment, i.e. four for smoke, and one each for bmi, and consumption of afat or vfat. Considering all effects with P-values < 0.10, smoke was associated with 12 adducts but the correlations were not always positive, as would be expected if smokers had consistently higher adduct levels. In fact, levels of seven adducts were lower in smokers than in nonsmokers, i.e. the three Cys34 oxidation products, the cysteinylation product of Cys34 that represents a global biomarker of oxidative stress 8, the cyanide adduct (also an oxidation product 23), and two unknown adducts (A5 & A34). The lower concentrations of oxidation products in smokers were unanticipated but may have resulted from smoking-associated hypoxia 24 or pertubations to the redox proteome 25. Levels of five adducts were elevated in smokers, i.e. the methylation product (A10), adducts of ethylene oxide (A14) and acrylonitrile (A18), the disulfides of Cys-Gly (-H2O) (A38) and N-acetylcysteine (A39), and an unknown adduct (A31). Interestingly, the methylation product was significantly elevated in smokers (P-value = 0.0003) in the multivariable model even after Bonferroni adjustment. Since some nicotine-derived N-nitrosamines [e.g. 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), and N′-nitrosonornicotine (NNN)] produce methylation products with DNA 26, this adduct could represent a similar modification of the T3 peptide at a site(s) other than Cys34. Among the other covariates, race was positively associated with five adducts (indicating higher levels in black subjects) and negatively associated with two adducts. Interestingly, associations with bmi, afat, and vfat were always in one direction. That is, all eight associations with bmi and five associations with vfat were negative (i.e. adduct levels decreased with increasing bmi or vfat), while all seven associations with afat were positive (i.e. adduct levels increased with increasing afat). Levels of two adducts were marginally higher in males than females.
Table 2.
Results of multivariable regression analyses for 34 T3 adducts or clusters. [P-values and directions of associations are shown for the following covariates: smoke (smoker=1; nonsmoker=0), race (black=1, white=0), gender (male=1, female=0), bmi (body mass index, kg/m2), afat (animal fat consumption, g/day), vfat (vegetable fat consumption, g/day). P-Values in bold face indicate significant effects after Bonferroni adjustment (P-value = 0.05/34 = 0.00147).] Arrows indicate associations that either increased (↑) or decreased (↓) with the predictor variable. (Full regression models and diagnostics are given in Table S5.)
| Adduct or cluster | smoke | race | gender | bmi | afat | vfat | Adj. R2 |
|---|---|---|---|---|---|---|---|
| Cys→Gly (A1) | 0.0146↓ | 0.147 | |||||
| Unknown (A5) | 0.0365↓ | 0.0450↓ | 0.0590↓ | 0.0056↑ | 0.211 | ||
| T3 Labile adduct (A6) | 0.0010↑ | 0.0059↓ | 0.256 | ||||
| Cys34→Gln crosslink (A9) | 0.0271↓ | 0.0753↓ | 0.231 | ||||
| Methylation (A10) | 0.0003↑ | 0.0074↑ | 0.0981↓ | 0.0543↓ | 0.301 | ||
| Cyanide (A11) | 0.0742↓ | 0.0379↓ | 0.215 | ||||
| Cys34 Sulfinic acid & sodiated adduct (A12+A19) | 0.0007↓ | 0.283 | |||||
| Ethylene oxide (A14) | <0.0001↑ | 0.0769↓ | 0.0629↑ | 0.892 | |||
| Cys34 Sulfonic acid (A15) | 0.0023↓ | 0.0612↓ | 0.269 | ||||
| Unknown (A16) | 0.0053↓ | 0.194 | |||||
| Acrylonitrile (A18) | <0.0001↑ | 0.643 | |||||
| S-Mercaptoacetic acid (A23) | 0.0145↑ | 0.0012↓ | 0.0138↑ | 0.398 | |||
| S-hCys (-H2O) (A28) | 0.0113↑ | 0.159 | |||||
| S-Cys & metal adducts (A29+A35+A37) | 0.0113↓ | 0.0448↑ | 0.0310↓ | 0.322 | |||
| Unknown (A31) | 0.0422↑ | 0.0853↑ | 0.137 | ||||
| Unknown (A34) | 0.0039↓ | 0.0051↓ | 0.323 | ||||
| S-Cys-Gly (-H2O) (A38) | 0.0026↑ | 0.0162↓ | 0.220 | ||||
| S-(N-AcetylCys) (A39) | 0.0025↑ | 0.0646↑ | 0.266 | ||||
| S-Cys-Gly (A40) | 0.0080↑ | 0.0007↓ | 0.272 | ||||
| S-Glutathione (A43) | 0.0050↓ | 0.197 |
The associations detected between adduct levels and smoking and other covariates indicate that Cys34 adductomics can be used in observational studies to discover population differences. In fact, levels of either the ethylene oxide adduct (A14) or acrylonitrile adduct (A18) perfectly discriminated between smokers and nonsmokers in our samples (AROC ≅ 1). The surprising unidirectional associations observed with bmi, afat and vfat also raise intriguing questions about connections between T3 adducts and the redox proteome 25. Since some of the associations with covariates involved unknown adducts (A6, A16, A31 & A34), follow-up studies will be needed to ascertain chemical structures and to pursue mechanistic understanding.
Possible Artifacts
Formation of analytical artifacts is well documented in proteomic research 27–29 and can be problematic when untargeted methods are used to screen for post-translational modifications. The most common artifacts are oxidation products 30 and metal adducts of Na+, K+, and Fe2+. To reduce artifact formation, we eliminated steps from our TQMS assay 9 that had been used for adduct enrichment, buffer exchanges, and peptide fractionation, and performed digestion in the absence of TCEP. Since Fenton chemistry is a major source of oxidation reactions 31, we used methanol as the primary solvent and added EDTA as a chelating agent to the digestion buffer. By replacing conventional ESI with NESI, we reduced electrospray generation of metal adducts 32,33. Since it is inevitable that some artifacts will be produced when collecting, storing and processing biospecimens, it is essential that the same methods be applied rigorously throughout the course of an investigation, including phlebotomy and blood processing, to reduce the likelihood of false-positive associations in population comparisons. The population differences in adduct levels detected in our study of plasma specimens stored for 13 y at −80°C (Table 2 and Table S3) suggest that artifact formation need not obscure meaningful comparisons in adductomic investigations.
Extending the Dynamic Range
We recognize that the 43 adducts detected in our validation study represent the most abundant Cys34 modifications. Indeed, given the many electrophilic species and precursor molecules that have been reported in cigarette smoke 34, we had expected to find more adducts in smoking subjects. In any case, the HRMS platform used for analyses (Orbitrap LTQ) is a decade old and two generations of instruments are currently available with increased sensitivity and much faster scan rates that permit more data-dependent MS2 spectra to be obtained. Thus, we expect that the list of detected adducts will increase with newer HRMS platforms.
Applying the Pipeline to Other Proteins
Although this investigation focused on Cys34 modifications for proof of principle, our adductomics pipeline can easily be extended to other HSA tryptic peptides containing nucleophilic hotspots, such as His146 and Lys199 6. In fact, the methodology is sufficiently general to permit untargeted adductomics of any protein that produces tryptic peptides with good sequence coverage of signature ions in MS2 spectra.
Supplementary Material
Acknowledgments
The authors appreciate the assistance of He Li who developed the original adductomics methodology, and of Wendy McKelvey who collected the plasma specimens and associated data that were used for validation. This work was supported by grant R33CA191159 from the National Cancer Institute of the U.S. National Institutes of Health. The authors also acknowledge support from grants U54ES016115 and R44ES022360 from the National Institute for Environmental Health Sciences and LRI4677 from the American Chemistry Council.
Footnotes
Supporting Information Available: (Table S1: Characteristics of validation samples, Table S2: Evidence for annotation of adducts, Table S3: Covariance parameters from mixed models for estimation of assay precision, Table S4: P-values from Wilcoxon tests of smoking status, race and gender, Table S5: Multivariable regression models and diagnostics, Figure S1: MS2 spectra for the T3 peptide and its modifications in validation samples, Figure S2: Selected ion chromatograms of T3 peptides in validation samples). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rappaport SM. PLoS One. 2016:e0154387. doi: 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. Environ Health Perspect. 2014;122:769–774. doi: 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappaport SM. Biomarkers. 2012;17:483–489. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liebler DC. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubino FM, Pitton M, Di Fabio D, Colombi A. Mass Spectrom Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 6.Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M. Chem Res Toxicol. 2008;21:824–835. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- 7.Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- 8.Nagumo K, Tanaka M, Chuang VT, Setoyama H, Watanabe H, Yamada N, Kubota K, Tanaka M, Matsushita K, Yoshida A, Jinnouchi H, Anraku M, Kadowaki D, Ishima Y, Sasaki Y, Otagiri M, Maruyama T. PLoS One. 2014;9:e85216. doi: 10.1371/journal.pone.0085216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, Rappaport SM. Mol Cell Proteomics. 2011;10:M110 004606. doi: 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlsson H, von Stedingk H, Nilsson U, Tornqvist M. Chem Res Toxicol. 2014;27:2062–2070. doi: 10.1021/tx5002749. [DOI] [PubMed] [Google Scholar]
- 11.Grigoryan H, Li H, Iavarone AT, Williams ER, Rappaport SM. Chem Res Toxicol. 2012;25:1633–1642. doi: 10.1021/tx300096a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirihara M, Asai Y, Ogawa S, Noguchi T, Hatano A, Hirai Y. Synthesis. 2007;21:3286–3289. [Google Scholar]
- 13.Lin YS, McKelvey W, Waidyanatha S, Rappaport SM. Biomarkers. 2006;11:14–27. doi: 10.1080/13547500500382975. [DOI] [PubMed] [Google Scholar]
- 14.Cai X, Perttula K, Pajouh SK, Hubbard AE, Nomura DK, Rappaport SM. Metabolomics: Open Access. 2014;4:131–139. [Google Scholar]
- 15.Keller BO, Sui J, Young AB, Whittal RM. Anal Chim Acta. 2008;627:71–81. doi: 10.1016/j.aca.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Liu Q, Zimmerman LJ, Ham AJ, Slebos RJ, Rahman J, Kikuchi T, Massion PP, Carbone DP, Billheimer D, Liebler DC. Mol Cell Proteomics. 2011;10:M110 006593. doi: 10.1074/mcp.M110.006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn EJ, Hughes WL, Jr, Weare JH. J Am Chem Soc. 1947;69:1753–1761. doi: 10.1021/ja01199a051. [DOI] [PubMed] [Google Scholar]
- 18.Liu P, O’Mara BW, Warrack BM, Wu W, Huang Y, Zhang Y, Zhao R, Lin M, Ackerman MS, Hocknell PK, Chen G, Tao L, Rieble S, Wang J, Wang-Iverson DB, Tymiak AA, Grace MJ, Russell RJ. J Am Soc Mass Spectrom. 2010;21:837–844. doi: 10.1016/j.jasms.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Rejtar T, Zhou ZS, Karger BL. Rapid Commun Mass Spectrom. 2010;24:267–275. doi: 10.1002/rcm.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olszowy PP, Burns A, Ciborowski PS. Anal Biochem. 2013;438:67–72. doi: 10.1016/j.ab.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, Zhu G, Li Y, Yang P, Dovichi NJ. Anal Chem. 2012;84:8715–8721. doi: 10.1021/ac3019608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fennell TR, MacNeela JP, Morris RW, Watson M, Thompson CL, Bell DA. Cancer Epidemiol Biomarkers Prev. 2000;9:705–712. [PubMed] [Google Scholar]
- 23.Fasco MJ, CR, Stack RF, O’Hehir C, Barr JR, Eadon GA. Chem Res Toxicol. 2007;20:677–684. doi: 10.1021/tx6003425. [DOI] [PubMed] [Google Scholar]
- 24.Sagone AL, Jr, Lawrence T, Balcerzak SP. Blood. 1973;41:845–851. [PubMed] [Google Scholar]
- 25.Go YM, Jones DP. J Biol Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht SS. Mutat Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 27.Auclair JR, Salisbury JP, Johnson JL, Petsko GA, Ringe D, Bosco DA, Agar NY, Santagata S, Durham HD, Agar JN. Proteomics. 2014;14:1152–1157. doi: 10.1002/pmic.201300450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Q, Yan X, Kilpatrick LE, Liang Y, Mirokhin YA, Roth JS, Rudnick PA, Stein SE. Mol Cell Proteomics. 2014;13:2435–2449. doi: 10.1074/mcp.O113.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piehowski PD, Petyuk VA, Orton DJ, Xie F, Moore RJ, Ramirez-Restrepo M, Engel A, Lieberman AP, Albin RL, Camp DG, Smith RD, Myers AJ. J Proteome Res. 2013;12:2128–2137. doi: 10.1021/pr301146m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boys BL, Kuprowski MC, Noel JJ, Konermann L. Anal Chem. 2009;81:4027–4034. doi: 10.1021/ac900243p. [DOI] [PubMed] [Google Scholar]
- 31.Mir SA, Bhat AS, Ahangar AA. International J PharmTech Research. 2014;6:759–768. [Google Scholar]
- 32.Karas M, Bahr U, Dulcks T. Fresenius’ journal of analytical chemistry. 2000;366:669–676. doi: 10.1007/s002160051561. [DOI] [PubMed] [Google Scholar]
- 33.Wilm M, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 34.Talhout R, Schulz T, Florek E, van Benthem J, Wester P, Opperhuizen A. Int J Environ Res Public Health. 2011;8:613–628. doi: 10.3390/ijerph8020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.