Abstract
In most free-living eukaryotes studied thus far, heme is synthesized from a series of intermediates through a well defined evolutionarily conserved pathway. We found that free-living worms, including the model genetic organism Caenorhabditis elegans, and parasitic helminths are unable to synthesize heme de novo, even though these animals contain hemoproteins that function in key biological processes. Radioisotope, fluorescence labeling, and heme analog studies suggest that C. elegans acquires heme from exogenous sources. Iron-deprived worms were unable to grow in the presence of adequate heme unless rescued by increasing heme levels in the growth medium. These data indicate that although worms use dietary heme for incorporation into hemoproteins, ingested heme is also used as an iron source when iron is limiting. Our results provide a biochemical basis for the dependence of worm growth and development on heme, and they suggest that pharmacologic targeting of heme transport pathways in worms could be an important control measure for helminthic infections.
Keywords: Caenorhabditis elegans, iron, metals, nematode, prophyrin
Helminthic infections are an enormous burden to public health and global agriculture. More than two billion people are affected by helminthiasis, including schistosomiasis, and plant-parasitic nematodes cause an estimated annual crop loss of eighty billion dollars (1, 2). There is an urgent need to find unique vulnerabilities in helminths because drug resistance by nematodes is already prevalent in livestock and other animals, and schistosomes resistant to praziquantel have been documented in places where this antihelminthic drug is copiously used (3, 4). Within a parasitized host, helminths exhibit distinct nutritional adaptations such that they acquire their food unidirectionally from the host to sustain their growth and reproduction (5). Thus, metabolic pathways essential for nutrient acquisition in worms could be exploited as potential drug targets to control helminthic infections.
Phylogenetic analysis of biosynthetic enzymes in the evolutionarily conserved multistep pathway for heme synthesis, δ-aminolevulinic acid dehydratases (ALAD) and porphobilinogen deaminases (PBGD), has suggested that Caenorhabditis elegans lacks orthologs for these enzymes and therefore may acquire tetrapyrroles nutritionally (6). Correspondingly, the trypanosomatid protozoa, Leishmania spp., appear to lack seven of the eight enzymes of the heme pathway with the exception being ferrochelatase (7). This defect in tetrapyrrole synthesis is manifested as a nutritional requirement for heme or its immediate precursor protoporphyrin IX. Early studies demonstrated that C. elegans, Caenorhabditis briggsae, and Rhabditis maupasi require cytochrome c or hemoglobin as a heme source for growth and reproduction (8–10). However, it is unclear why these nematodes require heme to grow and whether this nutritional necessity also exists in related helminths.
Hemes are critical cofactors for many biological processes, including oxidative metabolism (11), xenobiotic detoxification (12), the synthesis and sensing of diatomic gases (13), cellular differentiation (14), gene regulation at the level of transcription (15), protein translation (16) and targeting (17), and maintaining protein stability (18). Within cells, protoheme (iron-protoporphyrin IX) is synthesized via well defined intermediates that are highly conserved through evolution. Depending on the organelle and cell type, heme pathway intermediates are used in the synthesis of other tetrapyrrole compounds, including bilins, chlorophylls, and corrins (19, 20). The first universal precursor for the synthesis of heme is δ-aminolevulinic acid. Heme synthesis culminates when ferrochelatase catalyzes insertion of ferrous iron into the protoporphyrin IX ring to form protoheme (21). Protoheme is incorporated into numerous heme proteins or is modified further for the synthesis of other types of heme found in cytochrome c and terminal oxidases.
An analysis of worm genomes showed that worms lack orthologs to genes encoding enzymes for heme biosynthesis. Here we demonstrate that C. elegans, a free-living multicellular eukaryote, and medically relevant helminths are natural heme auxotrophs and suggest that C. elegans may serve as a model system for understanding the mechanisms of heme transport in eukaryotes.
Materials and Methods
Worm Culture and Growth Assays. Free-living worms were cultured in CeHR axenic liquid medium (Eric Clegg, personal communication; composition and preparation available on request). Worm growth (generation time 3.5 days), mobility, and development in CeHR medium were comparable to those of worms cultured on Escherichia coli in agar plates. We modified CeHR medium (henceforth called mCeHR medium) to eliminate all sources of exogenously added heme; basal growth medium comprised 20 μM hemin chloride (Frontier Scientific, Logan, UT) and 150 μM ferrous ammonium sulfate (Sigma). Worm strains were grown in mCeHR medium under aerobic conditions in tissue culture flasks at 20°C. We routinely obtained ≈3.7 × 106 worms after 3.5–4 days in a T75 flask with 30 ml of basal medium from an initial inoculum of ≈1.5 × 105 worms. For initial culturing of worms in axenic media, three generations of worms were sequentially bleached [1.1% Clorox bleach (sodium hypochlorite)/0.55 M NaOH] to eliminate any contaminating bacterial carryover from agar plates. In all dose–response experiments, worms were growth synchronized by treating the gravid hermaphrodite worms with bleach to dissolve adult worms. The eggs, resistant to bleach, were liberated from the carcasses and extensively washed with M9 buffer followed by overnight hatching in M9 buffer to synchronize growth. By harvesting at appropriate time intervals, synchronous larval stages and adult staged worms were collected for experimental manipulations. Identical numbers of synchronized L1 larvae were inoculated into media with different heme concentrations in 12- or 24-well culture plates. Worm growth was monitored each day and an aliquot was obtained for counting by microscopy, usually at days 3, 6, and 9. The worms in each well were counted twice and each growth condition was analyzed in triplicate. P values for statistical significance were calculated by using a one-way ANOVA with Student–Newman–Keuls multiple comparison test by using instat version 3.01 (GraphPad, San Diego).
Preparation of Hemin, Hemin Analogs, and [59Fe]Heme. Fresh stock solutions of hemin or hemin analogs (Frontier Scientific) were prepared immediately before use by dissolving required amounts in 0.3 M ammonium hydroxide. The pH of the stock solution was adjusted to 8.0 with 6 M HCl, and the solution was filter sterilized (pore diameter 0.45 μm). The upper limit of free iron was estimated to be 3.8 nM/1 μM hemin chloride by inductively coupled plasma–mass spectrometry (ICP-MS) analysis. For synthesis of [59Fe]heme, 50 ml of glacial acetic acid was stirred under a constant flow of N2 at 60°C followed by addition of 12 mg of protoporphyrin IX in pyridine for 30 min (22). To this mixture, 0.85 μCi of FeCl3 (specific activity 35.77 mCi/mg, Perkin–Elmer; 1 μCi = 37 kBq) was stirred in for an additional 3 h. Heme was extracted from this mixture with ethyl acetate followed by extensive washes with 4 M HCl and distilled water to remove unincorporated protoporphyrin IX and iron. The heme thus obtained was concentrated by evaporation of the ethyl acetate by using a RotaVapor and frozen at –20°C until further use. Total amount of [59Fe]heme synthesized by this method was measured by using a Packard Gamma Counter (≈21% efficiency). The purity of heme was determined by thin-layer chromatography using silica gel 60 matrix in an NH4OH-saturated chamber with 2,6-lutidine/water solvent. We estimated the specific activity of [59Fe]heme synthesized to be ≈2.8 × 106 dpm/nmol.
Metabolic Labeling, Heme Isolation, and TLC. Equal numbers of L1 worms were grown at 20°C in mCeHR medium with 20 μM hemin and supplemented with 9.4 × 106 dpm of either [59Fe]heme or 59FeCl3. Radiolabeled adult worms were harvested by first incubating them in M9 buffer for 30 min to empty the gut contents. Worms were then extensively washed with large amounts of M9 buffer/1 mM EDTA on a Gelman Metricel membrane (pore diameter 0.45 μm) that had been incubated with 20 μM hemin or FeCl3 to prevent nonspecific absorption of the radiotracer signal to the filter membrane. The experiment was done in parallel in the presence of 1 mM sodium azide (NaN3) to account for nonspecific binding of the radiolabel to the biological samples. We experimentally determined that this concentration of NaN3 was not lethal to worms. To isolate heme, worms were washed with M9 buffer and resuspended in ice-cold 1 M HCl to a final pH of 2.0. The acidified solutions were incubated on ice for 30 min to allow complete protein denaturation, and then an equal volume of ice-cold 2-butanone was added (23). The solutions were shaken and allowed to stand until the ketone (heme) and aqueous (worm debris) phases separated. The upper ketonic phase was removed and the heme was concentrated by rotary evaporation. The heme was resuspended in dimethylformamide, and equal volumes of samples and controls were spotted and resolved on silica gel 60 TLC plates. Plates were exposed to a PhosphorImager and the radiolabel signal corresponding to heme was excised and analyzed by using a γ counter. Counts (dpm) obtained were normalized for protein as determined by the bicinchoninic acid method (Sigma) performed on aliquot samples of intact worms. The specific activity of [59Fe]heme added to the growth medium was 4.7 × 104 dpm/nmol, and the specific activity of [59Fe]heme extracted from worms was 0.69 × 104 dpm/nmol. The most plausible explanation for this difference in specific activity of [59Fe]heme is dilution of the supplied radiolabel heme with preexisting unlabeled heme endogenous to the worm.
Pulse–Chase Analysis. Mixed populations of worms grown in mCeHR medium with 4 μM hemin were labeled for 16 h in the presence of either 40 μM zinc mesoporphyrin (ZnMP) or hemin. We empirically determined that 40 μM ZnMP labels the worms without affecting viability. Fluorescently labeled worms were washed with M9 buffer, dispensed into 12-well plates, and allowed to incorporate unlabeled hemin at concentrations of 40, 400, or 800 μM. At timed intervals, aliquots of worms were removed into the appropriate medium containing 10 mM NaN3, mounted on a slide, and immediately analyzed with a 543 He/Ne laser on a Zeiss 410 confocal microscope, or with a Peltier-cooled Retiga 1300 12-bit charge-coupled device camera fitted on to a Leica DMIRE2 epifluorescence/differential interference contrast microscope. Images were further analyzed with simplepci version 5.2 software (Compix, Cranberry Township, PA). Sensor gain and exposure times were kept constant during all image acquisition. No loss of fluorescence was observed when labeling experiments were performed in parallel in the presence of 1 mM NaN3 during the chase period and when worms were incubated in medium without hemin. To account for background autofluorescence in C. elegans, the sensor gain of the charge-coupled device camera or the laser was set to subtract any fluorescence obtained from control worms incubated in the presence of 40 μM hemin.
Other Methods. Other methods are described in Supporting Methods, which is published as supporting information on the PNAS web site.
Results
Analysis of publicly available worm genome databases revealed that these genomes lack obvious orthologs to heme biosynthesis pathway enzymes (6). We queried genome databases by using sequences of the human enzymes that catalyze the seven sequential steps to synthesize heme from the first universal precursor, δ-aminolevulinic acid. Expect (E) threshold values obtained from blast searches revealed nonsignificant hits only in the C. elegans database (Table 2, which is published as supporting information on the PNAS web site) as compared with genome databases from Saccharomyces cerevisiae, Drosophila melanogaster, and mouse, thus suggesting that C. elegans lacks orthologous genes for enzymes that catalyze heme synthesis (24, 25). These in silico observations were confirmed by measuring enzyme activity. Because free-living worms in the laboratory use E. coli as food, bacterial metabolites could confound identification of the source of heme and enzyme activity. Therefore, we grew worms axenically in mCeHR liquid medium in lieu of growth on Petri plates containing E. coli (see Materials and Methods). We used three physiologically distinct but phylogenetically related free-living bacteriovorous strains, C. elegans, Panagrellus redivivus, and Oscheius myriophila (Fig. 4, which is published as supporting information on the PNAS web site). Synchronized worms were grown aerobically at 20°C in mCeHR to the gravid adult stage and homogenized to obtain cytosol- and mitochondria-enriched fractions for analysis of heme biosynthesis enzymes. Under the assay conditions used, δ-aminolevulinic acid dehydratase and porphobilinogen deaminase activities were undetectable in worm lysates as compared with wild-type E. coli lysates (Table 1).
Table 1. Heme biosynthetic enzyme activities in free-living and parasitic worms.
| Enzyme activity, nmol/min per mg of protein*
|
|||||
|---|---|---|---|---|---|
| Organism | Host | ALAD | PBGD | FC | SDH |
| Helminths | |||||
| Caenorhabditis elegans† | Free | ND | ND | ND | 725 ± 160‡ |
| Panagrellus redivivus† | Free | ND | ND | ND | 3,419.3 ± 424.7‡ |
| Oscheius myriophila† | Free | ND | ND | ND | 1,567 ± 594.6‡ |
| Paragordius varius | Free§ | ND | ND | — | 715.9 ± 9.2 |
| Schistosoma mansoni | Human | ND | ND | ND | 515 ± 80.6 |
| Strongyloides stercoralis | Human | ND | ND | ND | 715.2 ± 53.8‡ |
| Ancylostoma caninum | Dog | ND | ND | ND | 266.7 ± 45.6‡ |
| Haemonchus contortus | Sheep/goat | ND | ND | ND | 30.3 ± 4.6‡ |
| Trichuris suis | Pig | ND | ND | ND | 7.6 ± 0.8‡ |
| Ascaris suum | Pig | ND | ND | ND | 24.2 ± 3.2‡ |
| Bacteria | |||||
| E. coli | NA | 1.47 ± 0.03 | 0.035 ± 0 | 16.36 ± 2.71 | 1,865 ± 318 |
| E. coli (ALAD mutant) | NA | 0.034 ± 0 | — | — | — |
| Yeast | |||||
| S. cerevisiae | NA | 0.97 ± 0.04 | 0.096 ± 0 | 27.42 ± 2.92‡ | 2,371.2 ± 32.1‡ |
| S. cerevisiae (FC mutant) | NA | 0.04 ± 0 | 0.032 ± 0 | ND | 915 ± 177‡ |
Wild-type and mutant E. coli and S. cerevisiae were used as positive controls for enzyme activity. ND, enzyme activity not detected under current assay conditions; NA, not applicable; —, not assayed; ALAD, δ-aminolevulinic acid dehydratase; PBGD, porphobilinogen deaminase; FC, ferrochelatase; SDH, succinate dehydrogenase.
Mean values (triplicates) of product formed ± SD.
Grown in axenic mCeHR medium.
Average activity found in combined crude mitochondria- and cytosol-enriched fractions.
P. varius adults live in fresh water but larvae develop in arthropods.
The protozoan Leishmania and certain microorganisms such as Haemophilus influenzae contain only part of the heme biosynthetic pathway (7, 19). Thus, it is possible that worms also have retained the ability to synthesize heme by using an intermediate of the heme pathway. To directly address this issue, we measured ferrochelatase activity, an inner mitochondrial membrane enzyme, which catalyzes the final step in the heme biosynthetic pathway. Because ferrochelatase from S. cerevisiae has been genetically and biochemically well characterized, we used yeast as a control source of this enzyme (21). Ferrochelatase activity was readily detected in wild-type yeast but was undetectable in combined mitochondria- and cytosol-enriched fractions from all three worm species and a S. cerevisiae ferrochelatase mutant with genetic disruption at the HEM15 locus (Table 1). Activity for succinate dehydrogenase, another inner mitochondrial membrane enzyme, was readily detectable in these fractions, indicating the presence of mitochondrial membranes. However, the inability to detect heme synthesis enzyme activities could be attributed to the presence of endogenous inhibitors in worm extracts. We found no inhibition of δ-aminolevulinic acid dehydratase, porphobilinogen deaminase, or ferrochelatase enzyme activities when C. elegans extracts were mixed in equal proportions with extracts from either E. coli or S. cerevisiae (Table 3, which is published as supporting information on the PNAS web site).
To address whether the lack of discernable heme enzyme activities also held for other worm species that are phylogenetically diverse, we examined five parasitic nematodes that have different host specificities (Strongyloides stercoralis, Ancylostoma caninum, Haemonchus contortus, Trichuris suis, and Ascaris suum), a nematomorph (Paragordius varius), and a trematode flatworm (Schistosoma mansoni) (Fig. 4). Irrespective of their host affiliations, enzyme activities were undetectable in all helminthic extracts in our assay conditions (Table 1). These measurements provide further support for the hypothesis that the helminths examined in this study do not have enzymes for heme synthesis.
C. elegans appears to have a large number of hemoprotein orthologs, including globin isoforms (26), guanylate cyclases (27), adenylate cyclases (28), catalases (29), cytochrome P450s (30), and respiratory cytochromes (31). Although hemes have been found in all phyla, certain microbial pathogens such as Borrelia burgdorferi and Treponema pallidum neither make heme nor contain hemoproteins (19, 32). Reduced minus oxidized absorption spectra of pyridine hemochromes revealed that C. elegans has discernable hemoproteins in worm fractions enriched either for cytosol or for membranes, including mitochondrial membranes (Fig. 1A). The purity of these fractions was determined by immunoblots probed with antisera for α-tubulin and ATP2, the β-subunit of the F1 sector of the mitochondrial F1Fo ATP synthase (33). With ATP2 used as a marker, <1% of the signal was found in the cytosol-enriched fraction, whereas ≈88% of ATP2 protein was found in the membrane-enriched fraction. Ultralow-temperature spectra of total worm homogenates revealed the presence of detectable amounts of respiratory cytochromes a, b, and c (Fig. 1B).
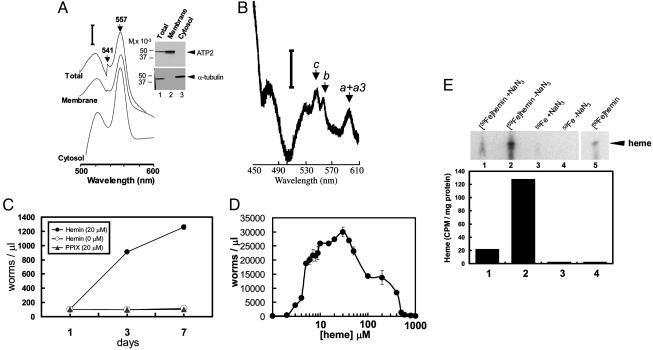
Fig. 1.
Heme auxotrophy of worms. (A) Dithionite-reduced minus ferricyanide-oxidized absorption spectra of pyridine hemochromes from total homogenate and membrane- and cytosolic-enriched fractions of C. elegans grown in axenic mCeHR medium supplemented with 20 μM hemin chloride. A peak at 557 nm and a trough at 541 nm indicate pyridine protohemochrome. All samples were reduced with 5 mM sodium dithionite or oxidized with 1 mM potassium ferricyanide. The vertical bar represents a ΔA of 0.005 for total homogenate, 0.012 for membrane fraction, and 0.02 for cytosolic fraction. (Inset) Immunoblot of the same samples (50 μg) that were separated by SDS/4–20% PAGE and probed with ATP2p antisera followed by chemiluminescent detection. This immunoblot was stripped to remove ATP2p antibodies and reprobed with α-tubulin antibody. (B) Ultralow-temperature spectrum of whole homogenate from C. elegans grown in mCeHR medium supplemented with 20 μM hemin. Only α bands are indicated for cytochromes c and b and cytochrome oxidase (a+a3). The vertical bar represents a ΔA of 1.0. (C) Aerobic growth of C. elegans in mCeHR medium supplemented with 0 or 20 μM hemin chloride or 20 μM protoporphyrin IX (disodium salt). Equal numbers of synchronized L1 larvae were used as primary inoculum in 24-well plates in triplicate and the cultures were analyzed quantitatively for growth at days 1, 3, and 7. (D) Biphasic response of C. elegans cultured in the presence of increasing amounts of hemin chloride (μM). Equal numbers of synchronized L1 larvae were grown in 24-well plates in mCeHR medium for 9 days and quantified (worms perμl) by microscopy. Each data point represents the mean ± SD from three separate experiments performed in triplicate. (E) Metabolic labeling in C. elegans cultured in the presence of heme. Synchronized L1 larvae were grown in mCeHR medium containing either 59FeCl3 or [59Fe]heme (9.4 × 106 dpm), and the worms were harvested as gravid adults. Heme was extracted and concentrated, and then resolved by TLC followed by detection with a PhosphorImager (Upper). Lane 5, [59Fe]heme control. Radiolabeled bands were quantified in a γ counter and cpm was normalized to total protein (Lower). To correct for nonspecific binding of the radiolabeled Fe and heme, parallel experiments were conducted in the presence of 1 mM sodium azide (samples 1 and 3).
To quantitatively determine the heme requirement of worms, C. elegans was cultured in the presence or absence of nutritional heme supplements. Worms grown in the absence of exogenous heme (supplemented as hemin chloride) were growth arrested at the L4 stage, whereas worms grown in the presence of heme grew robustly and reproduced over multiple generations (Fig. 1C). Similar heme-dependent growth was also observed for P. redivivus and O. myriophila (data not shown). This lack of growth in the absence of heme was reversible, because replenishing heme to the heme-depleted media resulted in normal growth rates of the arrested larvae (data not shown). However, unlike Leishmania, which can exogenously acquire either hemin or its immediate precursor protoporphyrin IX for growth (7), C. elegans was unable to use protoporphyrin IX (supplied as free acid or disodium salt) as a substitute for hemin (Fig. 1C). Cytochrome c or hemoglobin also sustained worm growth, supporting previous studies that have shown heme and vitamin B12, another tetrapyrrole, as necessary factors for C. briggsae development (8, 9, 34). Our observations that worms can develop to the L4 stage in the absence of exogenous heme in the growth medium suggest that either there are trace amounts of heme in the medium or, more plausibly, maternal heme stored in the egg during embryogenesis is able to sustain early larval growth.
Worms responded in a biphasic manner to heme when grown in the presence of various amounts of hemin (Fig. 1D). Although the optimal concentration of hemin for growth was 20 μM, worms continued to grow and reproduce at concentrations ranging from 1.5 to 500 μM hemin, albeit with significantly longer generation times and smaller brood size. Large amounts of hemin (≥800 μM) resulted in growth arrest at the L3 stage, possibly because hemes are cytotoxic because of peroxidase activity. To determine if C. elegans acquires heme directly from the growth medium, worms were metabolically labeled in the presence of equivalent amounts of 59Fe or [59Fe]heme, and worm homogenates were analyzed for the respective radiotracer. A specific signal was obtained in samples containing worms labeled with [59Fe]heme (Fig. 1E, lane 2). Thus, worms use [59Fe]heme directly from the growth medium to fulfill their heme auxotrophy and incorporate the tetrapyrrole into proteins. However, no radiolabeled signal was observed in heme extracted from worms when 59Fe by itself was used (Fig. 1E, lane 4). This observation corroborates our genomic and biochemical analysis (Tables 1 and 2) that C. elegans lack ferrochelatase, the terminal enzyme in heme biosynthesis. [59Fe]Heme uptake was negligible in worms that were metabolically inhibited in the presence of NaN3, an inhibitor of the mitochondrial respiratory chain.
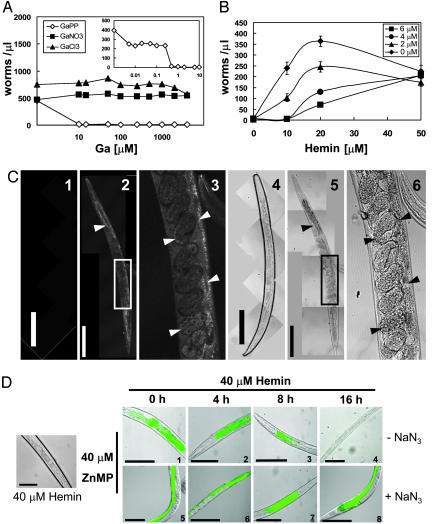
Studies with bacterial mutants that use heme and hemoglobin as an iron source have shown that non-iron metalloporphyrins act as heme analogs and gain entry into cells via high-affinity heme transport systems (35). After uptake, non-iron metalloporphyrins show various degrees of antibacterial activity, depending on their metal cofactor (35). To determine whether a heme uptake system exists in C. elegans, synchronized worms were grown in the presence of 4 μM hemin and various amounts of non-iron metalloporphyrins. We tested six different non-iron metalloporphyrins and found that gallium protoporphyrin IX (GaPP) was by far the most toxic heme analog. Compared with hemin, worms (P0) were growth retarded in the presence of 1 μM GaPP (800-fold sensitivity) (Fig. 2A), whereas F1 progeny, obtained from surviving P0 worms grown at lower concentrations of GaPP, were growth arrested at 0.25 μM GaPP (3,200-fold sensitivity). The ionic radii of Ga and Fe are very similar (0.62 versus 0.64 Å). Because Ga cannot undergo oxidation–reduction reactions like Fe, organisms that have heme uptake systems probably use and incorporate GaPP as a cofactor instead of heme, resulting in cytotoxicity (35). Neither gallium chloride nor gallium nitrate was able to mimic the toxicity of GaPP (even at concentrations >100-fold), suggesting that the antihelminthic activity of GaPP was not due to the adventitious release of Ga from GaPP (Fig. 2A). Notably, exposure of worms to GaPP resulted in developmental abnormalities that correlated in their severity to the levels of GaPP in the growth medium (Fig. 5, which is published as supporting information on the PNAS web site). These morphological phenotypes may reflect incremental inhibition of cellular pathways that are dependent on hemoproteins. Indeed, the toxic effect of GaPP was attenuated in the presence of increasing levels of hemin, indicating that GaPP likely exerts its antihelminthic effect via heme-dependent pathways (Fig. 2B).
Fig. 2.
Characterization of heme uptake in C. elegans. (A) Aerobic growth of C. elegans in mCeHR medium with 20 μM hemin supplemented with either gallium protoporphyrin IX (GaPP) or gallium salts. Synchronized L1 larvae were grown for 9 days in 24-well plates and quantified (worms per μl) by microscopy. Each data point represents the mean from a single experiment and each experiment was performed in triplicate. Inset depicts the GaPP analysis at lower concentrations for clarity. (B) Effect of heme on the cytotoxicity of GaPP. Synchronized L1 larvae were inoculated in 24-well plates containing mCeHR medium with 0, 2, 4, or 6 μM GaPP and increasing hemin (μM). The number of worms per μl was measured on day 9, and the data are presented as mean ± SD. (C) Fluorescent metabolic labeling of worms with either 40 μM hemin (images 1 and 4) or 40 μM ZnMP/4 μM hemin (images 2, 3, 5, and 6) for 3 h followed by confocal microscopy with a 546 laser (images 1–3) and differential interference contrast optics (images 4–6). Arrowheads indicate ZnMP fluorescence accumulation within intestinal cells and developing embryos. For clarity, the boxed portion of image 2 is magnified in images 3 and 6. (Bar, 100 μm.) (D) Worms were incubated with 40μM ZnMP/4μM hemin for 16 h followed by a chase with 40 μM hemin. Worms were analyzed by epifluorescence microscopy (TRITC channel) and differential interference contrast optics. Experiments were performed in the absence (images 1–4) or presence (images 5–8) of NaN3 during the chase periods to test for the nonspecific loss of ZnMP fluorescence. Photomicrograph 4 is shown at a lower power to depict the complete loss of ZnMP fluorescence. (Bar, 100 μm.) For C and D, four separate experiments were performed with a minimum of 50 worms per data point per experiment. The data are representative for >90% of worms analyzed.
To further elucidate heme uptake at the cellular level, we used metabolic labeling in live worms with ZnMP. This fluorescent heme analog is not a substrate for heme catabolic enzymes such as heme oxygenases (HOs), and thus a fluorescent signal can be amplified over time as ZnMP accumulates in cells (36, 37). Live worm imaging revealed a time-dependent accumulation of ZnMP in worms; fluorescence was detected in worm intestinal cells within 10 min of treatment with 40 μM ZnMP. Confocal microscopy showed ZnMP accumulation in multiple cell types in the adult worm, including the intestinal cells, eggs, and dividing embryos (Fig. 2C). We found discernable fluorescence signal in embryos within 130 min of incubation with ZnMP. This time point is likely an overestimate because ZnMP fluorescence is weak and is detectable only after substantial signal has accumulated over time. To correlate the specificity of ZnMP fluorescence with heme transport, worms were first fluorescently labeled by feeding them 40 μM ZnMP. They were then washed to remove ZnMP and incubated with 40 μM hemin. The fluorescence intensity diminished over time and was undetectable by 16 h (Fig. 2D). This loss in fluorescence was specific, because the signal did not diminish when worms were treated in parallel with a nonlethal dose of sodium azide.
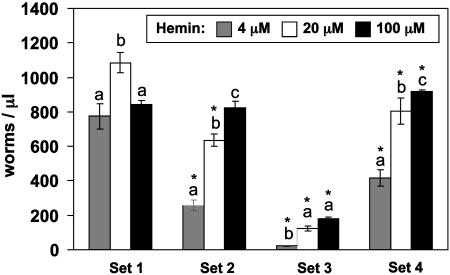
Bacteria and C. albicans that have high-affinity heme uptake systems use heme as an iron source when iron is limiting during infection within the host milieu (38, 39). HO degrades heme to release iron, biliverdin. and carbon monoxide; in some organisms, including mammals and cyanobacteria, biliverdin is subsequently converted to bilirubin by biliverdin reductase (40). Because the metabolism of heme and iron is interlinked, we determined how they are interrelated in worms. We grew C. elegans in either iron-deplete or -replete medium supplemented with low (4 μM), optimal (20 μM), or high hemin (100 μM). Worm growth was significantly slow in medium with 4 μM hemin but lacking exogenous iron (Fig. 3, sets 1 and 2). This diminution in growth was further accentuated in the presence of ferrozine, a membrane-impermeant iron chelator (Fig. 3, set 3). Conversely, iron supplementation reversed the effect of ferrozine on worm growth (Fig. 3, set 3 versus set 4), indicating that worms need both heme and iron to sustain growth and reproduction under nutrient-sufficient conditions. Importantly, 20 and 100 μM hemin significantly (P < 0.001) attenuated the growth-retarding effects of iron deprivation even in the presence of ferrozine (Fig. 3, set 3). In the absence of iron, hemin concentrations >100 μM in the growth medium did not result in any additional worm growth, plausibly because heme toxicity could mask the beneficial effects of heme as an iron source. This growth-promoting effect of hemin in the absence of iron was not due to trace amounts of free iron from hemin, because analysis of iron-dependent growth indicated that the amount of free iron in hemin (3.8 nM/μM) is insufficient to support worm growth (data not shown).
Fig. 3.
Worms use heme-iron under iron deprivation. Equal numbers of synchronized L1 larvae were grown in the presence of 4, 20, and 100 μM hemin, in basal mCeHR medium (set 1), or basal medium lacking exogenous iron (set 2), or as set 2 with 1 mM iron chelator ferrozine (set 3), or as set 3 with 486 μM ferrous ammonium sulfate (set 4). These values of ferrozine and iron were empirically determined by performing dose–response experiments and analyzing worm growth. The number of worms per μl was measured on day 9 and the data are presented as mean ± SD performed in triplicate. P < 0.001 between sets 1 and 3. Within each set, values with different letters are significantly different. * denotes significant differences with the corresponding heme concentrations in set 1.
Discussion
We have shown, using C. elegans as a model system, that worms are exceptional among known free-living eukaryotes because they lack the ability to synthesize heme. This conclusion is supported by genomic, biochemical, and nutritional analysis (Tables 1 and 2 and Fig. 1 C–E) and by previous studies which show that heme is a growth factor for C. elegans development (8, 9). This inability to make heme is surprising, given that other free-living metazoans make endogenous heme, and heme synthesis is catalyzed by enzymes encoded by at least seven separate genes that are not clustered in the genome. It is plausible that the ancestral worm lost the genes responsible for heme biosynthesis due to a lack of selective pressure because the progenitor had access to heme either from a parasitized host or from a symbiotic relationship with another organism. Recent studies have shown that the cattle tick Boophilus microplus, a bloodsucking arthropod, relies on blood meals to acquire heme (41), and pathogenic human filarial nematodes as well as certain insects harbor the bacteria Wolbachia, an intracellular symbiont that has a mutualistic relationship with its host such that the nematode acquires endobacterial-derived metabolites (42, 43). Indeed, the Wolbachia genome contains orthologs of genes for heme biosynthetic enzymes, suggesting that this endosymbiont has the ability to make heme.
The phylogenetic maximum parsimony tree (Fig. 4) shows that the loss of the heme pathway is common rather than exceptional among free-living and parasitic nematodes, and it can be found in higher taxa such as the Nematomorpha and Platyhelminthes, both of which have the potential for free-living and parasitic habits. Although the closest outgroup to nematodes is still controversial, if one accepts the Ecdysozoa theory, in which arthropods are a sister group to nematodes (44), the loss of the heme synthesis pathway would have occurred more frequently than with a more traditional grouping of helminths (45). A loss in the heme pathway would be consistent with a close phylogenetic relationship among helminths. Additional taxon sampling along with genomic and biochemical analyses may help clarify the disputed phylogenetic placement of helminths in the animal kingdom (46, 47). Evaluation of the complexity and phylogenetic importance of different molecular phylogenies versus the loss of the heme synthesis pathway cannot be made here. However, it is interesting to note that some other biochemical pathways also show distinct differences between classically simpler invertebrates (flatworms and nematodes) and higher invertebrates (arthropods and molluscs). These include an increased variety of different neuropeptides in higher invertebrates compared with lower (48) and the absence of a Toll-like pathway for immunity in nematodes that is present in insects (49, 50).
Our studies provide preliminary evidence that adaptation to heme auxotrophy in worms has enabled worms to use heme in its entirety as a prosthetic group under normal conditions and as an iron source when iron is low in the environment. We have been unable to identify any significant orthologs of HOs in the worm genome, although we found two putative orthologs of biliverdin reductase in the C. elegans and C. briggsae genomes. Because HOs from bacteria to humans are a diverse group of heme-catabolizing enzymes, it is possible that heme degradation in C. elegans is either catalyzed by a HO with low sequence homology to known HOs or by an entirely novel enzyme. Enzyme activities for HO and biliverdin reductase in homogenates from the trematode Schistosoma japonicum have been reported, thus raising the possibility that C. elegans may also have the ability to enzymatically degrade heme to obtain iron (51, 52).
Because worms lack the ability to make heme and therefore rely solely on exogenous heme for metabolic processes, these animals must have evolved specific mechanisms for intestinal heme absorption, trafficking, and sequestration. Perhaps, exceptional to worms, an intercellular heme transport system may be required to provide heme to other cell types beyond intestinal cells. Free heme is hydrophobic and is insoluble in the aqueous cellular milieu, and hemes have peroxidase activity that can damage biological macromolecules (53). Thus, in principle, intracellular pathways must exist for heme trafficking in eukaryotes. C. elegans may therefore serve as a unique animal model of an obligate heme auxotroph to genetically and cell-biologically delineate the pathways for heme homeostasis that have heretofore been elusive. Identification of molecules involved in heme homeostasis should permit selective drug-targeting of helminthic heme transport and heme-dependent cellular pathways that discriminate the parasite from its host and significantly reduce chronic morbidity and debilitation in affected individuals.
Supplementary Material
Acknowledgments
We thank Ray Fetterer, Ben Hanelt, Dolores Hill, Phil LoVerde, and Gerhard Schad for large quantities of hard-to-obtain worm samples for biochemical analysis; Jerry Bommer for ICP-MS analysis and expert technical advice on porphyrin chemistry; Eric Clegg for invaluable help with the CeHR medium; Carla Koehler for ATP2p antisera; W. Kelley Thomas for help with the SSU 18S rDNA gene sequencing strategy; Mark O'Brian and Jerry Kaplan for E. coli and yeast strains; the Caenorhabditis Genetics Center for worm strains; and Bruce Bowerman, Jonathan Gitlin, Dan Kosman, Mike Krause, Ian Mather, Mark O'Brian, and Tom Porter for helpful discussions. This work was supported by National Institutes of Health Grant DK68260 (to I.H.).
Author contributions: A.U.R., L.K.C., and I.H. designed research; A.U.R., L.K.C., E.L., and I.H. performed research; A.U.R., E.L., and I.H. contributed new reagents/analytic tools; A.U.R., L.K.C., E.L., and I.H. analyzed data; and I.H. wrote the paper.
Abbreviations: ZnMP, zinc mesoporphyrin; HO, heme oxygenase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY856093).
References
- 1.Colley, D. G., LoVerde, P. T. & Savioli, L. (2001) Science 293, 1437–1438. [DOI] [PubMed] [Google Scholar]
- 2.Chitwood, D. J. (2003) Pest Manage. Sci. 59, 748–753. [DOI] [PubMed] [Google Scholar]
- 3.Sangster, N. C. & Gill, J. (1999) Parasitol. Today 15, 141–146. [DOI] [PubMed] [Google Scholar]
- 4.Ismail, M., Botros, S., Metwally, A., William, S., Farghally, A., Tao, L. F., Day, T. A. & Bennett, J. L. (1999) Am. J. Trop. Med. Hyg. 60, 932–935. [DOI] [PubMed] [Google Scholar]
- 5.Halton, D. W. (1997) Int. J. Parasitol. 27, 693–704. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe, E. K. (2003) Chem. Biol. 10, 25–34. [DOI] [PubMed] [Google Scholar]
- 7.Sah, J. F., Ito, H., Kolli, B. K., Peterson, D. A., Sassa, S. & Chang, K. P. (2002) J. Biol. Chem. 277, 14902–14909. [DOI] [PubMed] [Google Scholar]
- 8.Hieb, W. F., Stokstad, E. L. & Rothstein, M. (1970) Science 168, 143–144. [DOI] [PubMed] [Google Scholar]
- 9.Vanfleteren, J. R. (1974) Nature 248, 255–257. [DOI] [PubMed] [Google Scholar]
- 10.Brockelman, C. R. & Jackson, G. J. (1978) J. Parasitol. 64, 803–809. [PubMed] [Google Scholar]
- 11.Wenger, R. H. (2000) J. Exp. Biol. 203, 1253–1263. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, D. F. (2004) Pharmacogenomics 5, 305–318. [DOI] [PubMed] [Google Scholar]
- 13.Rodgers, K. R. (1999) Curr. Opin. Chem. Biol. 3, 158–167. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima, O., Takahashi, S., Harigae, H., Furuyama, K., Hayashi, N., Sassa, S. & Yamamoto, M. (1999) EMBO J. 18, 6282–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun, J., Hoshino, H., Takaku, K., Nakajima, O., Muto, A., Suzuki, H., Tashiro, S., Takahashi, S., Shibahara, S., Alam, J., et al. (2002) EMBO J. 21, 5216–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J. J. & London, I. M. (1995) Trends Biochem. Sci. 20, 105–108. [DOI] [PubMed] [Google Scholar]
- 17.Lathrop, J. T. & Timko, M. P. (1993) Science 259, 522–525. [DOI] [PubMed] [Google Scholar]
- 18.Qi, Z., Hamza, I. & O'Brian, M. R. (1999) Proc. Natl. Acad. Sci. USA 96, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panek, H. & O`Brian, M. R. (2002) Microbiology 148, 2273–2282. [DOI] [PubMed] [Google Scholar]
- 20.Papenbrock, J. & Grimm, B. (2001) Planta 213, 667–681. [DOI] [PubMed] [Google Scholar]
- 21.Dailey, H. A. (2002) Biochem. Soc. Trans. 30, 590–595. [DOI] [PubMed] [Google Scholar]
- 22.Galbraith, R. A., Sassa, S. & Kappas, A. (1985) J. Biol. Chem. 260, 12198–121202. [PubMed] [Google Scholar]
- 23.Teale, F. W. (1959) Biochim. Biophys. Acta 35, 543. [DOI] [PubMed] [Google Scholar]
- 24.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 25.Altschul, S. F. & Gish, W. (1996) Methods Enzymol. 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 26.Kloek, A. P., McCarter, J. P., Setterquist, R. A., Schedl, T. & Goldberg, D. E. (1996) J. Mol. Evol. 43, 101–108. [DOI] [PubMed] [Google Scholar]
- 27.Riedl, C. A. & Sokolowski, M. B. (2004) Curr. Biol. 14, R657–8. [DOI] [PubMed] [Google Scholar]
- 28.Moorman, C. & Plasterk, R. H. (2002) Genetics 161, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petriv, O. I. & Rachubinski, R. A. (2004) J. Biol. Chem. 279, 19996–20001. [DOI] [PubMed] [Google Scholar]
- 30.Gotoh, O. (1998) Mol. Biol. Evol. 15, 1447–1459. [DOI] [PubMed] [Google Scholar]
- 31.Tsang, W. Y., Sayles, L. C., Grad, L. I., Pilgrim, D. B. & Lemire, B. D. (2001) J. Biol. Chem. 276, 32240–32246. [DOI] [PubMed] [Google Scholar]
- 32.Posey, J. E. & Gherardini, F. C. (2000) Science 288, 1651–1653. [DOI] [PubMed] [Google Scholar]
- 33.Grad, L. I. & Lemire, B. D. (2004) Hum. Mol. Genet. 13, 303–314. [DOI] [PubMed] [Google Scholar]
- 34.Lu, N. C., Hieb, W. F. & Stokstad, E. L. (1976) Proc. Soc. Exp. Biol. Med. 151, 701–706. [DOI] [PubMed] [Google Scholar]
- 35.Stojiljkovic, I., Kumar, V. & Srinivasan, N. (1999) Mol. Microbiol. 31, 429–442. [DOI] [PubMed] [Google Scholar]
- 36.Greenbaum, N. L. & Kappas, A. (1991) Photochem. Photobiol. 54, 183–192. [DOI] [PubMed] [Google Scholar]
- 37.Worthington, M. T., Cohn, S. M., Miller, S. K., Luo, R. Q. & Berg, C. L. (2001) Am. J. Physiol. 280, G1172–G1177. [DOI] [PubMed] [Google Scholar]
- 38.Stojiljkovic, I. & Perkins-Balding, D. (2002) DNA Cell Biol. 21, 281–295. [DOI] [PubMed] [Google Scholar]
- 39.Pendrak, M. L., Yan, S. S. & Roberts, D. D. (2004) Arch. Biochem. Biophys. 426, 148–156. [DOI] [PubMed] [Google Scholar]
- 40.Maines, M. D. (2000) Cell. Mol. Biol. (Noisy-le-grand) 46, 573–585. [PubMed] [Google Scholar]
- 41.Braz, G. R., Coelho, H. S., Masuda, H. & Oliveira, P. L. (1999) Curr. Biol. 9, 703–706. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, M. J. & Hoerauf, A. (1999) Parasitol. Today 15, 437–442. [DOI] [PubMed] [Google Scholar]
- 43.Henkle-Duhrsen, K., Eckelt, V. H., Wildenburg, G., Blaxter, M. & Walter, R. D. (1998) Mol. Biochem. Parasitol. 96, 69–81. [DOI] [PubMed] [Google Scholar]
- 44.Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387, 489–493. [DOI] [PubMed] [Google Scholar]
- 45.Brusca, R. C. & Brusca, G. J. (1990) Invertebrates (Sinauer, Sunderland, MA).
- 46.Blair, J. E., Ikeo, K., Gojobori, T. & Hedges, S. B. (2002) BMC Evol. Biol. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Copley, R. R., Aloy, P., Russell, R. B. & Telford, M. J. (2004) Evol. Dev. 6, 164–169. [DOI] [PubMed] [Google Scholar]
- 48.Masler, E. P. (2005) in Encyclopedia of Molecular Cell Biology and Molecular Medicine, ed. Meyers, R. A. (Wiley-VCH, Weinheim, Germany), Vol. 9, pp. 167–181. [Google Scholar]
- 49.Hodgkin, J. (2004) Nat Immunol. 5, 471–472. [DOI] [PubMed] [Google Scholar]
- 50.Fitzpatrick, M. J. & Sokolowski, M. B. (2004) Integr. Comp. Biol. 44, 28–36. [DOI] [PubMed] [Google Scholar]
- 51.Wen-qi, L. & Yong-long, L. (2004) Chin. J. Parasitol. Parasit. Dis. 22, 106–108. [PubMed] [Google Scholar]
- 52.Liu, W. Q., Li, Y. L. & Ruppel, A. (2001) Chin. J. Parasitol. Parasit. Dis. 19, 84–86. [PubMed] [Google Scholar]
- 53.Balla, G., Vercellotti, G. M., Muller-Eberhard, U., Eaton, J. & Jacob, H. S. (1991) Lab. Invest. 64, 648–655. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.