Abstract
Formaldehyde is a human carcinogen that readily binds to nucleophiles, including proteins and DNA. To investigate whether exogenous formaldehyde produces adducts in extracellular fluids, we characterized modifications to human serum albumin (HSA) following incubation of whole blood, plasma, and saliva with formaldehyde at concentrations of 1, 10 and 100 μM. The only HSA locus that showed the presence of formaldehyde modifications was Lys199. A N(6)-Lys adduct with added mass of 12 Da, representing a putative intramolecular crosslink, was detected in biological fluids that had been incubated with formaldehyde but not in control fluids. An adduct representing N(6)-Lys formylation was detected in all fluids, but levels did not increase above control values over the tested range of formaldehyde concentrations. An adduct representing N(6)-Lys199 acetylation was also measured in all samples. We then applied the assay to repeated samples of human plasma from 6 nonsmoking volunteer subjects (from Berkeley, CA), and single samples of serum from 15 workers exposed to airborne formaldehyde at about 1.5 ppm in a production facility and 15 control workers from Tianjin, China. Although all human plasma/serum samples contained basal levels of the products of N(6)-Lys formylation and acetylation, the putative crosslink product was not detected. Since the putative crosslink was observed in plasma incubated with formaldehyde at 1 μM, this suggests that the endogenous concentration of formaldehyde in serum was much lower than reported in the literature. Furthermore, concentrations of the formyl adduct were not higher in workers exposed to formaldehyde at about 1.5 ppm than in controls. Follow-up in vitro experiments with gaseous formaldehyde at 1.4 ppm detected the putative crosslink in plasma but not whole blood. This combination of results suggests that N(6) formylation occurs within cells with subsequent release of adducted HSA to the systemic circulation. Comparing across human samples, levels of N(6)-Lys199 formyl adducts were present at similar concentrations in subjects from California and China (about 1 mmol/mol HSA), but N(6)-Lys199 acetyl adducts were present at higher concentrations in Chinese subjects (0.34 vs. 0.13 mmol/mol HSA).
Keywords: adducts, formaldehyde, Lysine 199, blood, saliva, formylation, acetylation, covalent modification
1. Introduction
Formaldehyde is an economically important chemical used to manufacture plastics, resins and intermediates as well as to preserve tissues. The International Agency for Research on Cancer (IARC) regards formaldehyde as a human carcinogen based upon increased risks of respiratory-tract cancers and myeloid leukemias in exposed workers (IARC, 2012). The designation of formaldehyde as a respiratory carcinogen is consistent with this chemical’s strong electrophilicity, mutagenicity, and toxicity at the site of contact in biological systems. However, IARC’s classification of formaldehyde as a potential leukemogen is controversial because leukemia can involve effects to bone-marrow cells that are remote from sites of exogenous exposure (Heck and Casanova, 2004; Swenberg et al., 2013; Zhang et al., 2009). Given its high reactivity, rapid hydration in aqueous media (to form methanediol), and endogenous production via one-carbon metabolism, the mechanism by which inhaled formaldehyde would cause leukemia is a matter of scientific importance.
Endogenous production of formaldehyde presents particular difficulties in determining the potential for exogenous formaldehyde to cause systemic effects. If inhaled formaldehyde is to enter the systemic circulation, it must be absorbed in alveolar blood. As shown in Table 1, several authors have measured endogenous formaldehyde in human blood and cultured cells (Kato et al., 2001; Ke et al., 2014; Luo et al., 2001; Nagy et al., 2004), by applying a variety of analytical methods to small samples of each matrix. Reported formaldehyde concentrations were between 1 and 18 μM in cultured cells, between 4.7 and 22 μM in whole blood, and 37 μM in plasma. These endogenous concentrations are much larger than what would be anticipated from human inhalation of formaldehyde at concentrations of a few ppm, which are the highest levels that might be encountered in occupational settings (Tang et al., 2009). Assuming continuous exposure to formaldehyde at 1 ppm (1.23 mg/m3), a breathing rate of 1 m3/h, and a cardiac output of 294 l/h, the maximum blood concentration of formaldehyde (assuming complete absorption in the blood) would be 0.0042 mg/l = 0.14 μM. Such a small contribution of inhaled formaldehyde to the reported endogenous blood concentrations would be consistent with results from studies of rhesus monkeys and rats, where no increase was observed in blood concentrations following 6-h inhalation of formaldehyde at either 6 or 10 ppm (Casanova et al., 1988; Kleinnijenhuis et al., 2013).
Table 1.
Concentrations of endogenous formaldehyde reported in human blood and cultured human cells.
| Biological matrix | Method | Formaldehyde conc. (μM) | N | Notes | Reference |
|---|---|---|---|---|---|
| Whole blood | GC-MS | 87±4.7 | 6 | Mean±S.D. | (Heck et al., 1985) |
| Whole blood | LC-MS | 22 | 1 | (Nagy et al., 2004) | |
| Plasma | Derivatization + LC-fluorescence | 37 (29–45) | 2 | Median (Range) | (Luo et al., 2001) |
| Nasal epithelial cells | Derivatization + fluorescence | 3 | 2 | At LOD | (Neuss et al., 2010) |
| Cancer cells | SIFTCI-MS | 1.3 (0–4) | 6 | Median (Range) | (Kato et al., 2001) |
| HeLa cells | Derivatization + spectrophometry | 18 | 1 | (Ke et al., 2014) |
GC, gas chromatography; LC, liquid chromatography; LOD, limit of detection; MS, mass spectrometry; SIFTCI, selected-ion-flow-tube chemical ionization.
Because of its strong electrophilicity, formaldehyde readily binds to and forms crosslinks with glutathione and DNA (Lu et al., 2009; Shaham et al., 1996; Wang et al., 2009) and reacts with lysine residues to produce N(6)-formyllysine adducts with proteins in vitro and in vivo (Edrissi et al., 2013a; Edrissi et al., 2013b). Thus, another avenue by which inhaled formaldehyde could exert systemic effects would involve absorption by lung epithelial cells followed by release of modified glutathione, proteins and nucleic acids to the blood. In fact, immunoassays have detected putative antibodies against formaldehyde-HSA (human serum albumin) conjugates in the blood of exposed workers and smokers (Carraro et al., 1999; Ospina et al., 2011; Pala et al., 2008). Albumin is also abundant in respiratory fluids and saliva where reactions with inhaled formaldehyde would be expected (Aldini et al., 2008; Chung et al., 2013; Mellanen et al., 2001; Meurman et al., 2002; Mochca-Morales, 2000) with subsequent transfer to the blood.
Given the possibility that HSA adducts of blood and saliva could provide information regarding the disposition of inhaled formaldehyde after exposure, we characterized adducts of N(6)-Lys199 following in vitro reactions of formaldehyde with whole blood, plasma and saliva and also detected basal levels of N(6)-Lys199 acetylated adducts in the same specimens. We then measured these N(6)-Lys199 modifications in plasma or serum from volunteer subjects and from formaldehyde-exposed workers and control workers in Tianjin, China. (For this assay, plasma and serum specimens are essentially equivalent).
2. Materials and methods
2.1 Chemicals
Sodium borohydride (NaBH4), triethylammonium bicarbonate buffer (TEAB), porcine trypsin, HSA, ammonium acetate and acetoacetanilide were from Sigma-Aldrich (St. Louis, MO). Formalin (aqueous formaldehyde 37% by weight containing 10–15% methanol as stabilizer), methanol (LCMS grade), tris(2-carboxyethyl)phosphine (TCEP), formic acid (Optima®, LCMS Grade) and acetonitrile (Optima®, LCMS grade) were from Fisher Scientific (Pittsburgh, PA). Purified water (18.2 mΩ cm resistivity at 25°C) was prepared with a Milli-Q purification system (Millipore, Bedford, MA).
2.2 Human blood and saliva samples
Peripheral blood and saliva were collected with informed consent from groups of subjects according to human subject protocols at the participating institutions. Free formaldehyde was measured in blood that had been collected in heparin from a nonsmoking healthy Asian male. A portion of the blood specimen was immediately fractionated with Ficoll-Paque™ to obtain plasma, red blood cells (RBC), and peripheral blood mononuclear cells (PBMC). Blood for investigation of in vitro formaldehyde modifications to HSA was collected in EDTA from a nonsmoking healthy Caucasian male and immediately centrifuged at 2,000×g for 3 min to separate plasma from white blood cells and RBC. This subject also provided 1 ml of saliva that was immediately centrifuged at 18,000×g for 10 min using a filter (Amicon Ultra Centrifugal Filter, 0.5-ml capacity, 50 kDa MWCO, Millipore, Billerica, MA) to remove mucus and concentrate the proteins in about 20 μl.
Levels of HSA adducts from the general population were measured in plasma from 6 nonsmoking volunteer subjects in Berkeley, CA (3 males and 3 females), each of whom provided two or three peripheral blood specimens (in EDTA tubes) at intervals of three to four months. Blood samples were collected (after fasting) using EDTA tubes, which were inverted 8 times and centrifuged at 240×g for 10 min, to separate plasma from buffy coats and RBC. Subsequently, buffy coats were fractionated with Ficoll-Paque to obtain PBMC and granulocytes/RBC by centrifuging at 380×g for 40 min.
Blood samples were also obtained in 2012 from 15 formaldehyde-exposed (4 males and 11 females) and 15 unexposed workers (5 males and 10 females) in Tianjin, China. (All subjects were nonsmokers). Exposed subjects worked in a formaldehyde-production factory and control workers were employed in a nearby medical school and other local institutions in jobs that did not involve use of formaldehyde. Peripheral blood was collected without anticoagulant and was allowed to stand at room temperature for 30–60 min prior to centrifugation to separate serum from the clot. The serum was then aliquoted into vials and stored at −80 °C prior to analysis. Although individual exposure measurements were unavailable, formaldehyde exposure levels in the production factory were assessed with air samples collected annually at three locations representing the range of formaldehyde air concentrations in the workplace. Between 2012 and 2014 the estimated air concentrations ranged from a low value of 0.3 ppm to a high value of 10 ppm, which were at the upper end of reported exposure concentrations in Chinese factories (Tang et al., 2009). Based on all measurements, we estimate that workers in this factory were exposed to formaldehyde at the median air concentration of 1.5 ppm.
2.3 Time course of free formaldehyde in vitro
Whole blood, plasma, RBC (diluted 3:2 with PBS) and PBMC (7×106 cells/ml in PBS), were incubated with formaldehyde (prepared from formalin/PBS) at concentrations of 0, 25, 50 and 100 μM for 1, 2, 3, and 5 h at 37°C. Free formaldehyde was measured directly in plasma and – after centrifugation at 500×g for 2 min to remove cells – in supernatants of RBC, PBMC, and whole blood samples. The fluorimetric method of Li et al. (Li et al., 2007) was used by adding 20 μl of each blood fraction to a 96-well plate containing 100 μL of 4 M ammonium acetate, 40 μl of 0.2 M acetoacetanilide and 40 μl of ethanol. After incubation at 25 °C in the dark for 15 min, the relative fluorescence of the reagent blank and the sample solutions were measured at 470 nm with an excitation wavelength of 370 nm, using a FLx800 Fluorescence Microplate Reader (BioTek Instruments, Inc., Winooski, VT). Formaldehyde concentrations were calculated from a standard curve generated from serial dilutions of formaldehyde in PBS.
2.4 Adduct formation following incubation with formaldehyde solutions
Twenty microliters of whole blood, plasma or concentrated saliva (after spin filtering) was incubated for 3 h with formaldehyde solutions (prepared from formalin diluted with 10% aqueous methanol) at a concentration of 0, 1, 5, 10 or 100 μM in a final volume of 100 μl at 37 °C. After incubation, the blood was centrifuged at 2,000×g for 3 min to remove RBC from the plasma. To reduce Schiff bases resulting from modification by formaldehyde at 100 μM, NaBH4 was added to incubates at a final concentration of 0.5 M prior to digestion, as described by Aldini et al. (Aldini et al., 2006). Samples were stored at −80 °C prior to digestion. (Since reduced Schiff bases were not detected following incubation with 100 μM, this procedure was not repeated for samples incubated at lower concentrations.)
2.5 Adduct formation following incubation with gaseous formaldehyde
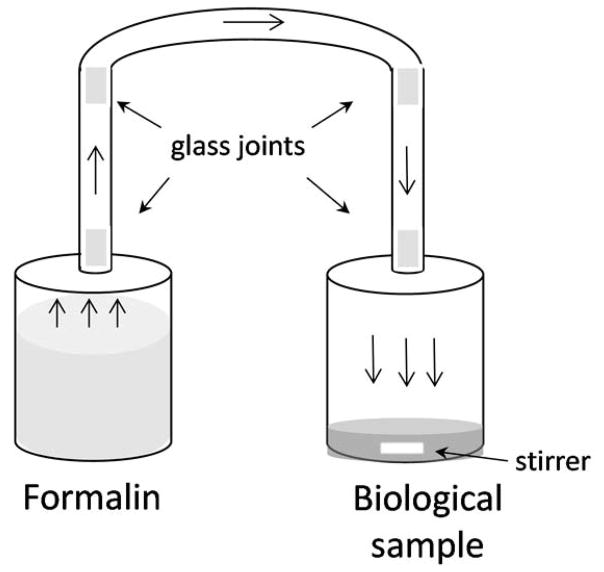
One hundred microliters of whole blood or plasma was incubated with gaseous formaldehyde for either 8 or 15 h using the apparatus illustrated in Figure 1. Basically, two glass 12-ml flasks were connected with all-glass unions to produce a static system with a total volume of 30 ml. One flask contained the biospecimen, which was stirred with a Teflon-coated bar, and the other flask contained 10 ml of a 5 mM solution of formaldehyde prepared from formalin. After adding the formalin solution and allowing the system to equilibrate overnight, the biospecimen was rapidly introduced into the other flask and the system was resealed for incubation at room temperature for either 8 or 15 h. The concentration of gaseous formaldehyde in the headspace was predicted from a relationship based on Betterton and Hoffmann (Betterton and Hoffmann, 1988), i.e. , where [CH2O]g is the concentration of formaldehyde gas in the headspace (ppm), [CH2(OH)2] is the concentration of the hydrated form of formaldehyde (methylene glycol) in solution, and H* is the intrinsic Henry’s Law constant for formaldehyde. Since the hydration constant for formaldehyde, representing the ratio of free formaldehyde to methylene glycol in solution, i.e. at 25 °C (Winkelman et al., 2002), the concentration of methylene glycol should have been essentially equal to the starting concentration of the formalin solution, i.e. [CH2(OH)2] ≅ 5 mM. Thus, given that H* = 3.44e03 μM/ppm at 25 °C (Allou et al., 2011), the headspace concentration of formaldehyde gas was predicted to be [CH2O]g = 1.4 ppm.
Figure 1.
Apparatus for incubating blood, plasma or saliva with gaseous formaldehyde, generated in the headspace above a 5 mM solution of formalin at room temperature. The estimated concentration of gaseous formaldehyde was 1.4 ppm and the total volume of the system was 30 ml.
2.6 Digestion of HSA with trypsin
Aliquots of 20 μl of plasma or concentrated saliva were diluted to 98 μl with 50 mM TEAB (pH 8.5) containing 5 mM TCEP and incubated for 15 min at 37° C in the dark to reduce protein disulfides. After adding 50 μg of trypsin in 2 μl (final volume 100 μl), samples were digested for 30 min at 37 °C using a pressurized system (Barocycler NEP2320, Pressure Biosciences Inc., South Easton, MA) programmed to generate 30 pressure cycles between ambient (15 s) and 1,380 bar (45 s). Trypsin was deactivated by adding formic acid to 1% and samples were stored at −80 °C prior to analysis.
2.7 Nanoflow liquid chromatography-mass spectrometry
Samples of digested blood fractions and saliva were diluted 1:100 and 1:10, respectively, into H2O/CH3CN/HCOOH (98/2/0.1; v/v/v) in clear glass autosampler vials (National Scientific, Rockwood, TN) and were analyzed using a Dionex UltiMate3000 nanoflow liquid chromatograph (nanoLC) that was connected in-line with an LTQ Orbitrap XL high resolution mass spectrometer, equipped with a Flex Ion nanoelectrospray ionization (nanoESI) source (Thermo Fisher Scientific, Waltham, MA). The nanoLC employed an Acclaim PepMap C8 column (0.75 μm×150 mm, 3-μm particle size, Thermo) and a 1-μl sample loop. The autosampler compartment was maintained at 4 °C and an injection volume of 1 μl was used at a flow rate of 300 nl/min. Solvent A was 99.9% water/0.1% formic acid and solvent B was 99.9% acetonitrile/0.1% formic acid (v/v). The elution program consisted of 2% B for 5 min, a linear gradient to 50% B over 50 min, 95% B for 5 min, and at 2% B for 6 min to reestablish initial conditions. Mass spectra were acquired in positive-ion mode, using the Orbitrap mass analyzer, with the following settings: spray voltage 2.1 kV, capillary temperature 200 °C, capillary voltage 35 V, scan range m/z = 250–1250, profile mode, automatic gain control (AGC) target 5×105 elementary charges, maximum inject time 500 ms, number of microscans set to 1, and mass resolving power 30,000 (at m/z = 400, measured at full width at half-maximum peak height). In the data-dependent mode, the six most intense ions exceeding an intensity threshold of 5×104 counts were selected from each full-scan mass spectrum for tandem mass spectrometry (MS/MS) analysis using collision-induced dissociation (CID). MS/MS spectra were acquired using the linear ion trap, in centroid mode, with the following settings: isolation width 3 m/z units, AGC target 1×104 elementary charges, maximum injection time 1000 ms, number of microscans set to 1, normalized collision energy 28%, and activation time 30 ms. To avoid the occurrence of redundant MS/MS measurements, real-time dynamic exclusion was enabled to preclude re-selection of previously analyzed precursor ions, with the following parameters: 2 repeat counts, repeat duration 30 s, exclusion list size 500, exclusion duration 90 s, and relative exclusion mass width ±20 ppm. Charge state screening and monoisotopic precursor ion selection were enabled to exclude singly charged ions and unassigned charge states from MS/MS analysis. The lock-mass option was enabled to provide real-time internal mass calibration during the analysis using 7 known background ions (Keller et al., 2008). Data acquisition was controlled using Xcalibur (version 2.0.7 SP1, Thermo) and Chromeleon Xpress (version 6.80, Thermo) software.
2.8 Peptide identification
Peptides were identified using turbo SEQUEST (Proteome Discoverer 1.3, Thermo) and MassMatrix (http://www.massmatrix.net) algorithms, searching against the primary sequence of HSA (Uniprot entry P02768). Trypsin was selected as the protease, allowing for a maximum of 2 missed cleavages. The mass tolerances for precursor and fragment ions were set to 10 ppm and 0.8 Da, respectively; all other parameters were set at default values. The variable modifications included in the list were covalent modifications of Arg, His, Lys and Cys to give hydroxymethyl (+30.0105 Da) or formyl derivatives (+27.9949 Da). Schiff bases (methylene imines, +12 Da) were predicted only for Arg and Lys residues. Isotopic distributions and MS/MS spectra of modified peptides were reviewed manually using the feature “peptide sequence fragmentation modeling” (Molecular Weight Calculator v.6.49, Matthew Monroe, www.alchemistmatt.com).
2.9 Relative quantitation of adducted HSA
Modification of HSA occurred at Lys199 and, therefore, was detected in the 8-mer peptide LK199CASLQK (#198–205) that resulted from a missed tryptic cleavage (Aldini et al., 2006). Adduct levels were normalized relative to a comparable 8-mer tryptic HSA peptide, i.e. AEFAEVSK (#226–233) to adjust each sample for the amount of HSA extracted and digested. Peak areas were calculated from the single ion chromatograms (SICs) extracted using the most abundant multiply charged ion for each peptide within a mass window of ±5 ppm (Figure S1 of Supplemental Information). Assuming comparable ionization efficiencies for LK199CASLQK adducts and AEFAEVSK, the relative adduct concentration of Lys(199) adducts was estimated as: . This method is analogous to that used by Grigoryan et al., who quantified HSA adducts, modified at Cys34, in tryptic digests relative to a ‘housekeeping peptide’, which was also derived from HSA. Using synthetic standards, the authors showed that the peak area ratios (Cys34 adduct/housekeeping peptide) were highly linear over a wide dynamic range of adduct concentrations (Grigoryan et al., 2016).
2.10 Statistical analyses
All pairs of assay duplicates were inspected for possible outliers. When a given pair had more than a 5-fold difference, each value was compared with the mean of all like observations and any value deviating by more than two standard deviations was excluded from analysis. Five exceptionally low values of formyl adducts in volunteer subjects (three in plasma and two in saliva) were excluded.
Statistical modeling and testing were performed with SAS for Windows (v. 9.3, SAS, Cary, NC). Differences in median adduct levels between populations were tested with Wilcoxon rank sums available in the NONPAR1WAY procedure. Random-effect models were fitted to the log-transformed adduct data from repeated blood samples of volunteer subjects using the MIXED procedure. Variance components were estimated by nesting multiple blood specimens within subjects and nesting duplicate assays within blood specimens. This resulted in three variance components, representing variation in (logged) adduct levels across subjects, within subjects and within assays (error term). The coefficient of variation representing technical replicates (CVe, in natural scale) was estimated as (Crow and Shimazu, 1988).
3. Results and discussion
3.1 Time course of free formaldehyde
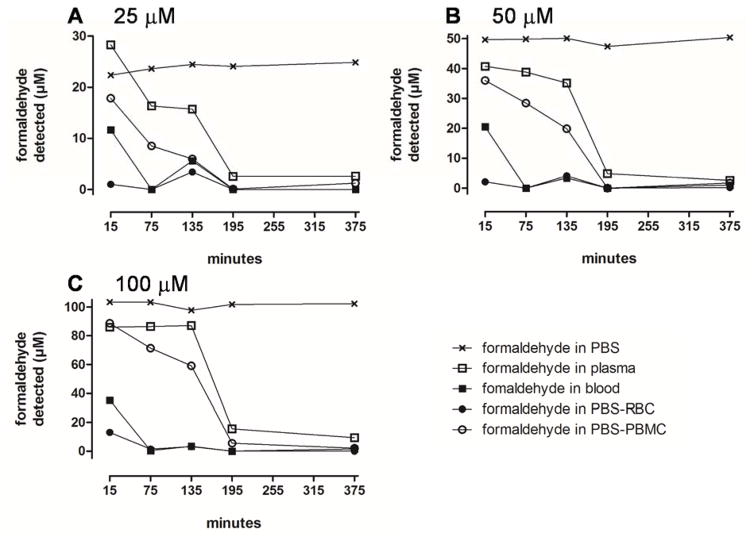
Figure 2 shows the time course of free formaldehyde in blood fractions incubated at concentrations between 25 and 100 μM. This concentration range was chosen to be consistent with reported concentrations of formaldehyde in blood and human cells (Table 1). Nevertheless, preliminary analysis of the blood used in our experiments detected no formaldehyde above the limit of detection for the fluorescence assay (approximately 3 μM). At the lowest tested concentration of 25 μM (Figure 2A), the curves suggest quasi-first-order (i.e. exponential) loss of formaldehyde in each blood fraction with rates decreasing in the order: RBC > whole blood > PBMC > plasma. As the initial formaldehyde concentration increased to 50 and 100 μM (Figures 2B and C), rates of reduction followed essentially the same order but displayed increasingly saturable kinetics. Notably, in RBC-containing fractions (RBC and whole blood) the rates of reduction in formaldehyde concentration were much greater than in RBC-free fractions (plasma and PBMC), consistent with previous findings (Malorny et al., 1965; Tran et al., 1972).
Figure 2.
Time course of free formaldehyde concentrations in different matrices incubated with formaldehyde at an initial concentration of 25 (A), 50 (B) and 100 μM (C). Matrices are indicated by the following symbols: × - PBS; □ - plasma; ■ - whole blood; ● - red blood cells; ○ - peripheral blood mononuclear cells.
3.2 Characterization of Lys199 modifications
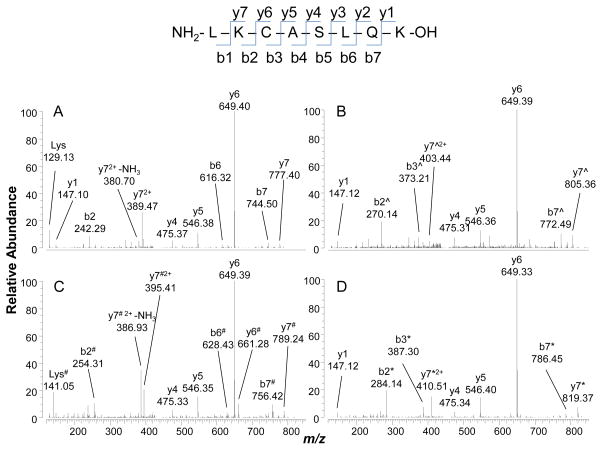
Shotgun proteomic analyses identified Lys199 as the only site of formaldehyde modifications in plasma, blood or saliva incubated with formaldehyde at or below 100 μM. As noted above, this residue is included in the peptide sequence LK199CASLQK originating from a missed cleavage during tryptic digestion. The unmodified peptide elutes at 6.4 min and is detected as a doubly charged precursor ion at m/z 445.7597, with a corresponding MS/MS spectrum characterized by a base peak at m/z 649.40 corresponding to y6+ ion (Figure 3A).
Figure 3.
Tandem mass (MS/MS) spectra of doubly charged precursor ions of the peptide LK199CASLQK. (A) Unmodified peptide, m/z 445.75971; (B) N(6)-formyl adduct, m/z 459.75657, with ^ indicating a nominal mass increase of 28; (C) Lys199-crosslink adduct, m/z 451.75966, with # indicating a nominal mass increase of 12; and (D) N(6)-acetyl adduct, m/z 466.76566, with * indicating a nominal mass increase of 42.
Three modifications of Lys199 were detected. The first, eluting at 12.6 min has a doubly charged precursor ions at m/z 459.7566 corresponding to the N(6)-formyl derivative (hereafter simply ‘formyl adduct’, added mass = 27.9949 Da). The high accuracy of Orbitrap mass measurements permitted unambiguous identification of this adduct, and excluded other possible structures such as double methylation (added mass 28.0313 Da). The MS/MS spectrum for this adduct (Figure 3B) displays prominent b2+ and y7+ ions, both of which show added masses of 28 Da consistent with Lys199 being the site of N(6)-formylation.
The second detected adduct was the N(6)-acetylated derivative of Lys199 (hereafter simply ‘acetyl adduct’, added mass = 42.0106 Da), represented by a doubly charged precursor ion at m/z 466.7657 eluting at 13.1 min. The MS/MS spectrum for this adduct (Figure 3D) also displays prominent b2+ and y72+ ions, both of which show added masses of 42 Da and, therefore, pinpoint Lys199 as the site of N(6)-acetylation.
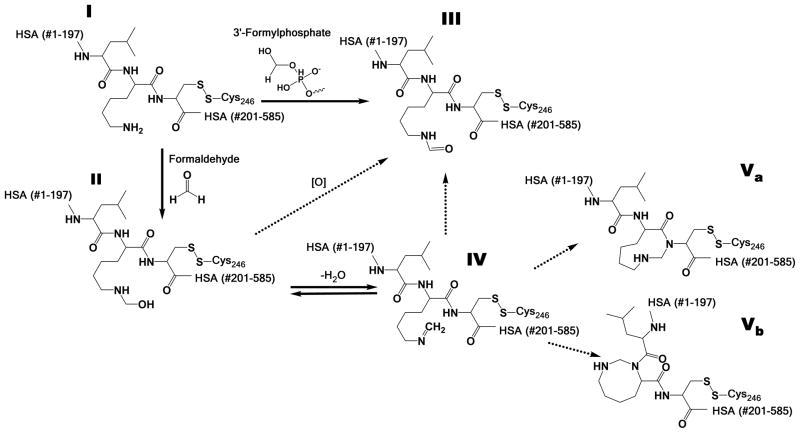
Since both formyl and acetyl adducts were found in all samples, with or without formaldehyde incubation, they appear to be normal constituents of human blood and saliva. It is not surprising that Lys199 is acetylated, since such modification has been reported following incubation of HSA with aspirin (Liyasova et al., 2010; Walker, 1976). Although N(6)-formylation of lysines has been reported in histones and other chromatin proteins from reactions involving both 3′-formylphosphate, a product of oxidative or nitrosative damage to DNA (Edrissi et al., 2013b; Jiang et al., 2007; Wisniewski et al., 2008) and endogenous formaldehyde (Edrissi et al., 2013b), this is apparently the first observation that Lys199 of HSA is formylated. We postulate in Scheme 1, which is similar to that of Edrissi et al. (Edrissi et al., 2013a) that the formyl adduct of Lys199 (structure III) arises from reaction of HSA (structure I) with formaldehyde, either via the hydroxymethyl intermediate (structure II) or the N(6)-methylene derivative (structure IV), as well as with 3′-formylphosphate derived from cellular processes.
Scheme 1.
Proposed scheme for N(6)-modifications to Lys199 of human serum albumin (HSA) (I). In the current study, formyl adduct (III) was observed in all human biospecimens. Putative cyclization of formaldehyde involving Cys200 (Va) and self-cyclization of Lys199 (Vb) are postulated from MS/MS spectra obtained from blood incubated with formaldehyde.
The third adduct was only observed in HSA from specimens (whole blood, plasma or saliva) that had been incubated with formaldehyde in vitro. This novel peptide elutes at 11.3 min and is represented by a doubly charged precursor ion at m/z 451.7597. Proteomic algorithms assigned this ion to the N(6)-methylene derivative of Lys199 based on a match of the precursor ion (m/z 451.7597) within 0.7 ppm. The N(6)-methylene derivative (Scheme 1, structure IV) has an added mass of 12 Da, which is consistent with important fragment ions from the MS/MS spectrum (Figure 2C), including b2+ and y72+ that pinpoint Lys199 as the site of modification. However, treatment of formaldehyde-modified HSA prior to digestion with a molar excess of NaBH4, a reducing agent used to confirm the presence of Schiff bases (Aldini et al., 2006), did not affect this modified peptide and thus discounts assignment of the detected adduct as the N(6)-methylene derivative (Scheme 1, structure IV). If the detected adduct had been an imine (structure IV), then it should have been reduced to N-methyl-Lys following addition of NaBH4 and no further post-digestion rearrangement would have been possible. Because a mass shift of 12 Da is also typical of formaldehyde crosslinks (Metz et al., 2006; Metz et al., 2004; Toews et al., 2008) we postulate that the detected adduct represents rearrangement of the methylene adduct to form an intramolecular crosslink (Scheme 1, structure IV). One clue as to the location of this putative crosslink is given by the MS/MS fragment ion at m/z 661.28 (Figure 3C), which corresponds to addition of 12 Da to y6+ and suggests involvement of Cys200 (structure Va of Scheme 1). Also, the fragment at m/z 254.31, corresponding to addition of 12 Da to b2+, points to intramolecular cyclization of Lys199 (structure Vb of Scheme 1). Although additional work will be required to confirm the actual structure(s) of the intramolecular crosslink(s), our data are consistent with the crosslinking mechanisms proposed by several authors (Metz et al., 2006; Metz et al., 2004; Toews et al., 2008)
3.3 Adduct formation following incubation with formaldehyde solutions
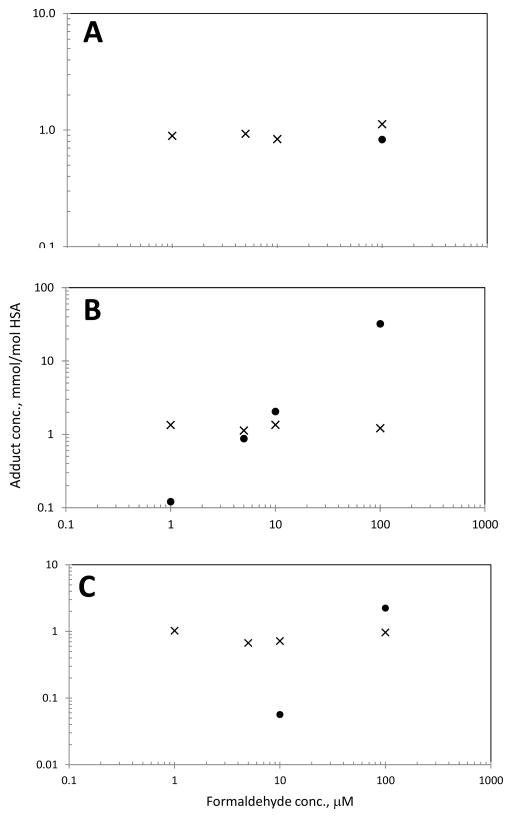
Figure 4 shows log-scale relationships between levels of formyl and putative crosslink adducts in duplicate samples of plasma, whole blood, and saliva incubated with formaldehyde at concentrations up to 100 μM. Although formyl adducts were readily observed in all samples, there was no trend towards higher adduct levels with increasing formaldehyde concentrations [Spearman correlations were negative for plasma and saliva and positive but not significant (P-value = 0.173) for whole blood]. Because the highest formaldehyde concentration used in these experiments was 100 μM, this result is not surprising given that Edrissi et al. did not detect increased levels of N(6)-formyl lysine in incubates of either L-lysine or TK6 cells at formaldehyde concentrations less than 1,000 μM (Edrissi et al., 2013a). Thus, our results are consistent with endogenous production of N(6)-formyl lysine that greatly exceeded the contribution of exogenous formaldehyde in tissues from animals exposed by inhalation (Edrissi et al., 2013a; Edrissi et al., 2013b).
Figure 4.
Relative concentrations of N(6)-formyl (×) and intramolecular-crosslink (●) adducts of Lys199 in whole blood (A), plasma (B), and saliva (C) after 3 h of incubation with formaldehyde solutions. (Mean values of duplicate samples are plotted.)
In plasma, the putative intramolecular crosslink was readily observed in all specimens incubated with formaldehyde at or above 1 μM (Figure 4B). However, in saliva this crosslink was only observed at or above 10 μM (Figure 4C) and in whole blood at 100 μM (Figure 4A). Since no crosslink was detected in blank matrices, it appears that the steady state concentration of formaldehyde should have been less than 1 μM in serum, and somewhat less than 100 μM in whole blood. This conjecture is supported by results from the time-course experiments (Figure 2) but appears to contradict earlier reports that formaldehyde was present at 37 μM in human plasma and between 22 and 87 μM in whole blood (Table 1). At the highest formaldehyde concentration tested in our study (100 μM), the level of the crosslinked adduct was 40 times greater in plasma than in blood (32.1 vs. 0.828 mmol/mol HSA). This suggests that virtually all (about 97%) of the formaldehyde in whole blood was scavenged by RBC, as indicated by measurements of free formaldehyde (Figure 2) and earlier observations (Malorny et al., 1965; Tran et al., 1972). Concentrations of the putative intramolecular crosslink did not change when the incubation time was increased from 3 to 15 h (data not shown), suggesting that the crosslinking reaction is relatively rapid and does not result from a slow conversion of other adducts such as Schiff bases.
Because albumin is ubiquitous in the body and is one of the most abundant proteins in secretions of the upper respiratory tract (Mellanen et al., 2001), saliva is a logical candidate for biomonitoring (Jeppsson et al., 2009; Kristiansson et al., 2004). Our results, summarized in Figure 4, indicate that comparable background concentrations of formyl adducts were detected in saliva and blood from the volunteer subject whose biospecimens were used for in vitro experiments.
3.4 Adducts detected in vivo
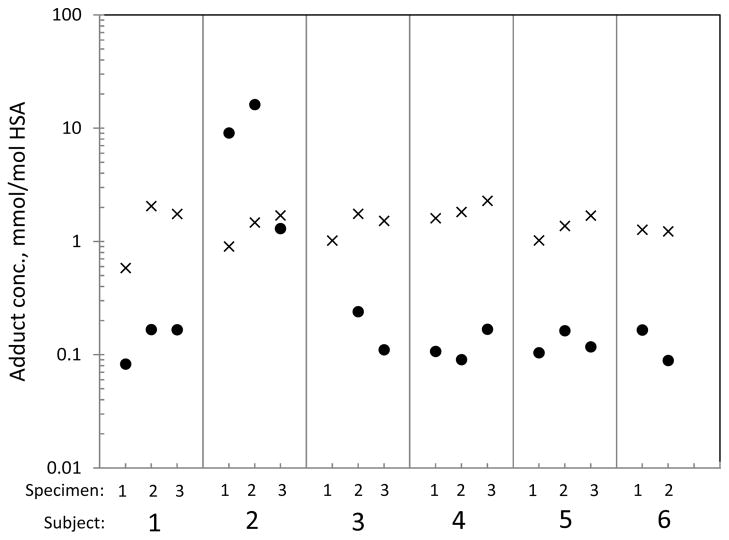
Formyl and acetyl adducts of Lys199 were detected in all plasma samples from 6 volunteer subjects while the putative intramolecular crosslink was not detected at all. As shown in Figure 5, the concentration of formyl adducts was typically 10 times that of the acetyl adducts across the 6 volunteer subjects (median values of 1.40 versus 0.133 mmol/mol HSA), with the notable exception of Subject #2, whose acetyl-adduct concentration (8.83 mmol/mol HSA) was much greater than that of the formyl adduct (1.36 mmol/mol HSA). On further examination, it was discovered that Subject #2 had taken aspirin during the days when the first two blood samples were collected but not on the date of the third sample. This pattern of aspirin consumption is reflected by levels of the acetyl adduct in Subject #2 that diminished 10-fold between the second and third blood specimens. (Note that the third specimen was obtained from this subject 29 days after the second sample to investigate the aspirin hypothesis). Although the connection between aspirin and N(6)-acetylation of Lys199 has been observed in vitro (Liyasova et al., 2010; Walker, 1976), this acetyl adduct of HSA has not been reported previously in human subjects with or without consumption of aspirin.
Figure 5.
Relative concentrations of N(6)-formyl (×) and N(6)-acetyl (●) adducts of Lys199 in plasma obtained from 6 healthy volunteer subjects on up to three occasions at intervals of 3–4 months. (The third specimen from Subject #2 was collected 29 d after the second specimen; mean values of single or duplicate samples are plotted).
Figure 5 shows that levels of formyl and acetyl adducts were very stable across the volunteer subjects and also within subjects over time for blood samples collected at intervals of three or four months (with the exception of the third specimen from Subject #2, which was collected after a 29-d interval as noted above). This finding is reasonable given that Lys199 adducts are probably eliminated with turnover of HSA, which has a mean residence time of 28 d in humans (Allison, 1960; Peters, 1970). After excluding Subject #2 from the model of acetyl adducts (due to consumption of aspirin), the between-subject and within-subject variance components estimated from random-effects models of (logged) concentrations of formyl and acetyl adducts were both zero, indicating trivial variability in adduct levels within and between subjects. Thus, the assay variability can be explained by technical error (i.e., duplicate assays of a given specimen) represented by the error variance. These log-scale error variances were 0.143 for formyl adducts (CVe = 0.392) and 0.183 for acetyl adducts (CVe = 0.449).
Formyl and acetyl adducts, but not the putative intramolecular crosslink, were also detected in serum from the 15 formaldehyde-exposed and 15 unexposed workers in Tianjin, China. The concentrations of formyl adducts were marginally smaller in exposed subjects (range: 0.61 – 2.44, median = 0.73 mmol/mol HSA) than in unexposed subjects (range: 0.47 – 3.82, median = 0.92 mmol/mol HSA) and thus were not increased by occupational exposure to formaldehyde at about 1.5 ppm. Also, in contrast to the volunteer subjects from Berkeley, CA, who had 10-fold greater median concentrations of formyl adducts (1.4 mmol/mol HSA) than acetyl adducts (0.13 mmol/mol HSA) (Figure 5), Chinese subjects had only 2.6-fold greater concentrations of formyl adducts (0.88 mmol/mol HSA) than acetyl adducts (0.34 mmol/mol HSA). The difference in levels of acetyl adduct between the two populations may reflect a genetic polymorphism for acetaldehyde-metabolizing enzymes (Iwahashi and Suwaki, 1998).
Since the putative intramolecular crosslink was not detected in either the plasma of volunteers or serum from formaldehyde-exposed workers and controls, we have no evidence that this HSA modification occurs in human blood either endogenously or following inhalation at air concentrations of about 1.5 ppm. To verify that formaldehyde gas can indeed react with HSA in biospecimens, we incubated duplicate samples of plasma and whole blood with formaldehyde gas at 1.4 ppm for either 8 or 15 h. The crosslinked adduct was detected in plasma exposed to formaldehyde gas for 8 h (estimated mean ± S.D. = 0.08 ± 0.03 mmol/mol HSA) and 15 h (0.75 ± 0.08 mmol/mol HSA). These levels of the intramolecular crosslink suggest that modification of plasma with gaseous formaldehyde at 1.4 ppm is roughly equivalent to incubation with formaldehyde solutions in the range of 1 to 5 μM (Figure 4B). Since the intramolecular crosslink was not detected in whole blood incubated with formaldehyde at 10 μM (Figure 4A), our inability to detect the intramolecular crosslink in serum derived from whole blood in Chinese workers exposed to about 1.5 ppm of formaldehyde (equivalent to 1 to 5 μM) is not surprising.
4. Conclusions
To our knowledge, this is the first mass-spectrometric characterization of formaldehyde-HSA adducts in human blood, plasma, and saliva. Following incubation of these biospecimens with formaldehyde, we identified a putative N(6)-Lys199 intramolecular crosslink (Scheme 1, Va and/or Vb). Further work is needed to confirm the structure of this putative adduct(s) as well as modification to produce the precursor (N(6)-methylene) adduct (Structure IV, Scheme 1), which was not detected either in vitro or in vivo. The putative intramolecular crosslink was detected after incubation of plasma with formaldehyde at or above 1 μM (Figure 4B), but was only observed in saliva at formaldehyde concentrations at or above 10 μM (Figure 4C), and in whole blood at 100 μM (Figure 4A). Likewise, this crosslink was detected in plasma but not whole blood incubated with 1.4 ppm of formaldehyde gas for 8 h. Through time-course experiments with free formaldehyde, we confirmed that most exogenous formaldehyde in whole blood was scavenged by RBC, as suggested by others (Malorny et al., 1965; Tran et al., 1972).
The Lys199 locus of HSA had previously been shown to form adducts with 4-hydroxy-2-nonenal (Aldini et al., 2006), hexahydrophthalic anhydride (Kristiansson et al., 2004), aspirin (Yang et al., 2007), vinyl sulfones (Regazzoni et al., 2016) and glucose (Iberg and Fluckiger, 1986). Here, we determined that HSA contained basal levels of adducts representing formylation and acetylation that varied across the subjects from Berkeley, California and Tianjin, China (Figures 5 and 6). Although adducts of endogenous formylation and acetylation have been reported previously at other protein loci (Edrissi et al., 2013a; Edrissi et al., 2013b; Jiang et al., 2007; Wisniewski et al., 2008; Zhao et al., 2010), this is apparently the first observation of Lys199 modifications to HSA in vivo. We also obtained preliminary evidence that acetylation of Lys199 was influenced by consumption of aspirin, as previously reported in vitro (Liyasova et al., 2010; Walker, 1976).
Finally, our inability to detect background levels of the N(6)-Lys199 intramolecular crosslink(s) in plasma - despite measuring this adduct in plasma incubated with formaldehyde at 1 μM (Figure 4B) – casts doubt on reports that human blood/plasma contains free formaldehyde at concentrations between 22 and 87 μM (Table 1). Likewise the fact that concentrations of the N(6)-formyl adduct of Lys199 did not increase in either whole blood, plasma or saliva incubated with formaldehyde at 100 μM (Figure 4) suggests that this HSA adduct did not arise from reactions in the serum or respiratory fluids. Rather, it appears that N(6) formylation occurs within cells with subsequent release of adducted HSA and, presumably, other modified proteins and small molecules to the systemic circulation. This conjecture is worthy of additional experimentation but is beyond the scope of our exploratory investigation.
Supplementary Material
Acknowledgments
The authors appreciate the efforts of Dr. Reuben Thomas, who assisted with collection of blood from volunteer subjects and of Dr. Cliona McHale who reviewed the manuscript, and the participation of all subjects who contributed blood specimens.
Funding Sources
This work was supported by a grant from the American Chemistry Council Long-range Research Initiative (to S.M.R.), by grant R33CA191159 9 to (S.M.R.) and by grant R01ES17452 from the National Institute of Environmental Health Science (to L.Z.).
ABBREVIATIONS
- AGC
automatic gain control
- CVe
coefficient of variation corresponding to the error variance
- Cys
cysteine
- GM
geometric mean
- HSA
human serum albumin
- Lys
lysine
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- nanoLC
nanoflow liquid chromatograph
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate buffered saline
- RBC
red blood cells
- SIC
single ion chromatogram
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflict of Interest
Much of the financial support for this work was derived from a grant from the American Chemistry Council, which is an industry trade association for American chemical companies, including those that manufacture and use formaldehyde.
References
- Aldini G, Gamberoni L, Orioli M, Beretta G, Regazzoni L, Maffei Facino R, Carini M. Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J Mass Spectrom. 2006;41:1149–1161. doi: 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
- Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M. Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species? Chem Res Toxicol. 2008;21:824–835. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- Allison AC. Turnover of erythrocytes and plasma proteins in mammals. Nature (London) 1960;188:37–40. doi: 10.1038/188037a0. [DOI] [PubMed] [Google Scholar]
- Allou L, El Maimouni L, Le Calve S. Henry’s law constant measurements for formaldehyde and benzaldehyde as a function of temperature and water composition. Atmospheric Environment. 2011;45:2991, e2998. [Google Scholar]
- Betterton EA, Hoffmann MR. Henry’s law constants of some environmentally important aldehydes. Environmental science & technology. 1988;22:1415–1418. doi: 10.1021/es00177a004. [DOI] [PubMed] [Google Scholar]
- Carraro E, Gasparini S, Gilli G. Identification of a chemical marker of environmental exposure to formaldehyde. Environmental research. 1999;80:132–137. doi: 10.1006/enrs.1998.3875. [DOI] [PubMed] [Google Scholar]
- Casanova M, Heck HD, Everitt JI, Harrington WW, Jr, Popp JA. Formaldehyde concentrations in the blood of rhesus monkeys after inhalation exposure. Food Chem Toxicol. 1988;26:715–716. doi: 10.1016/0278-6915(88)90071-3. [DOI] [PubMed] [Google Scholar]
- Chung MK, Regazzoni L, McClean M, Herrick R, Rappaport SM. A sandwich ELISA for measuring benzo[a]pyrene-albumin adducts in human plasma. Analyt Biochem. 2013;435:140–149. doi: 10.1016/j.ab.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow EL, Shimazu K, editors. Lognormal Distributions: Theory and Applications. Marcel Dekker; New York: 1988. [Google Scholar]
- Edrissi B, Taghizadeh K, Dedon PC. Quantitative analysis of histone modifications: formaldehyde is a source of pathological n(6)-formyllysine that is refractory to histone deacetylases. PLoS Genet. 2013a;9:e1003328. doi: 10.1371/journal.pgen.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrissi B, Taghizadeh K, Moeller BC, Kracko D, Doyle-Eisele M, Swenberg JA, Dedon PC. Dosimetry of N(6)-formyllysine adducts following [(1)(3)C(2)H(2)]-formaldehyde exposures in rats. Chem Res Toxicol. 2013b;26:1421–1423. doi: 10.1021/tx400320u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan H, Edmands W, Lu SS, Yano Y, Regazzoni L, Iavarone AT, Williams ER, Rappaport SM. Adductomics Pipeline for Untargeted Analysis of Modifications to Cys34 of Human Serum Albumin. Anal Chem. 2016;88:10504–10512. doi: 10.1021/acs.analchem.6b02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck H, Casanova M. The implausibility of leukemia induction by formaldehyde: a critical review of the biological evidence on distant-site toxicity. Regul Toxicol Pharmacol. 2004;40:92–106. doi: 10.1016/j.yrtph.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Heck HD, Casanova-Schmitz M, Dodd PB, Schachter EN, Witek TJ, Tosun T. Formaldehyde (CH2O) concentrations in the blood of humans and Fischer-344 rats exposed to CH2O under controlled conditions. Am Ind Hyg Assoc J. 1985;46:1–3. doi: 10.1080/15298668591394275. [DOI] [PubMed] [Google Scholar]
- IARC. Chemical Agents and Related Occupations, Volume 100 F, A Review of Human Carcinogens. International Agency for Research on Cancer; Lyon, France: 2012. [Google Scholar]
- Iberg N, Fluckiger R. Nonenzymatic glycosylation of albumin in vivo. Identification of multiple glycosylated sites. J Biol Chem. 1986;261:13542–13545. [PubMed] [Google Scholar]
- Iwahashi K, Suwaki H. Ethanol metabolism, toxicity and genetic polymorphism. Addict Biol. 1998;3:249–259. doi: 10.1080/13556219872065. [DOI] [PubMed] [Google Scholar]
- Jeppsson MC, Lindh CH, Kristiansson MH, Nielsen J, Jonsson BA. Methylhexahydrophthalic anhydride adducted albumin tryptic peptides in nasal lavage fluid. Inhalation toxicology. 2009;21:1013–1020. doi: 10.1080/08958370802715997. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Burke PJ, Koch TH, Bierbaum VM. Formaldehyde in human cancer cells: detection by preconcentration-chemical ionization mass spectrometry. Anal Chem. 2001;73:2992–2997. doi: 10.1021/ac001498q. [DOI] [PubMed] [Google Scholar]
- Ke YJ, Qin XD, Zhang YC, Li H, Li R, Yuan JL, Yang X, Ding SM. In vitro study on cytotoxicity and intracellular formaldehyde concentration changes after exposure to formaldehyde and its derivatives. Hum Exp Toxicol. 2014;33:822–830. doi: 10.1177/0960327113510538. [DOI] [PubMed] [Google Scholar]
- Keller BO, Sui J, Young AB, Whittal RM. Interferences and contaminants encountered in modern mass spectrometry. Anal Chim Acta. 2008;627:71–81. doi: 10.1016/j.aca.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis AJ, Staal YC, Duistermaat E, Engel R, Woutersen RA. The determination of exogenous formaldehyde in blood of rats during and after inhalation exposure. Food Chem Toxicol. 2013;52:105–112. doi: 10.1016/j.fct.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Kristiansson MH, Lindh CH, Jonsson BA. Correlations between air levels of hexahydrophthalic anhydride (HHPA) and HHPA-adducted albumin tryptic peptides in nasal lavage fluid from experimentally exposed volunteers. Rapid Commun Mass Spectrom. 2004;18:1592–1598. doi: 10.1002/rcm.1527. [DOI] [PubMed] [Google Scholar]
- Li Q, Sritharathikhun P, Motomizu S. Development of novel reagent for Hantzsch reaction for the determination of formaldehyde by spectrophotometry and fluorometry. Analytical sciences : the international journal of the Japan Society for Analytical Chemistry. 2007;23:413–417. doi: 10.2116/analsci.23.413. [DOI] [PubMed] [Google Scholar]
- Liyasova MS, Schopfer LM, Lockridge O. Reaction of human albumin with aspirin in vitro: mass spectrometric identification of acetylated lysines 199, 402, 519, and 545. Biochemical pharmacology. 2010;79:784–791. doi: 10.1016/j.bcp.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Ye W, Gold A, Ball LM, Swenberg JA. Formation of S-[1-(N2-deoxyguanosinyl)methyl]glutathione between glutathione and DNA induced by formaldehyde. Journal of the American Chemical Society. 2009;131:3414–3415. doi: 10.1021/ja808048c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li H, Zhang Y, Ang CY. Determination of formaldehyde in blood plasma by high-performance liquid chromatography with fluorescence detection. J Chromatogr B Biomed Sci Appl. 2001;753:253–257. doi: 10.1016/s0378-4347(00)00552-1. [DOI] [PubMed] [Google Scholar]
- Malorny G, Rietbrock N, Schneider M. the Oxidation of Formaldehyde to Formic Acid in the Blood, a Contribution to the Metabolism of Formaldehyde. Naunyn-Schmiedebergs Archiv fur experimentelle Pathologie und Pharmakologie. 1965;250:419–436. doi: 10.1007/BF00246893. [DOI] [PubMed] [Google Scholar]
- Mellanen L, Sorsa T, Lahdevirta J, Helenius M, Kari K, Meurman JH. Salivary albumin, total protein, IgA, IgG and IgM concentrations and occurrence of some periodontopathogens in HIV-infected patients: a 2-year follow-up study. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2001;30:553–559. doi: 10.1034/j.1600-0714.2001.300908.x. [DOI] [PubMed] [Google Scholar]
- Metz B, Kersten GF, Baart GJ, de Jong A, Meiring H, ten Hove J, van Steenbergen MJ, Hennink WE, Crommelin DJ, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjugate chemistry. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Meurman JH, Rantonen P, Pajukoski H, Sulkava R. Salivary albumin and other constituents and their relation to oral and general health in the elderly. Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics. 2002;94:432–438. doi: 10.1067/moe.2002.122345. [DOI] [PubMed] [Google Scholar]
- Mochca-Morales J. Nasal albumin in a population exposed for the first time to urban pollution. Archives of medical research. 2000;31:409–414. doi: 10.1016/s0188-4409(00)00091-6. [DOI] [PubMed] [Google Scholar]
- Nagy K, Pollreisz F, Takats Z, Vekey K. Atmospheric pressure chemical ionization mass spectrometry of aldehydes in biological matrices. Rapid Commun Mass Spectrom. 2004;18:2473–2478. doi: 10.1002/rcm.1648. [DOI] [PubMed] [Google Scholar]
- Neuss S, Moepps B, Speit G. Exposure of human nasal epithelial cells to formaldehyde does not lead to DNA damage in lymphocytes after co-cultivation. Mutagenesis. 2010;25:359–364. doi: 10.1093/mutage/geq013. [DOI] [PubMed] [Google Scholar]
- Ospina M, Costin A, Barry AK, Vesper HW. Characterization of N-terminal formaldehyde adducts to hemoglobin. Rapid Commun Mass Spectrom. 2011;25:1043–1050. doi: 10.1002/rcm.4954. [DOI] [PubMed] [Google Scholar]
- Pala M, Ugolini D, Ceppi M, Rizzo F, Maiorana L, Bolognesi C, Schiliro T, Gilli G, Bigatti P, Bono R, Vecchio D. Occupational exposure to formaldehyde and biological monitoring of Research Institute workers. Cancer detection and prevention. 2008;32:121–126. doi: 10.1016/j.cdp.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Peters T. Serum Albumin. AdvClinChem. 1970;13:37–111. doi: 10.1016/s0065-2423(08)60385-6. [DOI] [PubMed] [Google Scholar]
- Regazzoni L, Colombo S, Mazzolari A, Vistoli G, Carini M. Serum albumin as a probe for testing the selectivity of irreversible cysteine protease inhibitors: The case of vinyl sulfones. J Pharm Biomed Anal. 2016;124:294–302. doi: 10.1016/j.jpba.2016.02.056. [DOI] [PubMed] [Google Scholar]
- Shaham J, Bomstein Y, Meltzer A, Kaufman Z, Palma E, Ribak J. DNA--protein crosslinks, a biomarker of exposure to formaldehyde--in vitro and in vivo studies. Carcinogenesis. 1996;17:121–125. doi: 10.1093/carcin/17.1.121. [DOI] [PubMed] [Google Scholar]
- Swenberg JA, Moeller BC, Lu K, Rager JE, Fry RC, Starr TB. Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicologic pathology. 2013;41:181–189. doi: 10.1177/0192623312466459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Bai Y, Duong A, Smith MT, Li L, Zhang L. Formaldehyde in China: production, consumption, exposure levels, and health effects. Environment international. 2009;35:1210–1224. doi: 10.1016/j.envint.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Toews J, Rogalski JC, Clark TJ, Kast J. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal Chim Acta. 2008;618:168–183. doi: 10.1016/j.aca.2008.04.049. [DOI] [PubMed] [Google Scholar]
- Tran N, Laplante M, Lebel E. Abnormal oxidation of 14 C-formaldehyde to 14 CO2 in erythrocytes of alcoholics and nonalcoholics after consumption of alcoholic beverages. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 1972;13:677–680. [PubMed] [Google Scholar]
- Walker JE. Lysine residue 199 of human serum albumin is modified by acetylsalicyclic acid. FEBS Lett. 1976;66:173–175. doi: 10.1016/0014-5793(76)80496-6. [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng G, Balbo S, Carmella SG, Villalta PW, Hecht SS. Clear differences in levels of a formaldehyde-DNA adduct in leukocytes of smokers and nonsmokers. Cancer research. 2009;69:7170–7174. doi: 10.1158/0008-5472.CAN-09-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JGM, Voorwinde OK, Ottens M, Beenackers AACM, Janssen LPBM. Kinetics and chemical equilibrium of the hydration of formaldehyde. Chemical Engineering Science. 2002;57:4067–4076. [Google Scholar]
- Wisniewski JR, Zougman A, Mann M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bian C, Zhu L, Zhao G, Huang Z, Huang M. Effect of human serum albumin on drug metabolism: structural evidence of esterase activity of human serum albumin. J Struct Biol. 2007;157:348–355. doi: 10.1016/j.jsb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat Res. 2009;681:150–168. doi: 10.1016/j.mrrev.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.