Abstract
One of the central issues facing the emerging field of nanotechnology is cellular compatibility. Nanoparticles have been proposed for diagnostic and therapeutic applications, including drug delivery, gene therapy, biological sensors, and controlled catalysis. Viruses, liposomes, peptides, and synthetic and natural polymers have been engineered for these applications, yet significant limitations continue to prevent their use. Avoidance of the body's natural immune system, lack of targeting specificity, and the inability to control packaging and release are remaining obstacles. We have explored the use of a naturally occurring cellular nanoparticle known as the vault, which is named for its morphology with multiple arches reminiscent of cathedral ceilings. Vaults are 13-MDa ribonucleoprotein particles with an internal cavity large enough to sequester hundreds of proteins. Here, we report a strategy to target and sequester biologically active materials within the vault cavity. Attachment of a vault-targeting peptide to two proteins, luciferase and a variant of GFP, resulted in their sequestration within the vault cavity. The targeted proteins confer enzymatic and fluorescent properties on the recombinant vaults, both of which can be detected by their emission of light. The modified vaults are compatible with living cells. The ability to engineer vault particles with designed properties and functionalities represents an important step toward development of a biocompatible nanocapsule.
Keywords: capsule, nanoparticle
Vaults are large ribonucleoprotein particles that exist as naturally occurring nanocapsules with a thin (≈20 Å) protein shell surrounding an internal cavity of 5 × 107Å3 (1) (Fig. 1). The molecular mass of the vault is 12.9 ± 1 MDa. Cryoelectron microscopy (cryoEM) single-particle reconstruction has revealed the vault to be a hollow, barrel-like structure with two protruding caps and an invaginated waist with overall dimensions of 420 × 420 × 750 Å (2), larger in mass and size than some icosahedral viruses. Although many different roles have been proposed for the vault since its first description in 1986 (3), including nucleocytoplasmic transport and sequestration of molecular cargo, its cellular function remains unknown. The vault is the largest known cellular ribonucleoprotein particle, yet it has a relatively simple molecular composition with multiple copies of just three proteins, the 100-kDa major vault protein (MVP), the 193-kDa vault poly(ADP ribose) polymerase (VPARP), and the 290-kDa telomerase-associated protein 1, and one to three distinct untranslated RNA molecules (4). The MVP is presumed to be present in 96 copies per vault, based on the observed symmetry of the particle and the estimate that MVP accounts for ≈75% of the total protein mass in the particle. Expression of MVP in insect cells, by using the baculovirus expression system, revealed that this protein alone is capable of directing the formation of recombinant vault particles with a structure similar to endogenous particles (5).
Fig. 1.
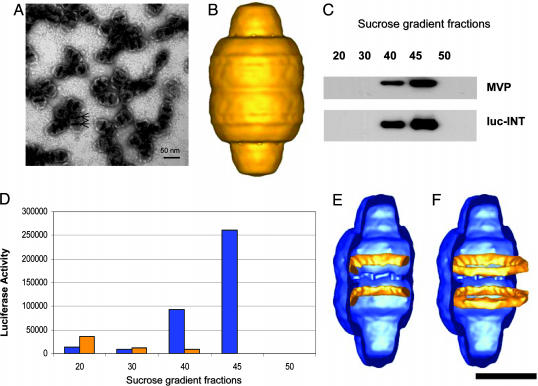
Purification, EM, and cryoEM single-particle reconstruction of the luc-INT vaults and localization of the luc-INT density. (A) An electron micrograph of negative-stained vault particles containing MVP and luc-INT. The arrows indicate the darkly staining bands across the central barrel. (Scale bar, 50 nm.) (B) The luc-INT vault reconstruction at a resolution of 28 Å based on 661 particle images collected on a CM120 cryotransmission electron microscope (FEI). (C and D) Sucrose gradient fractions were immunoblotted to detect the indicated protein species in the final step of luc-INT vault purification indicating their copurification. Likewise, luciferase activity peaks with the purified particles in the 40% and 45% fractions. Blue bars show the luc-INT vaults, and gold bars show luc-INT alone. (E and F) Difference density assigned to luc-INT (gold) superimposed on a reconstruction of a vault formed by MVP alone (blue). Both density maps are shown cropped lengthwise. The MVP vault reconstruction is that of HisT7-MVP (10). (F) The full luc-INT density rings (gold) are shown superimposed on the cropped MVP vault (blue). (Scale bar, 25 nm.)
A wide variety of strategies for encapsulation of biomaterials exist, including synthetic and natural polymers, hydrogels, peptides, liposomes, and viruses. However, the use of these materials in biological systems is often limited by poor biocompatibility, immune responses, lack of targeting specificity, and the inability to control packaging and release. We have been analyzing the use of recombinant vaults to overcome some or all of these limitations. Here, we demonstrate that, by using a vault-targeting peptide, a heterologous protein can be functionally sequestered within the central cavity of vault particles, and that these modified vaults are compatible with living cells.
Methods
Vault-Targeting Constructs and Purification. The luciferase-encoding DNA (6) was PCR-amplified from pGL3basic (Promega), fused to the vault interaction domain derived from VPARP (INT) (GenBank accession no. AF158255, amino acids 1471–1724) and inserted into the baculovirus expression vector, pFASTBAC (Invitrogen). The INT domain is defined as the 162-aa region at the C terminus of VPARP (amino acids 1563–1724), which is the smallest region identified for interaction with MVP. The expressed fusion domain is slightly larger (254-aa) but includes the full INT domain. The gene encoding a variant GFP, green lantern (GL) (Invitrogen) (7), was PCR-amplified, fused to either an INT domain (amino acids 1471–1724) or the N terminus of the cDNA-encoding rat MVP (GenBank accession no. U09870), and inserted into pFASTBAC. Using the Bac-to-Bac expression system (Invitrogen), recombinant insect viruses were generated. Sf9 cells were infected with the recombinant INT baculoviruses alone or in combination with recombinant tagged MVP baculoviruses, and recombinant vaults were purified as described in ref. 5. Immunoblot analyses were carried out by using either anti-MVP (8) or anti-VPARP (9) polyclonal antibodies. Note that the anti-VPARP antibody recognizes the INT domain of VPARP.
CryoEM Single-Particle Reconstruction. The luc-INT vault reconstruction at 28-Å resolution was based on 661 particle images collected on a CM120 cryotransmission electron microscope (FEI, Hillsboro, OR). CryoEM and single-particle reconstruction were performed as described in refs. 1, 2, and 10. The resolution was determined with the Fourier shell correlation 0.5 threshold criterion (11). The difference density was calculated by using the avs software package (Advanced Visual Systems, Waltham, MA) after filtering the two input reconstructions to the same resolution.
Luciferase Enzyme Assays. Purified luciferase (Promega) was diluted to 47 nM with 1× PBS (Fisher Scientific) before assaying. Luciferase assay substrate was prepared as described in ref. 12. Briefly, the substrate contained 25 mM Gly-Gly, 15 mM K2PO4, 15 mM MgSO4, 4 mM EGTA, 2 mM ATP, 1 mM DTT, 0.1 mM CoA, and 75 μM luciferin (pH 8.0). In a glass vial, 50 μl of either standard luciferase or sample solution was added, and 100 μl of the luciferase substrate was added in the dark a few seconds after data collection had begun. The glass vial was placed at the entrance slit of a single monochromator, which dispersed the captured luminescence onto an intensified charge-coupled device camera (PI-MAX, Princeton Instruments, Trenton, NJ). The camera was programmed to collect a spectrum every 500 ms for 5 min. The software was programmed to integrate the area under the luminescence peak from 540 to 575 nm for each spectrum that was collected. These areas were then plotted vs. time with excel (Microsoft) or igor pro software (WaveMetrics, Lake Oswego, OR).
For the preincubation experiments, 10 μl of 20 mM ATP or 10 μl of 750 μM luciferin was added to 50 μl of luc-int/MVP vaults. The mixture was incubated on ice for ≈40 min before being transferred to a glass vial. Luciferase substrate (90 μl) containing either no ATP or luciferin was added to the glass vial for data collection as described above.
Green Fluorescence Quenching. GL-INT vaults (0.01 mg/ml) in Mes buffer (20 mM, pH 6.5) were added to a glass vial, an equal volume of quencher solution (1 M KCl or KNO3 in 20 mM Mes buffer, pH 6.5) was added, and the intensity was monitored as a function of time. For the Stern–Volmer data, variable concentrations of Congo red in 20 mM Mes buffer (pH 6.5) were used, and the fluorescence intensity was monitored. The fluorescence was recorded by using a monochromator (HR-320, Instruments SA, Edison, NJ) and a gated intensified charge-coupled device camera (PI-MAX). The intensity was monitored by using the gated intensified charge-coupled device camera, where the integration time was 0.5 s and each integration followed directly after the previous integration. The same experiment was performed by using free GL-INT and GL-INT vaults. Samples were excited by using an argon ion laser (Innova 90C-5, Coherent Radiation, Palo Alto, CA). The excitation wavelength and power were 457 nm and 10 mW, respectively.
Uptake of GL-INT Vaults by HeLa Cells. HeLa cells were grown on four-well chamber slides (Lab Tek II, Nalge). Purified GL-INT vaults (10 μg) were added to the HeLa cell monolayer and incubated for 1 h at 37°C. The cells were washed with PBS, incubated with cholera toxin B/Alexa Fluor 594 conjugate (Molecular Probes) for 10 min at 12°C to stain cell membranes, washed a second time, and covered with glass coverslips, and live cells were visualized by using confocal microscopy (green HeNe, 594 nm; argon, 488 nm; 0.2-μm slices).
Results
Targeting Heterologous Proteins into the Central Cavity of Recombinant Vaults. INT was previously identified as being responsible for its interaction with MVP (9). We reasoned that, because this domain is responsible for VPARP assembly into vaults, INT could be used to target heterologous non-vault proteins into vault particles. We demonstrate the use of this targeting peptide with two proteins whose presence is revealed by light emission, the enzyme luciferase (6) and a variant of the fluorochrome-containing GFP (7). The INT domain was fused to the C terminus of luciferase. When the luciferase/INT fusion protein (luc-INT) was expressed in insect cells by using the baculovirus system, the modified protein retained enzymatic activity and remained in the soluble fraction. In contrast, coexpression of luc-INT and MVP in insect cells altered the luc-INT fractionation profile, resulting in its sedimentation at 100,000 × g, indicative of assembly into vaults. Analysis of the recombinant vaults (hereafter referred to as luc-INT vaults) after purification on a sucrose gradient demonstrated that both the luc-INT protein (Fig. 1C) and luciferase enzymatic activity cofractionate with MVP (Fig. 1D). An examination by negative-stain EM of the material purifying in the 40% and 45% sucrose fractions demonstrated abundant, regular vault-like particles (Fig. 1 A). The recombinant luc-INT vault particles appear similar to endogenous vaults except for the presence of two prominent, darkly staining stripes across the central barrel (Fig. 1 A, arrows).
The luc-INT protein was localized to the inner surface of the recombinant vault by using cryoEM single-particle reconstruction. Particle images of 661 luc-INT vaults were selected from cryomicrographs and processed to yield a 3D reconstruction at a resolution of 28 Å (Fig. 1B). The outer surface of this reconstruction has the characteristic features of other published vault reconstructions (1, 2, 10). Difference-mapping between reconstructions of the luc-INT vault and recombinant vaults formed from MVP alone revealed two internal bands of density attributed to multiple copies of the luc-INT protein (Fig. 1 E and F). The internal density bands are found ≈70 Å on either side of the central waist of the particle, adjacent to the location where the stripes were seen in the negative-stain images (Fig. 1 A). The cryoEM reconstruction and difference mapping indicate that the luciferase enzyme is packaged into the cavity of the vault particle and targeted to the inner surface of the vault by the INT domain derived from the VPARP vault protein.
Functional Sequesteration of Heterologous Proteins in Vitro and in Vivo. We compared the time dependence of chemiluminescence intensity of the luciferase reaction products from soluble luc-INT and luc-INT vaults by monitoring their emission at the 558-nm band maximum. The product spectra produced by the free, soluble luc-INT protein (Fig. 2A, blue trace) were virtually identical to those of the products produced by the enzyme sequestered in the luc-INT vaults (Fig. 2 A, red trace). When the luciferase substrates luciferin and ATP were added to a solution containing soluble luc-INT, the emission intensity of the product increased to a maximum within a few seconds and was followed by a gradual decrease over hundreds of seconds (Fig. 2B, blue trace). In contrast, when the substrates were added to a suspension of luc-INT vaults containing sequestered luciferase, the intensity had an initial rapid rise but then rose gradually to a maximum in ≈300 sec (Fig. 2B, red trace). When the luc-INT vaults were preincubated in the presence of ATP and then exposed to luciferin (Fig. 2C, red trace), the time dependence was similar to that of the soluble luc-INT protein, displaying a fast rise in the emission intensity. Alternately, when luc-INT vaults were preincubated with luciferin and then exposed to ATP (Fig. 2C, blue trace), the emission intensity exhibited a slow increase over ≈300 sec, comparable with the time dependence of vaults exposed to both substrates simultaneously. Thus, the delayed maximum light emission catalyzed by the sequestered luciferase may be attributed to slow accumulation of ATP into the vaults. Perhaps either the charge or the larger size of ATP (507 vs. 248 Da for the neutral luciferin) is the reason behind the time delay in reaching the sequestered enzyme.
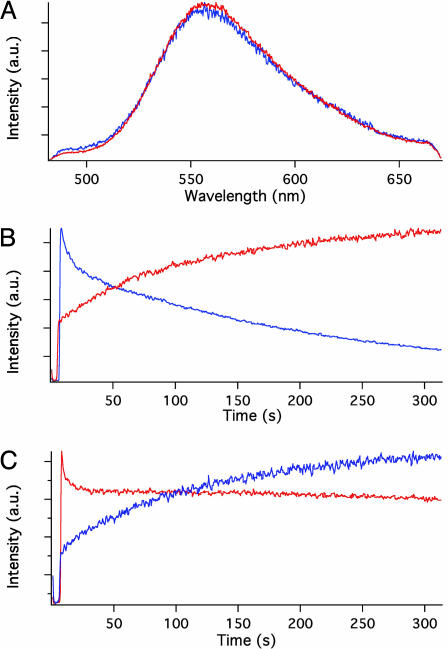
Fig. 2.
Analysis of the luc-INT enzyme reaction. (A) Superimposed emission spectra produced by the action of INT-derivatized luciferase in solution (blue trace) and engineered into the interior of vaults (red trace). (B) Time dependence of the reaction product emission intensity from a solution of INT-derivatized luciferase (blue trace) and from luciferase engineered into the interior of vaults (red trace). Note the slow decay of the intensity at long times caused by the activity of the soluble enzyme and the gradual rise at long times caused by the sequestered enzyme. (C) Time dependence of the emission intensity from suspensions of engineered vaults. Vaults that are preincubated in ATP solution (red trace) produce product in a time dependence similar to that from soluble enzyme (compare with blue trace in B), whereas vaults preincubated in luciferin solution (blue trace) produce product in a time dependence similar to that from vaults that were not pretreated (compare with red trace in B).
The generality of this approach was demonstrated by a second application with the GL protein, a variant of the GFP (7). GL-INT was expressed in insect cells by using the baculovirus system and was found to be highly fluorescent, with a band maximum at 510 nm. When the GL-INT protein is coexpressed with MVP in insect cells, recombinant vault particles containing both proteins, referred to as GL-INT vaults, can be purified. Evidence for this packaging is twofold. First, the sucrose gradient-purified vault pellet is bright green. Second, negative-stain EM demonstrated that these particles contain additional internal density similar to that of luc-INT vaults (data not shown).
Three spectroscopic studies verify that the GL-INT protein is packaged into vaults. First, fluorescence is observed only from the vaults themselves and not from the buffer; after multiple cycles of washing and ultracentrifugation followed by resuspension, fluorescence is observed from the resuspended vaults but not from the wash solvent. The fluorescence band maximum of the GL-INT vaults is 510 nm, identical to that of the soluble GL-INT protein. Second, Stern–Volmer kinetic analysis of quenching of the fluorescence of GL-INT vaults by Congo red revealed a very large quenching constant indicative of “superquenching” (13, 14) (Fig. 3A). This effect, a result of throughspace Förster quenching, occurs when one quencher molecule acts on closely spaced donor molecules. This quenching effect suggests that in the GL-INT vaults, multiple copies of the GL-INT protein are brought into close proximity within the central barrel of the vault. Third, and most importantly, GL-INT associated with vaults is protected from the outside environment. Ionic strength affects the GL absorption spectrum (15). The addition of KNO3 (16) or KCl to free GL-INT in solution causes immediate loss of fluorescence intensity (Fig. 3B, blue trace). In contrast, the addition of KCl to a suspension of GL-INT vaults causes a gradual decrease of the intensity (Fig. 3B, red trace). The quenching of GL-INT vaults by KCl requires >500 sec to reach the half-maximum value. The long quenching time indicates that GL-INT associated with vaults is protected from the quenching ions. These spectroscopic observations suggest that GL-INT is packaged within the central cavity of recombinant vault particles, consistent with our cryoEM observations for luc-INT associated with vaults.
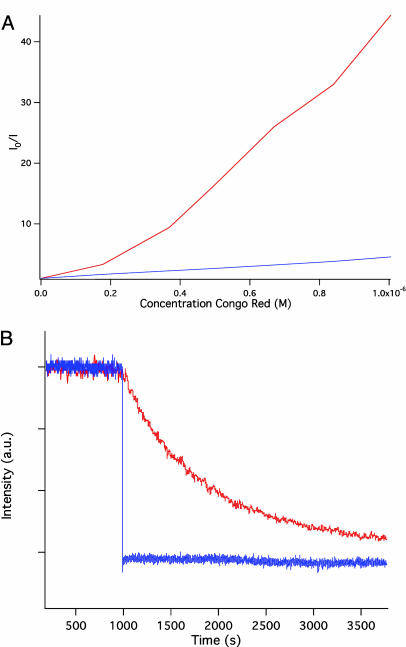
Fig. 3.
Analysis of the GL-INT fluorescent properties. (A) Stern–Volmer plot for GL-INT (blue) and GL-INT vaults (red). The fluorescence intensity without quencher (Io) divided by the intensity with quencher (I) is plotted against the Congo red concentration. Quenching is more efficient (demonstrated by the steep slope) when the fluorescent protein is sequestered within vaults. (B) Quenching of GL-INT fluorescence by KCl. The fluorescence of dissolved GL-INT is quenched immediately after addition of the salt at t = 1,000 sec (blue trace), but that of GL sequestered in vaults exhibits a much longer quenching time (red trace).
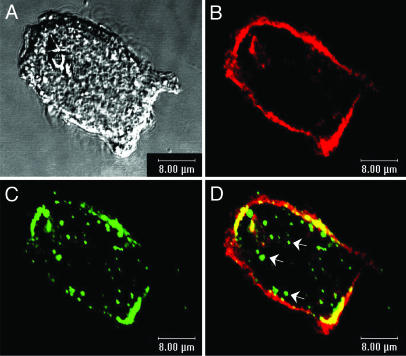
To determine whether recombinant vault nanocapsules are compatible with living cells and continue to sequester their contents within the cellular environment, we analyzed their uptake in cultured cells. Purified GL-INT vaults were added to cultured HeLa cells and incubated for 1 h, and their uptake was monitored by using confocal microscopy. A punctate pattern of green fluorescence was observed inside of the cells (Fig. 4). A similar punctate pattern was observed after uptake of GL-MVP vaults, formed from MVP tagged at the N terminus with GL (Fig. 5). In the case of GL-MVP vaults, the fluorescent protein is covalently linked to the vault. The comparable fluorescence patterns observed for both GL-INT and GL-MVP vaults suggest that the GL-INT vault particles remain intact after uptake into the cell.
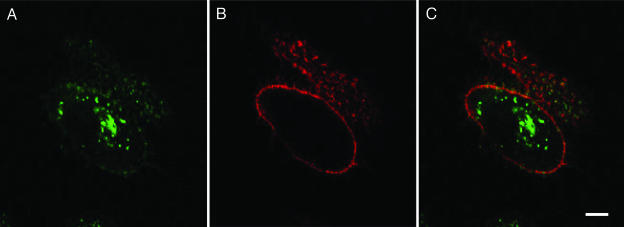
Fig. 4.
GL-INT vault uptake in HeLa cells. (A) GL-INT vaults (green) observed on the membrane and inside the cell. (B) Membrane staining (red) observed with cholera toxin B/Alexa Fluor 594 conjugate. (C) A merge of A and B showing green punctate fluorescence inside the cells. (Scale bar, 8 μm.) The HeLa cells did not display any green fluorescence in the absence of added GL-INT vaults.
Fig. 5.
GL-MVP vault uptake in HeLa cells. (A) DiC showing the HeLa cell. (B) Mostly membrane staining (red) seen with cholera toxin B/Alexa Fluor 594 conjugate. (C) Green fluorescent vaults seen on the membrane and inside the cells. (D) A merge of B and C showing vaults (green) inside the cells.
Discussion
These studies demonstrate that recombinant vaults can be produced with chemically active or fluorescent proteins sequestered within the particle cavity. The baculovirus system is robust, allowing for production and purification of 4–20 mg of vaults per liter culture of cells. The INT domain appears to be a general targeting sequence that should be able to direct a wide variety of recombinant proteins (and other molecules) into the vault. Although we have not determined the precise stoichiometry of the luc-INT protein in the vault, the INT domain was initially identified in a yeast two-hybrid screen that used MVP as the bait (9). Therefore, the minimum INT binding domain is a MVP monomer. Thus, the maximum number of INT binding sites per vault would be 96 because there are 96 MVPs per particle. Because the internal volume of the vault is 5 × 107 Å3, there is sufficient space for hundreds of proteins. Therefore, the maximum binding would depend on the size of the INT-tagged fusion protein, the spacing of the MVP binding sites, and any steric constraints that result from binding. The largest non-vault protein thus far targeted to the vault interior by using the INT domain is the ≈61-kDa firefly luciferase protein. Once targeted inside the vault, INT fusion proteins appear to be both functional and stably associated. This strategy could be used to confer unique properties onto vaults by targeting other molecules (e.g., metals, nucleic acids, polynucleotides, polymers, etc.) by virtue of fusing their protein binding domains onto the INT domain. Although the thin protein shell of the vault does not prevent the entry of small ligands, there appears to be a diffusion barrier, particularly for charged molecules. The entry of charged molecules into the vault cavity might require relatively slow conformational changes to take place within the vault protein shell. This behavior is consistent with recent in vitro assembly studies that demonstrate the vault protein shell to be a dynamic structure allowing the incorporation of the large vault associated proteins VPARP and telomerase-associated protein 1 into its interior (17).
The engineering of vault particles with designed properties and functionalities represents an important direction for the emerging field of bionanotechnology. This research establishes that intrinsic optical properties of proteins can be retained when they are confined within the vault nanocapsule. We expect that a wide range of proteins with other chemical properties may be sequestered within recombinant vaults by using this approach. We envision a number of applications for these engineered particles, including biologically based chemical sensors, bioreactors, and protein stabilizers, all of which can be targeted at the cellular level because of the biocompatibility of the capsule.
Acknowledgments
We thank Hedi Roseboro and Mike Torres for preparation of recombinant vaults and Dr. Laurent Bentolila for assistance with confocal microscopy performed at the University of California/California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Facility. This work was supported in part by National Science Foundation Grants MCB-0210690 (to L.H.R.), MCB-9722353 (to P.L.S.), and U.S. Public Health Service National Research Service Award GM07185 (to Y.G.).
Author contributions: V.A.K., E.J., S.R.-F., P.M., J.I.Z., B.D., P.L.S., and L.H.R. designed research; V.A.K., Y.G., Y.M., E.J., J.C.Z., S.R.-F., P.M., and L.H.R. performed research; V.A.K., Y.G., Y.M., E.J., J.C.Z., S.R.-F., P.M., J.I.Z., B.D., P.L.S., and L.H.R. analyzed data; and V.A.K., J.I.Z., B.D., P.L.S., and L.H.R. wrote the paper.
Abbreviations: cryoEM, cryoelectron microscopy; MVP, major vault protein; VPARP, vault poly(ADP-ribose) polymerase; INT, vault interaction domain derived from VPARP; luc-INT, luciferase/INT fusion protein; GL, green lantern.
References
- 1.Kong, L. B., Siva, A. C., Rome, L. H. & Stewart, P. L. (1999) Structure (London) 7, 371–379. [DOI] [PubMed] [Google Scholar]
- 2.Kong, L. B., Siva, A. C., Kickhoefer, V. A., Rome, L. H. & Stewart, P. L. (2000) RNA 6, 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kedersha, N. L. & Rome, L. H. (1986) J. Cell Biol. 103, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suprenant, K. A. (2002) Biochemistry 41, 14447–14454. [DOI] [PubMed] [Google Scholar]
- 5.Stephen, A. G., Raval-Fernandes, S., Huynh, T., Torres, M., Kickhoefer, V. A. & Rome, L. H. (2001) J. Biol. Chem. 276, 23217–23220. [DOI] [PubMed] [Google Scholar]
- 6.de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R. & Subramani, S. (1987) Mol. Cell. Biol. 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zolotukhin, S., Potter, M., Hauswirth, W. W., Guy, J. & Muzyczka, N. (1996) J. Virol. 70, 4646–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siva, A. C., Raval-Fernandes, S., Stephen, A. G., LaFemina, M. J., Scheper, R. J., Kickhoefer, V. A. & Rome, L. H. (2001) Int. J. Cancer 92, 195–202. [DOI] [PubMed] [Google Scholar]
- 9.Kickhoefer, V. A., Siva, A. C., Kedersha, N. L., Inman, E. M., Ruland, C., Streuli, M. & Rome, L. H. (1999) J. Cell Biol. 146, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikyas, Y., Makabi, M., Raval-Fernandes, S., Harrington, L., Kickhoefer, V. A., Rome, L. H. & Stewart, P. L. (2004) J. Mol. Biol. 344, 91–105. [DOI] [PubMed] [Google Scholar]
- 11.Orlova, E. V., Dube, P., Harris, J. R., Beckman, E., Zemlin, F., Markl, J. & van Heel, M. (1997) J. Mol. Biol. 271, 417–437. [DOI] [PubMed] [Google Scholar]
- 12.Dyer, B. W., Ferrer, F. A., Klinedinst, D. K. & Rodriguez, R. (2000) Anal. Biochem. 282, 158–161. [DOI] [PubMed] [Google Scholar]
- 13.Lu, L., Helgeson, R., Jones, R. M., McBranch, D. W. & Whitten, D. G. (2002) J. Am. Chem. Soc. 124, 483–488. [DOI] [PubMed] [Google Scholar]
- 14.Jones, R. M., Bergstedt, T. S., McBranch, D. W. & Whitten, D. G. (2001) J. Am. Chem. Soc. 123, 6726–6727. [DOI] [PubMed] [Google Scholar]
- 15.Ward, W. W., Prentice, H. J., Roth, A. F., Cody, C. W. & Reeves, S. C. (1982) Photochem. Photobiol. 35, 803–808. [Google Scholar]
- 16.Calhoun, D. B., Vanderkooi, J. M. & Englander, S. W. (1983) Biochemistry 22, 1533–1539. [DOI] [PubMed] [Google Scholar]
- 17.Poderycki, M. J., Kickhoefer, V. A., Kaddis, C. S., Raval-Fernandes, S., Loo, J. A. & Rome, L. H. (Submitted).