Abstract
Nuclear magnetic resonance (NMR) spectroscopy is a powerful tool to investigate the structure and dynamics of RNA, because many biologically important RNAs have conformationally flexible structures, which makes them difficult to crystallize. Functional, independently folded RNA domains, range in size between simple stem-loops of as few as 10–20 nucleotides, to 50–70 nucleotides, the size of tRNA and many small ribozymes, to a few hundred nucleotides, the size of more complex RNA enzymes and of the functional domains of non-coding transcripts. In this review, we discuss new methods for sample preparation, assignment strategies and structure determination for independently folded RNA domains of up to 100 kDa in molecular weight.
Keywords: RNA, Isotope labeling, Structure, NMR spectroscopy
1. Introduction
RNA has emerged as an increasingly important player in gene expression and its regulation, but the often elongated and only partially ordered structures responsible for its function make crystallization challenging. NMR of RNA is also challenging because proton density is much smaller than in proteins [1–4], and the chemical diversity of its monomeric units much reduced, making spectral overlap more severe [1,5]. RNA resonances also have shorter transverse relaxation times, since most protons in RNA are 13C-attached and the shape more anisotropic than globular proteins [3]. These challenges become increasingly severe for larger RNAs (>50–70 nt). As a consequence, the total number of RNA structures (excluding 51 DNA/RNA hybrids) deposited into the PDB accounts for only ~1% of the total PDB depositions, while the human genome codes for many more functional RNAs than proteins. Of all RNA structures in the PDB, 484 have a size of <155 nucleotides (nts) and were determined by NMR, including one from solid-state NMR spectroscopy [6] (Fig. 1). Remarkably, this constitutes about 40% of all RNA structures reported in the PDB, while x-ray crystallography accounts for about 55% of the total. It must be noted, of course, that structures determined by x-ray crystallography are often larger and much more complex and therefore structurally richer.
Fig. 1. PDB statistics of deposited RNA structures as of Dec 2016.
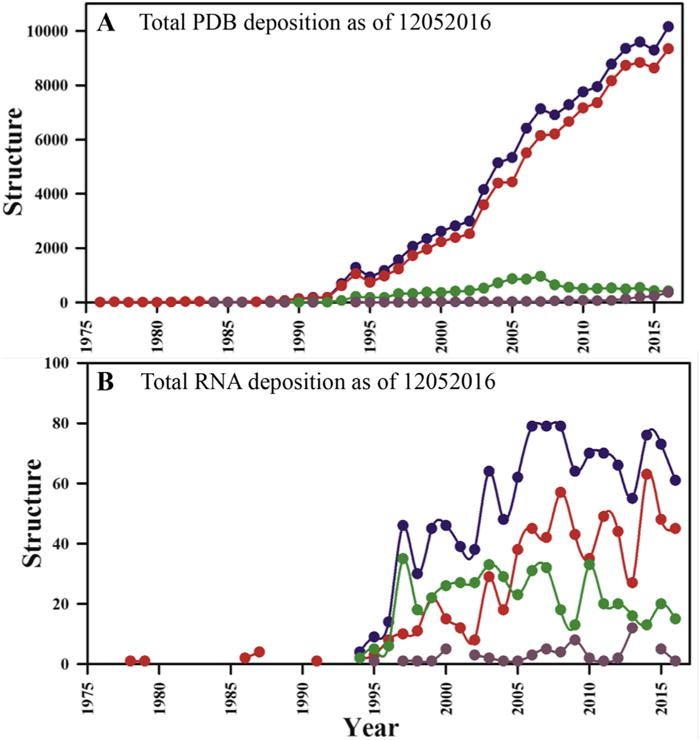
(A) Total number of PDB depositions per year and (B) total RNA structures deposited per year. Here, all reported coordinates are shown in blue, X-ray structures are in red, while NMR structures are in green. Structures determined with other techniques (e.g. electron microscopy; EM) are shown with dark brown filled circles. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Understanding the structural basis for the growing diversity of essential biological function of RNA requires an increase in the pace of high-resolution structure determination. In this review, we describe approaches to unravel the structure of folded RNAs that take advantage of recent developments in isotopic labeling schemes, higher magnetic field spectrometers and cryogenically cooled probes. We limit this review to techniques apt to investigate RNAs of up to about 200–300 nucleotides in size; larger RNAs can be studied by NMR, but formal structure determination with an acceptable density of experimental constraints remains an unmet challenge for RNAs greater than 100 nts. Many RNA-binding proteins recognize short single stranded RNAs of <10–15 nucleotides; or stem-loop or internal loop structures of 20–50 nts in size; most RNAs studied by NMR and deposited into the PDB belong to this group. Catalytically active small ribozymes, riboswitches and other important regulatory RNAs are less than 50 nts in size as well. More complex ribozymes and independently folded transcripts, such as tRNA, 5S RNA or 7SK RNA, and independent domain of non-coding transcripts as well, remain within the 200–300 nucleotide cutoff of this review.
2. Sample preparation
RNA can be prepared efficiently either by chemical synthesis or various in vitro enzymatic transcription protocols and purified by either polyacrylamide gel electrophoresis (PAGE) or, preferably, chromatographic methods [7,8]. Denaturing PAGE has been in use for many years [9,10]. It is very robust and provides nucleotide level resolution. The band corresponding to the RNA is excised from the gel and the RNA is extracted by electroelution, ethanol precipitation and extensive dialysis to refold the RNA and remove any leftover acrylamide. More recently, a set of chromatographic approaches has also been reported, which includes reverse-phase high-performance liquid chromatography (HPLC), anion-exchange HPLC, size exclusion chromatography and affinity based chromatography [11–14]. The chromatographic methods are faster, reduce sample losses and retain the RNA in its native conformation throughout, but it is not always possible to obtain sufficient purification of desired products from aborts.
2.1. Synthesis of small RNAs
2.1.1. Chemical synthesis
Small RNAs of up to 20 nts can be chemically synthesized and studied using two dimensional NMR. However, isotopic labeling may be required for highly repetitive sequences. Wenter et al. [15] reported a unique method for selective 13C labeling of RNA for the structure determination of protein-RNA complexes; however, their phosphoramidites are not commercially available [15]. One of the advantages of this approach for RNA synthesis is position selective labeling which is discussed at the end of the sample preparation section.
2.1.2. In-vitro transcription
RNAs of 20–60 nts can be prepared efficiently by in vitro enzymatic transcription [10] with T7, T3 or SP6 RNA polymerases (the first being the most commonly used) [16]. Transcription of RNAs of this size can be performed with chemically synthesized single-stranded or double stranded DNA templates comprising a polymerase promoter region. Two common promoter sequences for T7 are used in this laboratory, called class II and class III [17]; other RNA polymerases (T3 and SP6), require different promoter sequences. In vitro transcription based on T7 RNA polymerase often ends with 3′-inhomogeneity, which can be greatly reduced by incorporation of ribozyme sequences in the template in cis and trans [18–20] or by chemically incorporating 2′-O-CH3 nucleotides at the end of the template to reduce jitter of the polymerase [21,22].
2.1.3. Small ribozymes
An alternative approach places hammerhead (HH) ribozymes (or other self-cleaving small ribozymes) at the 5′-end of the DNA template to obtain efficient co-transcriptional cleavage during in-vitro transcription (Fig. 2A) [18] to release the desired sequence. Hammerhead ribozymes are a class of small self-cleaving/catalytic RNAs that perform self-cleavage at a well-established site within a folded structure. However, this method cannot be used to make RNAs <10 nts, especially AU-rich sequences, because the 5′-end of the target RNA must base-pair with the HH ribozyme and it is difficult to purify the cleaved RNA from aborted products.
Fig. 2. Methods for small RNA preparation.
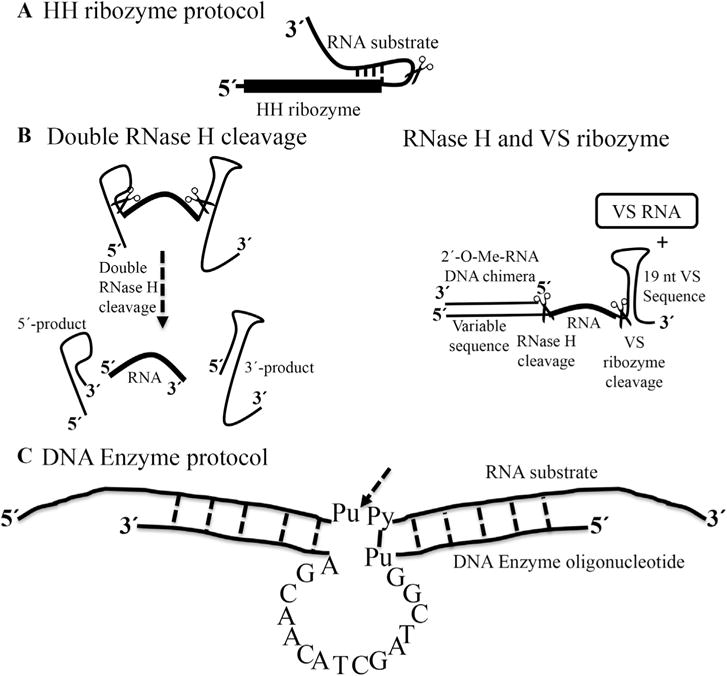
(a) Preparation of RNA based on the incorporation of an HH ribozyme at the 5′-end of the target RNA. The first 10–12 residues of the target RNA are complementary to a guide sequence in the HH ribozyme. The scissors indicates the cleavage site [18]. (b) Alternative methods; double RNase H cleavage and RNase H/VS ribozyme cleavage protocols [23] Both methods use a chimeric DNA sequence to anneal to the RNA template before RNase H cleavage. (c) Diagram of a DNA Enzyme of the “10–23” family shown with single letters core and two flanking regions of 5–12 nts on either side, designed to base-pair to the RNA substrate. The arrow indicates the cleavage site between unpaired purine (Pu) and paired pyrimidine (Py) in the RNA substrate [25].
Elegant works by Duss et al. to produce a small isotopically labeled RNA uses double sequence-specific RNase H cleavages and combined RNase H and Varkud satellite (VS) ribozyme cleavage working in trans [23] (Fig. 2B). The VS ribozyme is an RNA sequence of 154 nts derived from the VS RNA, which can perform both RNA cleavage reaction or the reverse, RNA ligation, depending on conditions. During the first RNase H cleavage, the 3′-end of the RNA is protected by the stem loop structure, to prevent any degradation due to the presence of impurities in the buffer or co-purified with the recombinant proteins. The first double RNase H cleavage approach requires two 2′-O-Me-RNA/DNA chimeras specifically hybridizing to the desired cleavage site on the RNA, while the latter approach based on RNase H/VS ribozyme cleavage requires one 2′-O-Me-RNA/DNA chimera and VS ribozyme in trans [24]. This method is not limited to small RNAs, but can be used to prepare longer RNAs by segmental labeling, as discussed in later paragraphs.
Finally, an RNA-cleaving DNA Enzyme (DNAzyme) [25,26] can also be used to prepare isotopically labeled RNAs of various lengths. The DNAzyme contains a small catalytic core of 15 nts that anneals to RNA targets by base pairing of two flanking arms of 6–12 nts (Fig. 2C) and cleaves the RNA at a specific position upon addition of divalent ions [27]. Altogether, these different methods provide alternatives to chemical synthesis to produce short single stranded RNAs that are typically studied in complex with proteins.
2.2. Divide and conquer
Structure determination of RNAs longer than about 50 nt, requires non uniform isotopic labeling strategies. The synthetically simplest approach to study large RNAs where spectral overlap and fast relaxation are limiting is by using a divide and conquer approach. Long extended structures such as stem-loops often have no interactions between separate segments, and even complex multi-stem-loop structures do not always have interactions between distinct stem-loops [28,29], as can be established by comparison of spectra of isolated fragments, or from linewidth/relaxation rates (in the absence of long range interactions, domains tumble independently, leading to inefficient relaxation and sharp NMR lines, as observed in multi-domain proteins [30]). In this case, there is no reason to pursue any of the much more demanding and less efficient labeling strategies described below.
We illustrate this simple but effective strategy using our recent work on a 110 nts RNA thermometer from Neisseria meningitides [31], of which 70 nts comprise the functional and folded core. This approach allowed us to obtain complete assignments and a large number of experimental constraints that would have been impossible if we had studied the complete RNA due to its large asymmetry leading to short T2 relaxation and spectral overlap. We divided this RNA core of around 70 nucleotides into three overlapping segments (Fig. 3A), which generate the complete RNA when the base-pairs are superimposed and correspond to the upper helix, lower stem and overlapping middle stem, respectively. Overlay of the NOESY spectra of the three segments on the spectrum of the full RNA revealed a highly transferable pattern of chemical shifts and NOESY cross-peaks for both the exchangeable and non-exchangeable sections of the NMR spectrum (Fig. 3B), demonstrating that there are no interactions between distant elements of the RNA. Spectral assignments and experimental constraints were obtained on the smaller fragments and transferred to generate the structure of the full RNA.
Fig. 3. NMR Assignment strategy for an RNA thermometer of ~70 nucleotides.
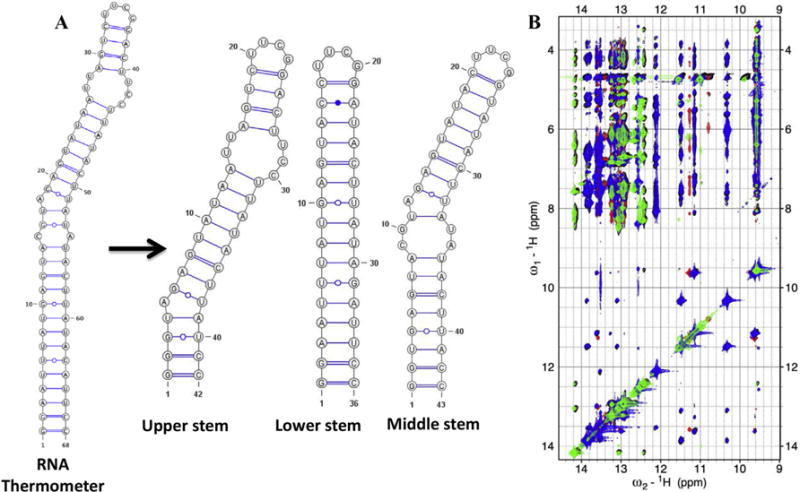
(A) Three smaller constructs were prepared; the first corresponds to the upper stem, the second overlaps with the lower stem while the third is representative of the middle region of the RNA. (B) An overlay of the imino regions of 2D 1H–1H NOESY spectra. The black color represents the spectrum for the complete RNA, while green, blue and red are from the fragments, upper, lower and middle stem, respectively. The spectra of the fragments match the full RNA almost perfectly. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Nucleotide-specific labeling strategies
Once the size of the RNA exceeds about 50 nts, spectral overlap in the sugar region makes resonance assignment challenging and slow asymmetric tumbling (leading to efficient the 1H–13C dipolar relaxation) increases proton and carbon linewidths making coherence preservation difficult. Unlike proteins, full deuteration is not a viable option for large RNAs, because there are too few NH’s to establish any structural property beyond base pairing. Selective base labeling by residue or resonance type leads to simpler NMR spectra and only requires one of many labeled nucleotide triphosphates (NTPs) prepared by chemical or chemi-enzymatic methods, mixed in with the remaining unlabeled NTPs during in vitro enzymatic transcription [31–35]. For example, multiple nucleotide-specific 13C, 15N labeled and specific deuterated nucleotide labeled samples were prepared to investigate the structures of the 56 nts tRNApro; the 68 nts U5 primer binding site (U5-PBS) domain of murine leukemia virus (MLV) RNA; the 63 nts pseudoknot from MLV and the 68 nts core of the RNA thermometer from Neisseria meningitidis [31–33,36].
This approach may include the preparation of RNA with site-specific (H6/H8, H1′, H2′, D3′, D4′, D5′/D5″ and D5) ribose [37,38] (Fig. 4) and base deuteration [2,28], which simplify the spectra and reduces dipolar relaxation considerably. The most extensive overlap in RNA spectra occurs for ribose protons. The H2′ to H5″ protons resonate between 4 and 5 ppm, and their carbons at 62–85 ppm, respectively. Deuteration of the H3′, H4′ and H5′/5′ positions in the sugar and H5 position in C/U can be done straightforwardly using commercially available NTPs [37,38]. This method was successfully applied to determine several interesting RNA structures [31,39].
Fig. 4. Effect of site-specific ribose deuteration on the quality of the NMR spectra of an RNA thermometer.
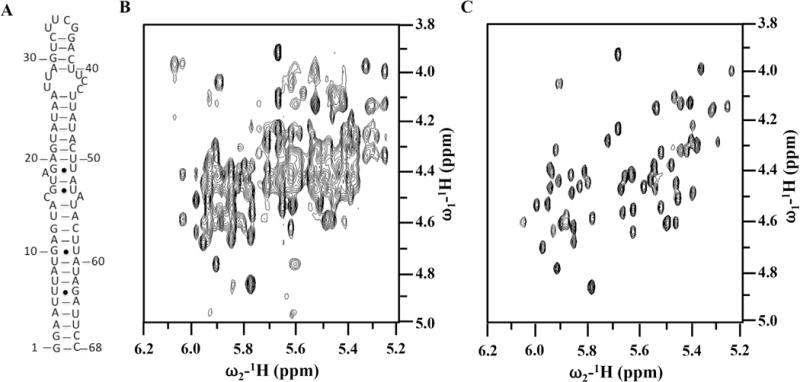
(A) secondary structure of the ‘core’ thermometer. (B) and (C) are 2D 1H–1H NOESY spectra recorded in D2O for completely protonated (B) or with H6/H8, H1′, H2′, D3′, D4′, D5′/D5′ and D5 ribose deuteration, respectively, at 25 °C and 600 MHz. Spectral simplification is remarkable.
Site-specific deuteration for spectral simplification of the base region of the NMR spectrum was also done for purine H8 and pyrimidine H5’s [2,28]. Purine C8 deuteration distinguishes adenine H8 from guanine H8 and adenine H2 [2], whereas deuteration of the C5 position of pyrimidines remove the strong H5–H6 cross-talk in 2D 1H–1H NOESY and TOCSY spectra [28]. For example, in the 48 nts K10 transport signal RNA, many cross-peaks otherwise obscured by the H5–H6 protons were rendered visible by selective deuteration [40], and we have had similar very positive experiences with several other RNAs in the 50–70 nts size range.
The above-mentioned methods have been extensively used and extended by Summers’s group [28,36,41,42] to accomplish the assignment of very large RNAs from the HIV genome. Perdeuteratated, fully protonated and partially deuterated samples were often combined with segmental labeling schemes (as described next) to overcome the ambiguous assignment problems in large RNAs, such as the 101 nts moloney murine leukemia virus (MoMuLV) RNA [36,42], the 131 nt double kissing hairpin of the MoMuLV 5′-untranslated region RNA [43] and the 155 nts region of the HIV-1 RNA packaging signal [28,29].
Unlike just 5 or 10 years ago, there are now a very large number of selectively labeled nucleotides that have been prepared and are often commercially available from Cambridge Isotope Laboratories, Sigma-Aldrich and Silantes. The development of these methods was very important and availability of selective nucleotide labels does not limit the spectroscopy. The Dayie’s group [34,44–47] used chemi-enzymatic synthesis of NTPs exploiting enzymes from pentose phosphate pathway to synthesize many specifically labeled nucleotide [48]. This approach can, in principle, be used to generate any type of specific labeling pattern, though these NTPs are not yet commercially available. The group also reported the preparation of selectively labeled 6-13C-pyrimidine RNA phosphoramidites, which can be used to incorporate 6-13C-uridine/cytidine residues site-specifically in the RNA [35,49].
2.4. Segmental labeling
This is the most general method to simplify an RNA without affecting its structure and is required when there are interactions between distant regions of an RNA that make divide and conquer impossible. Although this method does not solve the relaxation problem, it reduces the number of overlapping resonances in heteronuclear NMR experiments and can be used to obtain specific labels (e.g. post-transcriptional modifications) or labels for paramagnetic relaxation enhancement studies (PRE) [50]. The early approaches of Kim et al. [51] and Tzakos et al. [52] used T4 RNA ligase to link together free 3′-hydroxyl and 5′-phosphate groups and HH ribozyme to segmentally label an RNA (Fig. 5A–B). Two different transcription reactions were performed, the samples were purified and the two segments were ligated using T4 RNA ligase. The 5′-HH ribozyme generates a free 5′-hydroxyl terminus for the 5′-fragment, while a 3′-HH ribozyme yields the 3′-fragment with monophosphates, ready to be ligated. This process was streamlined by the construction of a plasmid containing a T7-promoter followed by the 3′-fragment and its 3′-HH ribozyme in cis. The 3′-HH ribozyme is connected to the HH-ribozyme of the 5′-fragment via a flexible sequence followed by the 5′-segment of the RNA. Transcription off the linearized plasmid from this construct yields two RNA fragments with desired 3′-monophosphate for the 3′-region and 5′-hydroxyl group for the 5′-region, respectively [52]. NMR structural studies of domain II of the internal ribosomal entry site (IRES) of Hepatitis C virus (HCV) [53] and the 74 nts BC1-DTE RNA were performed with this approach [52].
Fig. 5. Segmental labeling of RNA.
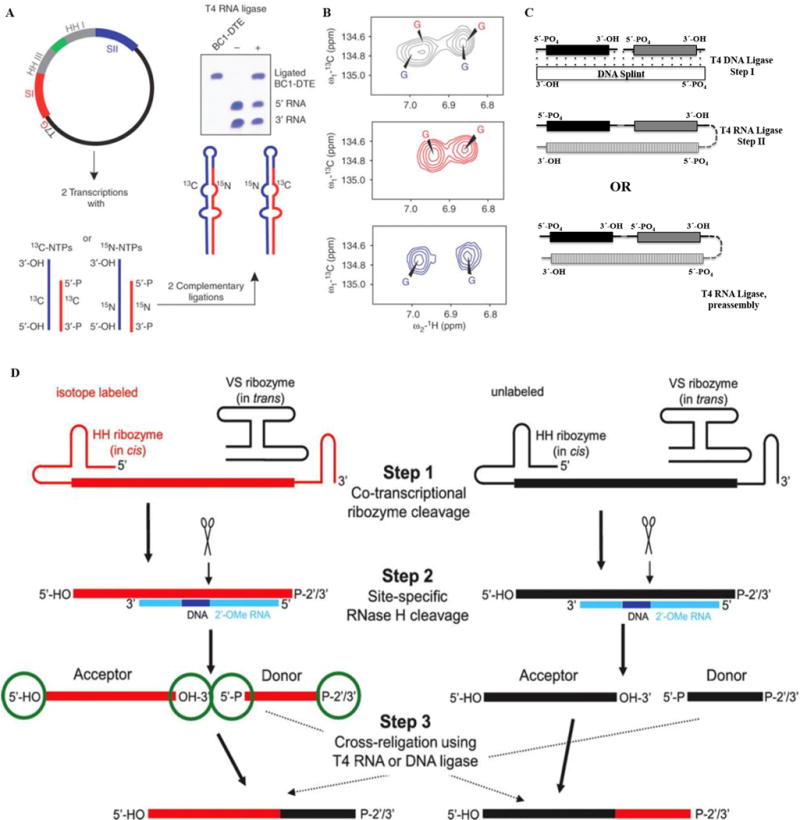
(A–B) Overview of the preparation of a segmentally isotope labeled RNA. Here (A) shows the construction of a plasmid where 3′-HH and 5′-HH are engineered. Panel (B) shows the simplification of aromatic regions of 1H, 13C-TROSY spectra for the RNA, the top panel shows spectra for uniformly 13C/15N-labeled RNA (black), middle for 5′13C- and 3′15N-labeled RNA (red) and the bottom spectrum corresponds to 5′15N- and 3′13C- labeled RNA (blue). The figure is taken from Tzakos et al. [54]. (C) Segmental labeling approach based on T4 DNA ligase, DNA splint and T4 RNA ligase adapted from Nelissen et al. [55]. The segmentally labeled RNA can be prepared either in single or multi-step ligations to obtain the desired product. Here, dark black, grey and vertically patterned fill colors represent three RNA fragments to be joined either in a two-step ligation based on T4 DNA ligase/DNA splint/T4 RNA ligase or single step ligation by T4 RNA ligase. Each fragment can be differently labeled or unlabeled. (D) A newly developed segmental labeling approach from Duss et al. [24] based on HH/VS ribozymes, 2′-O-methyl RNA/DNA chimera, RNase H and T4 DNA or T4 RNA ligases. In the first step, HH/VS ribozyme cleavage occurs co-transcriptionally, followed by site-specific RNase H cleavage in step II. T4 DNA/RNA ligase based ligation is done in step III. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Another approach uses T4 DNA ligase and T4 RNA ligase 1 ligations [55] and multiple segmental labeling can be achieved in a single step ligation reaction (Fig. 5C). This innovative method can swiftly generate RNAs with multiple choices of labeling.
Finally, the method developed by Duss et al. [24] can also be utilized to segmentally label RNA. This method is an extension of what has been discussed before, i.e. RNase H and VS ribozyme cleavage to generate shorter fragments. Here, a HH ribozyme is included upstream of the variable sequence site (Fig. 2B). After ribozyme and RNase H cleavage steps, the acceptor and donor from differently labeled/unlabeled RNAs are cross-ligated using T4 RNA ligase. This method provides a much better yield than previous approaches and requires less time to prepare segmentally label samples (Fig. 5D).
2.5. Position selective labeling
A very powerful but very laborious position-selective labeling of RNA (PLOR) method addresses the limitations of base selective labeling methods [56]. A hybrid solid-liquid phase transcription method and automated robotic platform was used for the synthesis of RNAs with position selective labeling and was demonstrated for the 71 nts aptamer domain of an adenine riboswitch. Various single position isotope labels and 19F labeled RNA samples were prepared to study the conformational states of riboA71, but the method remains to be used widely because of its complexity and the specialization of the experimental apparatus. While in principle this method is extremely powerful, in practice segmental labeling remains far from accessible to most groups.
Altogether, advances in synthetic strategies allow the preparation of samples with a wide range of labeling patterns and segmental labeling strategies, but these methods are still relatively inefficient, laborious and sequence dependent. The reagents (NTPs) are relatively expensive and the protocols require high levels of specialized skills and familiarity with the protocols.
3. Resonance assignments
The poor dispersion in chemical shift makes it difficult to obtain unambiguous resonance assignments in RNA [1,2] especially in the ribose proton region (Fig. 6). The use of 13C/15N/2H and selective/segmentally labeled sample made it possible to utilize heteronuclear triple resonance scalar correlated experiments, as for proteins [57], and establish intranucleotide correlations between base and sugar, which dramatically reduces the ambiguity present in NOESY experiments, but these approaches provide much more limited information than comparable experiments in proteins. Furthermore, many experiments have been proposed to facilitate assignments of RNA but they rapidly lose sensitivity with increasing molecular weight. Thus, resonance assignments for RNAs >40–50 nucleotides rely primarily on NOE-based experiments. The bulk of information for RNA spectral assignments originates in NOESY spectra, but different labeling strategies suggest different assignment protocols, as discussed below [1,16,58].
Fig. 6. Chemical shifts for nuclei in RNA from the BMRB data bank.
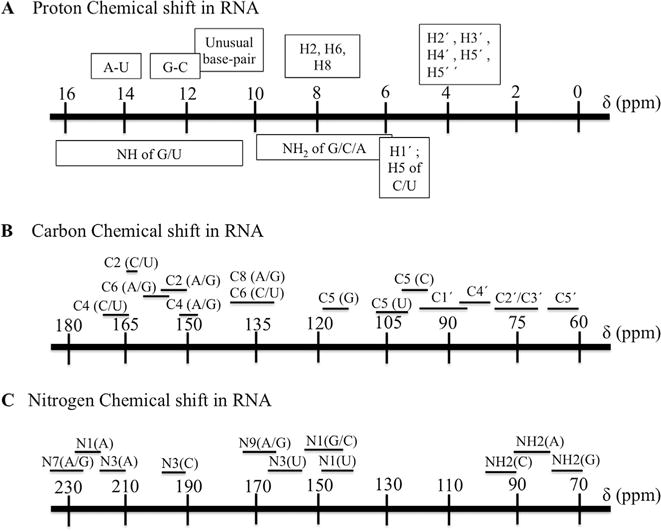
Proton (A), carbon (B) and nitrogen chemical shift ranges (C). The most extensive overlap is seen for the sugar H2′, H3′, H4′, H5′, H5′.
Resonance assignments in RNA are performed through the identification of sequential base to ribose NOE with patterns typical of helical regions. A consistent set of NOE patterns establishes a sequential walk (Table 1), within which the sequence can be read since Purine and Pyrimidine can be easily distinguished; Ura/Cyt can be differentiated from the carbon 5 shift on the base; the AdeH2 resonance is unique. These connections can either be observed in 2D spectra if the sample is unlabeled or in 13C-edited 3D NOESY’s. We typically start assigning exchangeable protons in the base pairs; then use helical NOE patterns to establish sequential connectivities with non-exchangeable protons; finally tackle the remaining single stranded nucleotides, or nucleotides in non-canonical structures, by a process of elimination and taking advantage of sequential NOE connectivities that are always present between neighboring nucleotides and within each nucleotide, regardless of secondary structure.
Table 1.
Typical NOE patterns observables in 1H–1H NOESY spectra for a double helix in the A-form RNA conformation typical of RNA helices [1,59].
| 1H–1H NOE | Sequential | Intra-nucleotide |
|---|---|---|
| H1′-H8 | Weak | Weak |
| H2′-H8 | Strong | Weak |
| H3′-H8 | Medium | Medium |
| H2′-H1′ | Weak | Strong |
| H6–H5 | Weak | Strong |
The following steps are taken to assign an RNA spectrum.
Exchangeable imino (and amino) resonances from 1H–1H NOESY’s (or the 15N-edited 3D versions) recorded in water identify the base pairs (each base paired nucleotide provides a protected NH and several NH2 peaks to the spectrum). 1H-15N correlations can be observed in 2D 1H-15N HMQC [60] and 3D 1H-15N NOESY-HMQC spectra [61] to assign the 15N resonances and distinguish individual bases from each other.
Cross-peaks between exchangeable and non-exchangeable protons are very useful for assignments and important for structure determination. The NOE cross peak between Ura NH and Ade H2 is very strong in a base pair, but NOEs are often seen from Gua NH to H5 and H6 in Cyt as well. Cross-strand NH2 to H1′ peaks are also observed in A-form RNA, analogous to the Ade H2 to H1′ peaks characteristic of this helical structure.
Sequential resonance assignments require 1H–1H NOESYs recorded in D2O (and their 13C-edited 3D versions), plus 1H–1H TOCSYs. TOCSY spectra identify the H6–H5 peaks of Cyt and Ura and establish the sugar conformation in RNA based on different intensities of cross peaks for A- or B-form sugar conformations. Through bond base-sugar correlations can be observed with HCN [62], HCNH [63] and HCNCH [64,65] experiments, and are particularly useful in regions with non-helical conformation.
The 31P resonances can sequentially correlate neighboring nucleotides H3′-P-H4′/H5′/H5′ peaks and require 1H-31P HETCOR/HETTOC spectra [66–68], or their 3D analogues (1H,13C,31P HCP/PCH/PCCH-TOCSY/HPHCH) [69–73] spectra recorded in D2O, but these experiments rapidly lose sensitivity for RNAs larger than 20–30 nucleotides because of fast relaxation of the 31P nuclei, especially at higher magnetic fields, due to large chemical shift anisotropy.
The severe overlap of ribose proton resonances requires 13C/15N/2H and selective/segmentally labeling for RNAs larger than about 40 nts. Assignments are facilitated by correlated experiments to identify each of the ribose spin systems; namely 3D HCCH-COSY [74] and 3D HCCH-TOCSY [69,75] and variations of these experiments.
Through bond H2–H8 correlations can be obtained by HCCH-TOCSY/1H-13C HMQC recorded in D2O. HCCH-based experiments optimized for the bases are very useful to assign the ribose, Cyt/Ura H5/H6 and adenine spin systems [60,69,75]. A TROSY relayed HCCH-COSY pulse sequence can be used for correlating H2–H8 resonances of adenine in the large RNA [76].
Through-bond exchangeable and non-exchangeable base protons can be correlated by 3D HNCCH/HCCNH [77,78] spectra in water. These experiments are rather insensitive for larger RNAs. An alternative is based on triple-resonance two-dimensional H5(C5C4N)H experiments where through-bond H5 to imino/amino protons in pyrimidines can be identified for 13C, 15N-labeled RNA [79].
An illustrative study (RNA thermometer from N. meningitidis)
This is an RNA stem-loop of about 70 nucleotides whose secondary structure is illustrated in Fig. 5A. Base paired residues an exchangeable protons were identified from imino-imino, Cytidine amino-Guanine NH1 and Adenine H2-Uridine H3 patterns in 2D 1H–1H-NOESYs (100–150 ms mixing time) acquired at both 7 °C and 15 °C. Sequential assignments of non-exchangeable protons followed typical H1′-H6/H8 NOE patterns observed in 2D 1H–1H NOESYs acquired in D2O at 25 °C (120 ms mixing time) (Fig. 7). Further validation and structural restraints were provided by H2′-H6/H8 and H1′–H2′ NOEs of samples incorporating rNTPs deuterated at the ribose H3′, H4′, and H5′/5″ and pyrimidine base H5. Pyrimidine H5/H6 resonances were identified from 1H–1H TOCSYs (70 ms) acquired in D2O. Correlated 15N and 13C chemical shifts were determined from 1H-15N HSQC, 1H-13C constant time (CT) and non-constant time HSQC, and 1H-13C edited NOESY-HSQC experiments (100–150 ms mixing time). Non-exchangeable protons at the H1′, H2′, H5, H6, H8, and H2 positions; the exchangeable imino protons and the corresponding nitrogen atoms, the exchangeable NH2 protons, and H3′ proton resonances were all obtained, but carbon resonances were not fully assigned because increased relaxation broadened many peaks in the correlated spectra. Altogether, we could assign ~56% of non-exchangeable protons (most of H1′, H2′, H5, H6, H8, H2, and ~41% of H3′) and ~46% of the exchangeable protons excluding H2′ hydroxyls (~82% of imino protons), and ~68% of the imino nitrogens.
Fig. 7. Typical H1′-H6/H8 assignment ‘walk’ for an RNA thermometer.
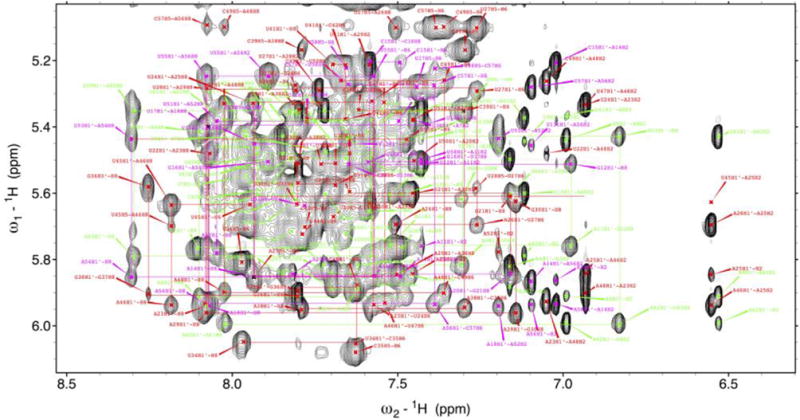
Spectral assignments were confirmed using the smaller constructs of Fig. 5 which are shown with various colored lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Structure determination
Structure determination requires collecting the NMR structural constraints, followed by structure calculation, refinement and validation as outlined below.
4.1. Structural constraints
-
(a)
H-bonds: Hydrogen bonds (H-bonds) define the secondary and tertiary structures of nucleic acids. They are powerful, highly informative constraints that are relatively easy to obtain (compared to proteins), numerous (compared to proteins) and that restrict the conformational search significantly. Although it is generally true that the density of constraints in RNA is lower than in proteins, the number of hydrogen bonding constraints is generally larger by a factor of 2 or 3 per residue, and these constraints are much more powerful than NOEs because they correspond to narrow distance ranges and constrain the bases to linear and highly planar arrangements [80].
The presence of a hydrogen bond can be identified by detection of imino protons, which are well-protected from exchange with solvent when base paired, and confidently established by scalar couplings across H-bonds in HNN-COSY type experiments [81]. These two bond couplings of 6–7 Hz allow magnetization transfer to correlate chemical shifts across base pairs. They are generally not necessary (though advisable) in Watson-Crick paired established by NOESY walks, but should be used to validate non-canonical pairs, base triples, or tertiary interactions. The utility was shown in several cases, such as the adenine–sensing riboswitch [82] where tertiary interactions were mapped using HNN-COSY experiment; a guanine binding riboswitch where an uridine imino proton to N3 nitrogen of hypoxanthine is observed [83]; the observation of non-canonical interactions in a frameshifting pseudoknot with adiabatic HNN-COSY [84]. The sensitivity of these experiments has been optimized by the use of BEST relaxation enhancement and TROSY for longer RNAs [85].
However, both set of experiments only detect relatively stable base pairs and often miss weaker base pairs broadened by exchange as the base pairs ‘breathe’ and open with thermal fluctuations. Dallman et al. [86] have developed a very sophisticated way of detecting J-coupling across H-bonds in weak base pairs that uses a long-range HNN-COSY experiment to detect A:U base pairs where the adenosine N1 nucleus is linked to the H2 proton and the uracil N3 nucleus is linked to the H5 proton. This experiment has 8-fold sensitivity improvement. In addition, a sensitivity improved pyrimidine H(CC)NN-COSY experiment allows to simultaneously detect A:U and G:C base pairs. This experiment uses an out-and back band-selective magnetization transfer from the H5 of U/C to C5 to C4 to N3 and across the H-bond. These experiments allow the detection of weak H-bonds, but by no means eliminate all false negatives. Another interesting experiment observes H-bonds of backbone phosphate groups in non-canonical structures, based on the long range 1H,31P HSQC experiment [87].
In our case study, a total of 110 H-bonding constraints were used for structure calculation. These constraints were primarily based on the observation of JNN couplings from HNN-COSY experiments; however, restraints for weak base pairs (A-U, A-C, and U-U base pairs) were also incorporated based on characteristic NOE patterns observed in 1H–1H NOESYs. Potential A-U base pairs showing typical cross-strand H1′-H2, sequential H1′-H2, and strong AH2-UH3 imino NOEs were considered base paired despite the lack of HNN-COSY transfer, due to the broad cross-peaks caused by fraying of the bases.
-
(b)
NOE constraints: For structure determination of RNA by NMR, proton to proton distances of up to 5–6 Å play the major role. We classify distance constraints as strong (0–3.0 Å), medium (0–4.0 Å), weak (0–5.0 Å) and very weak (3.5–6.5 Å) based on the comparison of peak intensities with peaks corresponding to known interproton distances.
In principle, >60 distances per residue can be measured in RNA, but many are uninformative, being constrained by the covalent structure to within narrow ranges. Nevertheless, it is important to collect these constraints for completeness and to confidently identify informative NOEs; we consider this task to be an important metric of the completeness of the spectral analysis. Some intranucleotide NOEs are also very informative, as they establish the glycosidic angle (e.g. NOE’s involving the base and H1′ protons), sugar conformation (base and H2′, H3′ protons) and β/γ backbone torsion angles (base and H5′-H5″ protons). These NOEs are not difficult to obtain and should be examined with particular care.
The most important information for RNA structural information comes from sequential sugar-to base, base-to-base and sugar-to-sugar distances that establish the base stacking and indirectly define the conformation of the phosphodiester backbone. These NOEs account for approximately 20% of all NOEs observable in RNA. A high-quality NMR-derived RNA structure should include about 10 internucleotide sequential constraints, although as many as 15–20 per residue are observable in shorter RNAs of 30–40 nucleotides. Fewer constraints would leave the structure under-determined as demonstrated by an extensive simulation 20 years ago [5] and confidence in the structural conclusions decreases rapidly below these thresholds. A combination of base-type selective labeling and 2D filter/edited NOESY should be used to increase the number of internucleotide NOEs, especially in large RNAs [88].
Longer-range NOEs in RNA are much fewer than sequential interactions, very seldom more than 10 per residue, though 5 are not impossible to observe, but they are extremely important to establish the correct helical structure, the presence of unusual base pairs and especially any long range tertiary interactions. Many such constraints involve base resonances (especially NH’s, NH2 and H2) but some can also involve sugar protons.
For the example of the RNA thermometer, we were able to obtain a total of 920 NOEs, which included 212 intra-residue, 531 sequential and 177 non-sequential NOE restraints derived from NOESY spectra. These correspond to about 8 sequential NOE per nucleotide, and about 3 long distance NOEs per nucleotide.
-
(c)
Dihedral angle constraints: RNA structure is uniquely defined by 6 backbone dihedral angles (α, β, γ, δ, ε and ζ), five ribose torsion angles (ν0–ν4) and one glycoside torsion angle (χ) per nucleotide but the sugar conformation is described in terms of the amplitude of non-planarity of one of the ribose carbons, and its identity because of the sugar stereochemistry. The sugar conformation and population of the major C2′-endo and C3′-endo conformers, which are most common, are estimated from the observed peak splitting in DQF-COSY or peak intensities in 1H–1H TOCSY. For double helices, it is safe to assume A-form torsion angles for β, γ, and ε, but nucleotides with atypical NOE patterns or those that appear to experience conformational exchange are left unrestrained. Dihedral angles can be established from a careful analysis of correlated experiments [1], but these experiments are difficult to execute beyond approximately 40 nucleotides. Some torsion angles can also be constrained from NOE intensities to base protons and the glycoside torsion angle (χ) is constrained in the anti or syn conformation on the basis of H1′-H5/H6/H8 NOE intensities.
-
(d)
Long-range distance constraints (PRE): Paramagnetic relaxation enhancement (PRE), arising from unpaired electrons, generates pronounced and long-distance effects in NMR spectra [50,89]. The unpaired electrons (paramagnetic species) introduced into an RNA (or protein in complex with RNA) increases significantly the relaxation rates of NMR resonances in their vicinity. The effect is proportional to the inverse sixth power of the distance between the paramagnetic label and the reporter nucleus (up to about 30 Å). Comparison of the intensity of peaks in spectra recorded with the oxidized and reduced species provides relative internuclear distances, which can be incorporated in structure calculation and refinement in the same way of NOE-based distance constraints.
In the late 90s, our group attached a proxyl spin label tag by derivitizing a 4-thiouracil base to supplement intermolecular distance restraints when studying the interaction of a stem-loop RNA with an RNA-binding protein [90]. This approach remains the most popular spin label tag for detecting PREs in RNA, but a recent review includes other paramagnetic tags used for RNA and proteins; spin-labeling strategies for RNP complexes, and common ways of utilizing PRE information for RNA and RNP complexes [91].
-
(e)
Global orientational constraints (RDCs): Residual dipolar couplings (RDCs) arise when a molecular system is partially ordered and report on the inter-nuclear bond vector orientation with respect to the external magnetic field [92,93]. They carry local angular information and define the global orientation of the molecular system [94]. RDC’s are particularly useful for structural studies of RNA, because the usually elongated shape of RNA require global information to better define the structure. RDC information can also be used to observe low-population intermediates, as recently demonstrated with HIV-TAR RNA [95], and as has been done with proteins for some time [96–98].
For long, highly anisotropic duplex RNAs, RDCs might be observed due to their intrinsic magnetic susceptibility anisotropy, which leads to spontaneous partial orientation [99]. In general, however, the RNA sample needs to be oriented in an alignment medium to generate RDC’s of measurable magnitude. Out of many alignment media developed for macromolecular systems, Pf1 filamentous phage suits nucleic acids very well, because its highly negative-charged nature avoids sample precipitation caused by strong interaction with the medium [100]. Other media, like bicelles and polyacrylamide gels doped with negatively charged reagents, together with poly (ethylene) glycol ether provide more options for alignment of RNA [101–103]. However, our group found that RNA tends not to align independently in different media due to its uniform electrostatics and elongated shape, limiting the usefulness of using multiple alignment media [104].
Several practical reasons (fixed bond length reduces the complexity of data interpretation, large 1H gyromagnetic ratio provides large RDC value hence accuracy, and well-resolved distribution of resonances), compel the use of only one-bond 1H-15N and 1H-13C bond vectors. In RNA, these correspond to imino 1H-15N, aromatic 1H-13C and ribose 1H1′-13C1′ pairs in RNA; the other sugar resonances are often too over-lapped for this use, with some exceptions. For small to medium size RNAs, where spectral overlap is not severe and line width is relatively narrow, 1H-15N and 1H-13C RDCs can be simply measured using coupled HSQCs, by subtracting the splitting measured in an isotropic sample (scalar coupling J only) from the splitting measured in the anisotropic sample (the sum of scalar coupling J and residual dipolar coupling D). As the size of RNA increases, however, the already-crowded 2D spectra will contain twice as many resonances in a coupled HSQC. Then IPAP experiments (in phase-antiphase [105]) in which the up-field and down-field components are displayed in separate spectra by adding or subtracting interleaved experiments with different phase cycles, alleviate the spectral crowding. Due to the large carbon spectral widths and differing 13C-13C coupling constants, it is best to record aromatic and ribose RDCs in separate experiments and to measure RNA 1H-13C RDC values in the direct dimension [106–108]. It is advisable to use PEG (C12E5/hexanol) media [103] rather than phage Pf1 for RNA-protein complexes, since RNA-binding proteins tend to interact with negatively charged phage Pf1.
For larger RNA systems with dominant TROSY effects, experiments selecting the sharpest component of the downfield doublets are required. A recently developed TROSY-based method called ARTSY (Amide RDCs by TROSY Spectroscopy) [109,110] is intensity-based, and overcomes the lineshape distortions caused by undesired homo-nuclear coupling that generates errors in frequency-resolved experiments. It was firstly designed for amide 1DNH in large perdeuterated proteins, but several modified versions of the pulse program to detect base-paired 1DNH and aromatic 1DCH in nucleic acids have been released on Bax’s website (https://spin.niddk.nih.gov/bax/pp/). The J splitting is extracted from the relative intensity observed in two spectra where 1JNH (1JCH) dephasing is either active during the full interval (attenuated experiment) or only during the half interval (reference experiment) in a TROSY INEPT transfer. Unlike the conventional way of recording 1H-13C correlation spectra in D2O, ARTSY allows RNA samples to be examined in H2O and takes advantage of band-selective BEST method and reduced viscosity compared to D2O, 1H-15N and 1H-13C correlations are enhanced and the spectral resolution is also improved. This method is especially promising for collecting RDC data on RNP complexes. 1H-15N RDCs for proteins and RNA and aromatic 1H-13C RDCs for RNA can be collected in a single sample, eliminating the errors arising from sample differences.
RDC values can be incorporated as initial restraints in structural calculation or at later stages of structural refinement. They can also be used to validate a structure by comparing experimental and back-calculated RDCs from the calculated structure. In practice, for RNA structure, the RDC information is often included at the refinement stage due to the difficulty of defining an accurate alignment tensor prior to structure calculation (limited measurable bond vectors and non-randomly distributed orientations). Therefore, the practical strategy to refine an RNA structure with RDC data is to use grid search methods that float the alignment tensor by varying rhombicity values and the corresponding magnitude around the initially estimated value, calculate structure to compare the difference between experimental and predicted RDC’s from the structures, until satisfactory correlation between experimental and back-calculated RDCs is reached [111,112].
-
(f)
Long range distance and orientational constraints define global properties of RNA structure: The use of RDC and PRE in structure calculation and refinement increases accuracy and precision. We demonstrated this approach in studying an RNA-protein complex [112,113]. The inclusion of RDC data during refinement defined the relative orientation of the two helical stems, and the new structure satisfied a larger set of experimental data than without RDC’s. In this recent work by our group on the investigation of long-range interaction in complex of pre-miRNA-20b and Rbfox protein, we attached a paramagnetic spin-label tag to a bulged thiouridine and were able to detect an interaction between the C-terminal tail of Rbfox RRM and the distant lower stem of pre-miRNA-20b (Fig. 8), providing further insight into the structural basis of inhibition of precursor miR-20b processing by Rbfox2 [114]. In the case study of the RNA thermometer, RDCs were measured using the ARTSY pulse sequence [109]. A total of 135 RDCs were used for the structure calculation, which is a large number of additional and highly informative constraints.
Fig. 8.
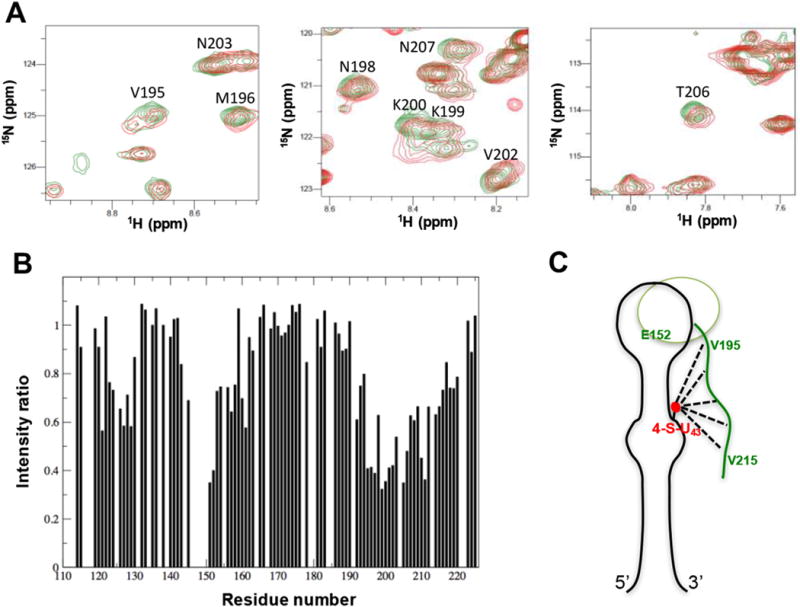
Long-range interactions between the C-terminal tail of Rbfox2 RRM and the lower stem of pre-miR-20b detected by PRE. (A) Overlays of 1H-15N HSQC spectra of 15N-labeled Rbfox2 RRM in complex with paramagnetic spin-labeled pre-miR-20b before (red) and after (green) reduction of the spin label introduced at position U43. Some resonances from the C-terminal tail of the Rbfox2 RRM, which are broadened by the paramagnetic spin label of pre-miR-20b, are annotated. (B) Intensity ratios of NH cross-peaks from the Rbfox2 RRM in complex with pre-miR-20b, between paramagnetic and diamagnetic forms. Residues from the β2β3 loop (around Glu152) and the C-terminal region (Val195–Val215) show significant depressions, indicating long-range contacts between the bottom part of the RNA and the protein. (C) Cartoon representation showing how the highly conserved C-terminal tail of the Rbfox2 RRM can reach the bottom part of the stem-loop to provide additional intermolecular interactions. The figure is adapted from the supplementary information in Ref. [114]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The most beneficial application of RDC’s and PRE’s occurs in defining the global tertiary fold of large RNAs by defining the relative orientation and position of different RNA helices that compose this structure. While this method has been demonstrated for tRNA many years ago [115], it has yet to be applied to describe higher order RNA structures and conformational rearrangements that accompany the assembly and disassembly of cellular RNPs. We anticipate this approach could lead to very significant insight on the structural basis of RNA function and the mechanism of RNP assembly and disassembly.
-
(g)
Fluorine probes: If an RNA is labeled with 19F at specific sites during in-vitro enzymatic transcription with appropriate NTPs, it is possible to obtain additional experimental constraints. 19F is nearly 100% naturally abundant and nearly as sensitive as proton (83% of 1H). The van der Waals radius of fluorine is also comparable to 1H (1.4 Å for 19F against 1.2 Å for 1H). Several groups have reported the synthesis of 2-fluoroadenosine-5′-triphosphate (2F-ATP) [116], 5-fluorouridine-5′-triphosphate (5F-UTP) and 5-fluorocytidine-5′-triphosphate (5F-CTP) [117,118] for use in in vitro enzymatic transcription. Kreutz et al. [119] reported the use of 2′-F labeled ribose in the RNA to investigate secondary structure. This approach is worth investigating further to generate new sources of structural information for increasingly large RNAs.
4.2. Structural calculation and refinement
CYANA [120], Xplor-NIH [121] or CNS [122] are used for structural calculations. All protocols initiate with a simulated annealing protocol starting with 50–100 initial random-generated structures. Relative weights of experimental constraints and force field are to be set carefully. In Xplor-NIH, force constants for bond and angular (angles and improper torsions) are set to 1.0 kcal mol−1 Å2 and 1.0 kcal mol−1 rad−2, respectively during initial structure calculation. The target function used in simulated annealing includes NOE-derived and hydrogen bond distance constraints (final force constant ~50 kcal mol−1 Å2) as well as dihedral torsion angle (final force constant ~ 200 kcal mol−1 rad−2) restraints. A quartic van der Waals repulsion term (final force constant of 4 kcal mol−1 Å2 and a van der Waals radius scale factor of 0.8), a torsion angle database potential (RAMA; with final weighting of 1) and impropers (final scale factor of 1; with a maximum deviation of 5°) are used to describe non-bonded interactions.
The simulated annealing procedure starts with high temperature torsion angle dynamics (10,000 steps at 3500 K). The bath temperature is cooled from 3500 to 298 K in steps, introducing the van der Waals terms and incrementally raising the force constants for angles, impropers, dihedral angles, NOEs, van der Waals repulsion and RAMA. Cooling is done over 100 steps; at each step, torsion angle dynamics is ran for 10 ps. The molecule undergoes two sequential Powell minimizations, first in torsion angle, then in Cartesian space after the final cooling step. The calculations are repeated multiple times with different random seeds for the assignment of initial velocities; the lowest energy structures without violations of distance (>0.3 Å) or torsion angle (>5′) restraints are retained for further refinement.
Structure refinement for these converged structures is also based on torsion angle dynamics and Cartesian coordinate minimization. The refinement protocol includes further potential terms for correcting both local and overall geometry: planarity restraints for base pairs (weight factor = 50), the ORIE database for relative positioning of base pairs (final scale factor = 0.2), and residual dipolar couplings for orienting the entire molecule (final force constant = 0.75 kcal mol−1 Hz−1), when available. During the first rounds of refinement, the RDC tensor is allowed to vary in order to find the optimum values of Da and R; during the final structure calculations, these values are fixed based on the optimal values. Initial high temperature dynamics using only the phosphate-phosphate non-bonded terms is followed by high temperature torsion angle dynamics (10,000 steps at 3,000 K). The bath temperature is gradually reduced from 3000 K to 298 K over 200 steps with increases in force constants for angles, impropers, dihedral angles, NOEs, van der Waals repulsion, RAMA, ORIE, and RDCs. Cooling occurs over 200 steps and, at each step, torsion angle dynamics is ran for 0.4ps followed by Powell minimizations in torsion angle and Cartesian space. The calculation is repeated for 100–200 structures and the 10 lowest energy structures without violations in distance (>0.3 Å) or torsion angle (>5°) restraints are retained for analysis.
A new force-field (RNA-ff1) has been recently introduced to overcome the steric clashes and conformational ambiguity often observed when NMR and crystallographic structures are compared [123]. This force field should improve covalent geometry, backbone conformation and MolProbity validation criteria for clashes. It should also display better base-pair step conformational properties in the RNA.
5. Concluding remarks
NMR structural biology of RNA is now tackling structures of significant complexity and size. A panel of relatively large RNA structures (>65 nts) determined by NMR in the last 10 years is shown in Fig. 9. In several cases, these were determined using experimental information beyond NMR. For example, for the CssA thermometer [31], J-K region of EMCV IRES RNA [124] and yeast U2/U6 complex [125], small angle X-ray scattering (SAXS) data were used to validate the structure or in the structure calculation itself. In the case of the 131 nts conserved retroviral RNA packaging element, cryo-EM was used along with NMR constraints to determine/validate the NMR structure [43].
Fig. 9. NMR spectroscopy of large RNA Structures (> 65 nt).
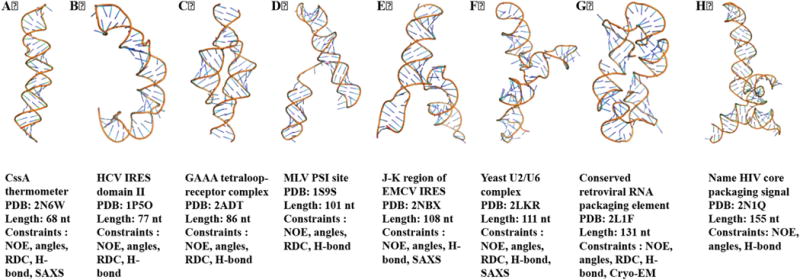
RNA structures determined using NMR spectroscopy in last fifteen years are shown here, from the 70 nts thermometer to the 155 nts conserved retroviral RNA packaging element.
This review provides a standard set of protocols and guidelines for structure determination that can be used to determine RNA structures by NMR, but large RNAs require additional technical developments for examples, NMR experiments such as TROSY/CRINEPT [126,127] for increased resolution/sensitivity; other structural tools i.e., SAXS [31], SANS [128,129] and electron paramagnetic resonance (EPR) [130]. The measurement time of 3D and higher dimension NOESY experiments could be reduced by applying rapid data collection [131–135] and non-uniform sampling methods [136]. These developments promise to allow high quality structure determination of much larger RNAs than that has been possible in the past, but their promise remain to be implemented: up to now, the number of constraints per nucleotide of large RNAs that have been modeled remain small. The further development of hybrid approaches linking NMR with computational modeling [95], SAXS/SANS [31,128,129], EPR [130] and cryo-EM [43] is needed to extend the limit of RNA to be studied with NMR spectroscopy. Solid state NMR can also aid with RNA assignments and high-resolution structural studies, particularly when applied in conjunction with segmentally labeled RNA [6]. Altogether, while technical challenges to extending the size limitations of NMR are significance, opportunities and biological significance of the problems will continue to motivate technological progress.
Acknowledgments
The studies and techniques reported in this review were supported by grants from NIH 1R01 GM103834 and NSF (MCB 1613678) to GV. RPB acknowledges the University Grants Commission-Faculty Recharge Program (UGC-FRP) of the Government of India for a UGC Assistant Professor position.
Abbreviations
- 1DNH
one bond dipolar coupling between nitrogen and proton
- 1DCH
one bond dipolar coupling between carbon and proton
- 1JNH
one bond scalar coupling between nitrogen and proton
- 1JCH
one bond scalar coupling between carbon and proton
- 2D
two-dimensional
- 3D
three-dimensional
- Å
angstrom
- ARTSY
Amide RDCs by TROSY Spectroscopy
- ATP
adenosine-5′-triphosphate
- BEST
band selective short transient
- BMRB
biological magnetic resonance data bank
- CNS
crystallography & NMR system
- COSY
correlation spectroscopy
- CRINEPT
cross-correlated relaxation-enhanced polarization transfer
- CTP
cytidine-5′-triphosphate
- CYANA
Combined assignment and dynamics algorithm for NMR applications
- D2O
deuterium oxide
- DNA
deoxyribonucleic acid
- DQF-COSY
double quantum filtered COSY
- EM
electron microscopy
- EPR
electron paramagnetic resonance
- ff
force-field
- H-bond
hydrogen bond
- HCV
Hepatitis C virus
- HETCOR
heteronuclear COSY
- HETTOC
heteronuclear TOCSY
- HH
hammerhead
- HIV
human immunodeficiency virus
- HPLC
high-performance liquid chromatography
- HMQC
heteronuclear multiple quantum coherence
- HSQC
heteronuclear single quantum coherence
- INEPT
Insensitive nuclei enhanced by polarization transfer
- IPAP
inphase-antiphase
- IRES
internal ribosomal entry site
- K
kelvin
- kDa
kilo Dalton
- kcal
kilo calorie
- MHz
mega hertz
- miRNA/miR
microRNA
- pre-miR
precursor microRNA
- MLV
murine leukemia virus
- MoMuLV
moloney murine leukemia virus
- ms
millisecond
- NMR
nuclear magnetic resonance
- NOE
nuclear overhauser effect
- NOESY
nuclear overhauser effect spectroscopy
- nt
nucleotide
- NTP
nucleotide triphosphates
- PAGE
polyacrylamide gel electrophoresis
- PDB
protein data bank
- PEG
polyethylene glycol
- Pf1
filamentous phage 1
- PRE
paramagnetic relaxation enhancement
- PLOR
position-selective labeling of RNA
- ps
picosecond
- RNA
ribonucleic acid
- RRM
RNA recognition motif
- RNP
ribonucleoprotein
- RDC
residual dipolar coupling
- SANS
small angle neutron scattering
- SAXS
small angle X-ray scattering
- TOCSY
total correlation spectroscopy
- tRNA
transfer RNA
- TROSY
transverse relaxation optimized spectroscopy
- UTP
uridine-5′-triphosphate
- VS
Varkud satellite
References
- 1.Varani G, Aboulela F, Allain FHT. NMR investigation of RNA structure. Prog Nucl magnetic Reson Spectrosc. 1996;29:51–127. [Google Scholar]
- 2.Varani G, Tinoco I., Jr RNA structure and NMR spectroscopy. Q Rev biophysics. 1991;24(4):479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 3.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Method Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 4.Tzakos AG, Grace CR, Lukavsky PJ, Riek R. NMR techniques for very large proteins and rnas in solution. Annu Rev biophysics Biomol Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 5.Allain FH, Varani G. How accurately and precisely can RNA structure be determined by NMR? J Mol Biol. 1997;267(2):338–351. doi: 10.1006/jmbi.1996.0855. [DOI] [PubMed] [Google Scholar]
- 6.Marchanka A, Simon B, Althoff-Ospelt G, Carlomagno T. RNA structure determination by solid-state NMR spectroscopy. Nat Commun. 2015;6:7024. doi: 10.1038/ncomms8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46(1):113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann RK, Bindereif A, Schon A, Westhof E. Handbook of RNA Biochemistry. Wiley, VCH; 2005. [Google Scholar]
- 9.Varani G, Wimberly B, Tinoco I., Jr Conformation and dynamics of an RNA internal loop. Biochemistry. 1989;28(19):7760–7772. doi: 10.1021/bi00445a036. [DOI] [PubMed] [Google Scholar]
- 10.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic acids Res. 1987;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duss O, Lukavsky PJ, Allain FHT. Isotope Labeling and Segmental Labeling of larger RNAs for NMR structural studies. Isotope Labeling Biomol Nmr. 2012;992:121–144. doi: 10.1007/978-94-007-4954-2_7. [DOI] [PubMed] [Google Scholar]
- 12.Easton LE, Shibata Y, Lukavsky PJ. Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. Rna-a Publ Rna Soc. 2010;16(3):647–653. doi: 10.1261/rna.1862210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keel AY, Easton LE, Lukavsky PJ, Kieft JS. Large-scale native preparation of in vitro transcribed RNA. Method Enzymol. 2009;469:3–25. doi: 10.1016/S0076-6879(09)69001-7. [DOI] [PubMed] [Google Scholar]
- 14.Zlobina M, Sedo O, Chou MY, Slepankova L, Lukavsky PJ. Efficient large-scale preparation and purification of short single-stranded RNA oligonucleotides. Biotechniques. 2016;60(2):75–83. doi: 10.2144/000114383. [DOI] [PubMed] [Google Scholar]
- 15.Wenter P, Reymond L, Auweter SD, Allain FH, Pitsch S. Short, synthetic and selectively 13C-labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic acids Res. 2006;34(11):e79. doi: 10.1093/nar/gkl427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen ED, Durbin RK, Risman SS, McAllister WT. Specific contacts between the bacteriophage T3, T7, and SP6 RNA polymerases and their promoters. J Biol Chem. 1991;266(1):645–651. [PubMed] [Google Scholar]
- 17.Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol Biol. 2011;703:29–41. doi: 10.1007/978-1-59745-248-9_3. [DOI] [PubMed] [Google Scholar]
- 18.Ferre-D’Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic acids Res. 1996;24(5):977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo HC, Collins RA. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J. 1995;14(2):368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields TP, Mollova E, Marie LSte, Hansen MR, Pardi A. High-performance liquid chromatography purification of homogenous-length RNA produced by trans cleavage with a hammerhead ribozyme. Rna. 1999;5(9):1259–1267. doi: 10.1017/s1355838299990945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. Rna. 1999;5(9):1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudisser S, Tinoco I., Jr Solution structure of Cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G.A mismatches. J Mol Biol. 2000;295(5):1211–1223. doi: 10.1006/jmbi.1999.3421. [DOI] [PubMed] [Google Scholar]
- 23.Duss O, Diarra Dit Konte N, Allain FH. Cut and paste RNA for nuclear magnetic resonance, paramagnetic resonance enhancement, and electron paramagnetic resonance structural studies. Methods Enzym. 2015;565:537–562. doi: 10.1016/bs.mie.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Duss O, Maris C, von Schroetter C, Allain FH. A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic acids Res. 2010;38(20):e188. doi: 10.1093/nar/gkq756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997;94(9):4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyce GF. RNA cleavage by the 10–23 DNA enzyme. Methods Enzym. 2001;341:503–517. doi: 10.1016/s0076-6879(01)41173-6. [DOI] [PubMed] [Google Scholar]
- 27.Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37(38):13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 28.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. RNA structure. Structure of the HIV-1 RNA packaging signal. Sci (New York, N Y) 2015;348(6237):917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane SC, Van V, Frank HM, Sciandra CA, McCowin S, Santos J, Heng X, Summers MF. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci U S A. 2016;113(46):13033–13038. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnwal RP, Lee SD, Moore C, Varani G. Structural and biochemical analysis of the assembly and function of the yeast pre-mRNA 3′ end processing complex CF I. Proc Natl Acad Sci U S A. 2012;109(52):21342–21347. doi: 10.1073/pnas.1214102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnwal RP, Loh E, Godin KS, Yip J, Lavender H, Tang CM, Varani G. Structure and mechanism of a molecular rheostat, an RNA thermometer that modulates immune evasion by Neisseria meningitidis. Nucleic acids Res. 2016;44(19):9426–9437. doi: 10.1093/nar/gkw584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller SB, Yildiz FZ, Lo JA, Wang B, D’Souza VM. A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature. 2014;515(7528):591–595. doi: 10.1038/nature13709. [DOI] [PubMed] [Google Scholar]
- 33.Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480(7378):561–564. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dayie TK, Thakur CS. Site-specific labeling of nucleotides for making RNA for high resolution NMR studies using an E. coli strain disabled in the oxidative pentose phosphate pathway. J Biomol NMR. 2010;47(1):19–31. doi: 10.1007/s10858-010-9405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longhini AP, LeBlanc RM, Becette O, Salguero C, Wunderlich CH, Johnson BA, D’Souza VM, Kreutz C, Dayie TK. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic acids Res. 2016;44(6):e52. doi: 10.1093/nar/gkv1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337(2):427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 37.Scott LG, Tolbert TJ, Williamson JR. Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzym. 2000;317:18–38. doi: 10.1016/s0076-6879(00)17004-1. [DOI] [PubMed] [Google Scholar]
- 38.Tolbert TJ, Williamson JR. Preparation of specifically deuterated and C-13-labeled RNA for NMR studies using enzymatic synthesis. J Am Chem Soc. 1997;119(50):12100–12108. [Google Scholar]
- 39.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex, vol. 351, pg 371, 2005. J Mol Biol. 2006;360(3):742–742. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 40.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol. 2010;17(6):703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Sci (New York, N Y) 2011;334(6053):242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431(7008):586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 43.Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Mantey A, Iyalla K, Loeliger K, D’Souza V, Khant H, Schmid MF, Garcia EL, Telesnitsky A, Chiu W, Summers MF. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol. 2010;404(5):751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thakur CS, Brown ME, Sama JN, Jackson ME, Dayie TK. Growth of wildtype and mutant E. coli strains in minimal media for optimal production of nucleic acids for preparing labeled nucleotides. Appl Microbiol Biotechnol. 2010;88(3):771–779. doi: 10.1007/s00253-010-2813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thakur CS, Dayie TK. Asymmetry of (13)C labeled 3-pyruvate affords improved site specific labeling of RNA for NMR spectroscopy. J Biomol NMR. 2011;51(4):505–517. doi: 10.1007/s10858-011-9581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thakur CS, Dayie TK. Asymmetry of 13C labeled 3-pyruvate affords improved site specific labeling of RNA for NMR spectroscopy. J Biomol NMR. 2012;52(1):65–77. doi: 10.1007/s10858-011-9582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur CS, Luo Y, Chen B, Eldho NV, Dayie TK. Biomass production of site selective 13C/15N nucleotides using wild type and a transketolase E. coli mutant for labeling RNA for high resolution NMR. J Biomol NMR. 2012;52(2):103–114. doi: 10.1007/s10858-011-9586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarado LJ, LeBlanc RM, Longhini AP, Keane SC, Jain N, Yildiz ZF, Tolbert BS, D’Souza VM, Summers MF, Kreutz C, Dayie TK. Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem a Eur J Chem Biol. 2014;15(11):1573–1577. doi: 10.1002/cbic.201402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wunderlich CH, Juen MA, LeBlanc RM, Longhini AP, Dayie TK, Kreutz C. Stable isotope-labeled RNA phosphoramidites to facilitate dynamics by NMR. Methods Enzym. 2015;565:461–494. doi: 10.1016/bs.mie.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Clore GM. Practical aspects of paramagnetic relaxation enhancement in biological macromolecules. Methods Enzym. 2015;564:485–497. doi: 10.1016/bs.mie.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim I, Lukavsky PJ, Puglisi JD. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc. 2002;124(32):9338–9339. doi: 10.1021/ja026647w. [DOI] [PubMed] [Google Scholar]
- 52.Tzakos AG, Easton LE, Lukavsky PJ. Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J Am Chem Soc. 2006;128(41):13344–13345. doi: 10.1021/ja064807o. [DOI] [PubMed] [Google Scholar]
- 53.Kim I, Lukavsky PJ, Puglisi JD. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc. 2002;124(32):9338–9339. doi: 10.1021/ja026647w. [DOI] [PubMed] [Google Scholar]
- 54.Tzakos AG, Easton LE, Lukavsky PJ. Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat Protoc. 2007;2(9):2139–2147. doi: 10.1038/nprot.2007.306. [DOI] [PubMed] [Google Scholar]
- 55.Nelissen FH, van Gammeren AJ, Tessari M, Girard FC, Heus HA, Wijmenga SS. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic acids Res. 2008;36(14):e89. doi: 10.1093/nar/gkn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Holmstrom E, Zhang J, Yu P, Wang J, Dyba MA, Chen D, Ying J, Lockett S, Nesbitt DJ, Ferre-D’Amare AR, Sousa R, Stagno JR, Wang YX. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522(7556):368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl magnetic Reson Spectrosc. 1999;34(2):93–158. [Google Scholar]
- 58.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog Nucl magnetic Reson Spectrosc. 1998;32:287–387. [Google Scholar]
- 59.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley-Interscience; 1986. [Google Scholar]
- 60.Bax A, Ikura M, Kay LE, Torchia DA, Tschudin R. Comparison of different modes of 2-dimensional reverse-correlation Nmr for the study of proteins. J magnetic Reson. 1990;86(2):304–318. [Google Scholar]
- 61.Nikonowicz EP, Pardi A. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in C-13/N-15 labeled nucleic-acids. J Mol Biol. 1993;232(4):1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- 62.Sklenar V, Peterson RD, Rejante MR, Feigon J. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J Biomol NMR. 1993;3(6):721–727. doi: 10.1007/BF00198375. [DOI] [PubMed] [Google Scholar]
- 63.Tate S, Ono A, Kainosho M. An alternative triple-resonance method for the through-bond correlation of intranucleotide H1′ and H8 Nmr signals of purine nucleotides - application to a DNA dodecamer with fully C-13/N-15-Labeled deoxyadenosine residues. J Am Chem Soc. 1994;116(13):5977–5978. [Google Scholar]
- 64.Sklenar V, Peterson RD, Rejante MR, Wang E, Feigon J. 2-Dimensional triple-resonance Hcnch experiment for direct correlation of ribose H1′ and base H8, H6 protons in C-13, N-15 labeled Rna oligonucleotides. J Am Chem Soc. 1993;115(25):12181–12182. [Google Scholar]
- 65.Farmer BT, Muller L, Nikonowicz EP, Pardi A. Unambiguous resonance assignments in C-15, N-15-Labeled nucleic-acids by 3d triple-resonance Nmr. J Am Chem Soc. 1993;115(23):11040–11041. [Google Scholar]
- 66.Sklenar V, Miyashiro H, Zon G, Miles HT, Bax A. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. Febs Lett. 1986;208(1):94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 67.Kellogg GW. Proton-detected hetero-tocsy experiments with application to nucleic-acids. J magnetic Reson. 1992;98(1):176–182. [Google Scholar]
- 68.Kellogg GW, Schweitzer BI. 2-Dimensional and 3-dimensional P-31-Driven Nmr procedures for complete assignment of backbone resonances in oligodeoxyribonucleotides. J Biomol NMR. 1993;3(5):577–595. doi: 10.1007/BF00174611. [DOI] [PubMed] [Google Scholar]
- 69.Wijmenga SS, Heus HA, Leeuw HA, Hoppe H, van der Graaf M, Hilbers CW. Sequential backbone assignment of uniformly 13C-labeled RNAs by a two-dimensional P(CC)H-TOCSY triple resonance NMR experiment. J Biomol NMR. 1995;5(1):82–86. doi: 10.1007/BF00227472. [DOI] [PubMed] [Google Scholar]
- 70.Marino JP, Prestegard JH, Crothers DM. Correlation of adenine H2/H8 resonances in uniformly C-13 labeled rnas by 2d Hcch-tocsy - a new tool for H-1 assignment. J Am Chem Soc. 1994;116(5):2205–2206. [Google Scholar]
- 71.Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C. A 3-dimensional triple-resonance H-1,C-13,P-31 experiment - sequential through-bond correlation of ribose protons and intervening phosphorus along the Rna oligonucleotide backbone. J Am Chem Soc. 1994;116(14):6472–6473. [Google Scholar]
- 72.Heus HA, Wijmenga SS, Vandeven FJM, Hilbers CW. Sequential backbone assignment in C-13-Labeled Rna via through-bond coherence transfer using 3-dimensional triple-resonance spectroscopy (H-1, C-13, P-31) and 2-dimensional hetero tocsy. J Am Chem Soc. 1994;116(11):4983–4984. [Google Scholar]
- 73.Varani G, Aboulela F, Allain F, Gubser CC. Novel 3-dimensional H-1-C-13-P-31 triple-resonance experiments for sequential backbone correlations in nucleic-acids. J Biomol NMR. 1995;5(3):315–320. doi: 10.1007/BF00211759. [DOI] [PubMed] [Google Scholar]
- 74.Tate S, Ono A, Kainosho M. Sequential backbone assignment in C-13-Labeled DNA by the H-1, C-13, P-31 triple-resonance experiment, Hcp-Cch-Cosy. J Magn Reson Ser B. 1995;106(1):89–91. doi: 10.1006/jmrb.1995.1016. [DOI] [PubMed] [Google Scholar]
- 75.Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C. Sequential correlation of anomeric ribose protons and intervening phosphorus in Rna oligonucleotides by a H-1,C-13,P-31 triple-resonance experiment - Hcp-Cch-tocsy. J Biomol NMR. 1995;5(1):87–92. doi: 10.1007/BF00227473. [DOI] [PubMed] [Google Scholar]
- 76.Simon B, Zanier K, Sattler M. A TROSY relayed HCCH-COSY experiment for correlating adenine H2/H8 resonances in uniformly 13C-labeled RNA molecules. J Biomol NMR. 2001;20(2):173–176. doi: 10.1023/a:1011214914452. [DOI] [PubMed] [Google Scholar]
- 77.Simorre JP, Zimmermann GR, Pardi A, Farmer BT, Mueller L. Triple resonance HNCCCH experiments for correlating exchangeable and nonexchangeable cytidine and uridine base protons in RNA. J Biomol NMR. 1995;6(4):427–432. doi: 10.1007/BF00197641. [DOI] [PubMed] [Google Scholar]
- 78.Sklenar V, Dieckmann T, Butcher SE, Feigon J. Through-bond correlation of imino and aromatic resonances in C-13-,N-15-labeled RNA via heteronuclear TOCSY. J Biomol NMR. 1996;7(1):83–87. doi: 10.1007/BF00190460. [DOI] [PubMed] [Google Scholar]
- 79.Wohnert J, Ramachandran R, Gorlach M, Brown LR. Triple-resonance experiments for correlation of H5 and exchangeable pyrimidine base hydrogens in (13)C,(15)N-labeled RNA. J magnetic Reson. 1999;139(2):430–433. doi: 10.1006/jmre.1999.1797. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y, Kortemme T, Robertson T, Baker D, Varani G. A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys. Nucleic acids Res. 2004;32(17):5147–5162. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J Am Chem Soc. 1998;120(33):8293–8297. [Google Scholar]
- 82.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic acids Res. 2007;35(15):5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic acids Res. 2007;35(2):572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cornish PV, Giedroc DP, Hennig M. Dissecting non-canonical interactions in frameshift-stimulating mRNA pseudoknots. J Biomol NMR. 2006;35(3):209–223. doi: 10.1007/s10858-006-9033-x. [DOI] [PubMed] [Google Scholar]
- 85.Farjon J, Boisbouvier J, Schanda P, Pardi A, Simorre JP, Brutscher B. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J Am Chem Soc. 2009;131(24):8571–8577. doi: 10.1021/ja901633y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dallmann A, Simon B, Duszczyk MM, Kooshapur H, Pardi A, Bermel W, Sattler M. Efficient detection of hydrogen bonds in dynamic regions of RNA by sensitivity-optimized NMR pulse sequences, Angew. Chem Int Ed Engl. 2013;52(40):10487–10490. doi: 10.1002/anie.201304391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duchardt-Ferner E, Ferner J, Wohnert J. Rapid identification of noncanonical RNA structure elements by direct detection of OH…O=P, NH…O=P, and NH2…O=P hydrogen bonds in solution NMR spectroscopy. Angew Chem Int Ed Engl. 2011;50(34):7927–7930. doi: 10.1002/anie.201101743. [DOI] [PubMed] [Google Scholar]
- 88.Peterson RD, Theimer CA, Wu H, Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J Biomol NMR. 2004;28(1):59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 89.Otting G. Protein NMR using paramagnetic ions. Annu Rev Biophys. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]
- 90.Ramos A, Varani G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J Am Chem Soc. 1998;120(42):10992–10993. [Google Scholar]
- 91.Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FH. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl magnetic Reson Spectrosc. 2011;58(1–2):1–61. doi: 10.1016/j.pnmrs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 92.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev biophysics Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 93.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev biophysics. 2000;33(4):371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 94.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzym. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 95.Borkar AN, Bardaro MF, Jr, Camilloni C, Aprile FA, Varani G, Vendruscolo M. Structure of a low-population binding intermediate in protein-RNA recognition. Proc Natl Acad Sci U S A. 2016;113(26):7171–7176. doi: 10.1073/pnas.1521349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neudecker P, Robustelli P, Cavalli A, Walsh P, Lundstrom P, Zarrine-Afsar A, Sharpe S, Vendruscolo M, Kay LE. Structure of an intermediate state in protein folding and aggregation. Sci (New York, NY) 2012;336(6079):362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 97.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Sci (New York, NY) 2010;329(5997):1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 98.De Simone A, Aprile FA, Dhulesia A, Dobson CM, Vendruscolo M. Structure of a low-population intermediate state in the release of an enzyme product. eLife. 2015;4 doi: 10.7554/eLife.02777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol. 1997;4(9):732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 100.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzym. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 101.Meier S, Haussinger D, Grzesiek S. Charged acrylamide copolymer gels as media for weak alignment. J Biomol NMR. 2002;24(4):351–356. doi: 10.1023/a:1021609207024. [DOI] [PubMed] [Google Scholar]
- 102.Losonczi JA, Prestegard JH. Improved dilute bicelle solutions for high-resolution NMR of biological macromolecules. J Biomol NMR. 1998;12(3):447–451. doi: 10.1023/a:1008302110884. [DOI] [PubMed] [Google Scholar]
- 103.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122(32):7793–7797. [Google Scholar]
- 104.Bardaro MF, Jr, Varani G. Independent alignment of RNA for dynamic studies using residual dipolar couplings. J Biomol NMR. 2012;54(1):69–80. doi: 10.1007/s10858-012-9655-0. [DOI] [PubMed] [Google Scholar]
- 105.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J magnetic Reson. 1998;131(2):373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 106.Leeper TC, Athanassiou Z, Dias RL, Robinson JA, Varani G. TAR RNA recognition by a cyclic peptidomimetic of Tat protein. Biochemistry. 2005;44(37):12362–12372. doi: 10.1021/bi0510532. [DOI] [PubMed] [Google Scholar]
- 107.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCVIRES domain II determined by NMR. Nat Struct Biol. 2003;10(12):1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 108.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7(12):1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 109.Ying J, Wang J, Grishaev A, Yu P, Wang YX, Bax A. Measurement of (1)H-(15)N and (1)H-(13)C residual dipolar couplings in nucleic acids from TROSY intensities. J Biomol NMR. 2011;51(1–2):89–103. doi: 10.1007/s10858-011-9544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fitzkee NC, Bax A. Facile measurement of (1)H-(1)5N residual dipolar couplings in larger perdeuterated proteins. J Biomol NMR. 2010;48(2):65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clore GM, Gronenborn AM, Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J magnetic Reson. 1998;131(1):159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 112.Bayer P, Varani L, Varani G. Refinement of the structure of protein-RNA complexes by residual dipolar coupling analysis. J Biomol NMR. 1999;14(2):149–155. doi: 10.1023/a:1008360331296. [DOI] [PubMed] [Google Scholar]
- 113.Allain FH, Howe PW, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16(18):5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Y, Zubovic L, Yang F, Godin K, Pavelitz T, Castellanos J, Macchi P, Varani G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream dicer. Nucleic acids Res. 2016;44(9):4381–4395. doi: 10.1093/nar/gkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J Am Chem Soc. 2000;122(46):11561–11562. [Google Scholar]
- 116.Scott LG, Geierstanger BH, Williamson JR, Hennig M. Enzymatic synthesis and 19F NMR studies of 2-fluoroadenine-substituted RNA. J Am Chem Soc. 2004;126(38):11776–11777. doi: 10.1021/ja047556x. [DOI] [PubMed] [Google Scholar]
- 117.Hennig M, Scott LG, Sperling E, Bermel W, Williamson JR. Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J Am Chem Soc. 2007;129(48):14911–14921. doi: 10.1021/ja073825i. [DOI] [PubMed] [Google Scholar]
- 118.Hennig M, Munzarova ML, Bermel W, Scott LG, Sklenar V, Williamson JR. Measurement of long-range 1H-19F scalar coupling constants and their glycosidic torsion dependence in 5-fluoropyrimidine-substituted RNA. J Am Chem Soc. 2006;128(17):5851–5858. doi: 10.1021/ja060165t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kreutz C, Kahlig H, Konrat R, Micura R. Ribose 2′-F labeling: a simple tool for the characterization of RNA secondary structure equilibria by 19F NMR spectroscopy. J Am Chem Soc. 2005;127(33):11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
- 120.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 121.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J magnetic Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 122.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 123.Bermejo GA, Clore GM, Schwieters CD. Improving NMR structures of RNA. Structure. 2016;24(5):806–815. doi: 10.1016/j.str.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Imai S, Kumar P, Hellen CU, D’Souza VM, Wagner G. An accurately pre-organized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat Struct Mol Biol. 2016;23(9):859–864. doi: 10.1038/nsmb.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE. Structure of the yeast U2/U6 snRNA complex. Rna. 2012;18(4):673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Riek R, Pervushin K, Wuthrich K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem Sci. 2000;25(10):462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- 127.Pervushin K. Impact of transverse relaxation optimized spectroscopy (TROSY) on NMR as a technique in structural biology. Q Rev biophysics. 2000;33(2):161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 128.Falb M, Amata I, Gabel F, Simon B, Carlomagno T. Structure of the K-turn U4 RNA: a combined NMR and SANS study. Nucleic acids Res. 2010;38(18):6274–6285. doi: 10.1093/nar/gkq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gabel F. Small-angle neutron scattering for structural biology of protein-RNA complexes. Methods Enzym. 2015;558:391–415. doi: 10.1016/bs.mie.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 130.Duss O, Yulikov M, Allain FH, Jeschke G. Combining NMR and EPR to determine structures of large RNAs and protein-RNA complexes in solution. Methods Enzym. 2015;558:279–331. doi: 10.1016/bs.mie.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 131.Pervushin K, Vogeli B, Eletsky A. Longitudinal (1)H relaxation optimization in TROSY NMR spectroscopy. J Am Chem Soc. 2002;124(43):12898–12902. doi: 10.1021/ja027149q. [DOI] [PubMed] [Google Scholar]
- 132.Hiller S, Fiorito F, Wuthrich K, Wider G. Automated projection spectroscopy (APSY) Proc Natl Acad Sci U S A. 2005;102(31):10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Atreya HS, Szyperski T. Rapid NMR data collection. Methods Enzym. 2005;394:78–108. doi: 10.1016/S0076-6879(05)94004-4. [DOI] [PubMed] [Google Scholar]
- 134.Frydman L, Scherf T, Lupulescu A. The acquisition of multidimensional NMR spectra within a single scan. Proc Natl Acad Sci U S A. 2002;99(25):15858–15862. doi: 10.1073/pnas.252644399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barnwal RP, Rout AK, Chary KV, Atreya HS. Rapid measurement of pseudocontact shifts in paramagnetic proteins by GFT NMR spectroscopy. Open Magnetic Reson J. 2008;1:16–28. [Google Scholar]
- 136.Hyberts SG, Arthanari H, Wagner G. Applications of non-uniform sampling and processing. Top Curr Chem. 2012;316:125–148. doi: 10.1007/128_2011_187. [DOI] [PMC free article] [PubMed] [Google Scholar]


