Abstract
While α-linked amino acids in the L-form are exclusively utilized in mammalian protein building, β-linked and D-form amino acids also have important biological roles. Unfortuately, the structural elucidation and separation of these different amino acid types in peptides has been analytically challenging to date due to the numerous isomers present, limiting our knowledge about their existence and biological roles. Here we utilized an ultrahigh resolution ion mobility spectrometry (IMS)-MS platform to separate amyloid β (Aβ) peptides containing L-aspartic acid, D-aspartic acid, L-isoaspartic acid, and D-isoaspartic acid residues which span α-and β-linked amino acids in both D- and L-forms. The results illustrate how IMS-MS could be used to better understand age-related diseases or protein folding disorders resulting from amino acid modifications.
Due to difficulties separating protein and peptide isomers containing α- and β-linked amino acids, significant gaps exist in our understanding of their presence and possible biological roles. Isoaspartic acid (isoAsp) is a naturally occurring β-linked amino acid that results from the degradation of aspartic acid (Asp) and asparagine (Asn). Asp and isoAsp have the same mass and are very difficult to separate with current analytical tools, especially when their epimeric L- and D-forms are considered. However, even with the limited separations available, these amino acids (and their epimers) are known to play an important role in antibiotics, bacterial envelopes and long-lived proteins (LLPs)1, such as crystallin in eye lenses, myelin in nerve cells, and both Tau and Aβ in the brain2. The presence of isoAsp in these systems is thought to impact proteolytic stability and cause changes in the protein’s conformation (e.g. misfolding) that affect activity, degradation and turnover. However, this is not true in all systems as peptides containing β-linked amino acids can also exhibit increased stability by folding into α-helices or hairpin structures with higher variability than α-linked amino acid peptide structures and greater resistance to proteolytic degradation3, 4. Further studies are thus needed to determine when stabilization or destabilization results from isoAsp and other β-linked amino acids. Racemization of Asp and Asn from the L-form to the D-form is another important consideration in peptide stability studies. This transformation mainly occurs through a cyclic succinimide intermediate and results in four isomer products: L-Asp, D-Asp, L-isoAsp and D-isoAsp (Figure 1). Since these four isoforms can greatly influence protein structure, full characterization of Asp variants is needed, especially in LLPs, to better understand disease etiology and develop potential therapeutic treatments.
Figure 1.
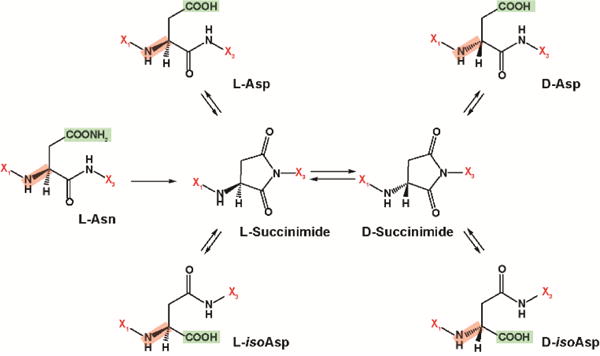
The isomerization pathway and structures for L-aspartic acid (L-Asp), D-aspartic acid (D-Asp), L-isoaspartic acid (L-isoAsp) and D-isoaspartic acid (D-isoAsp). X1 and X3 denote amino acids before and after each specific residue shown. The key -NH2 group was highlighted in red for L- and D-differentiation, and -COOH groups (and -COONH2 for Asn) were highlighted in green to illustrate Asp isomerization.
Tau and Aβ proteins are the hallmarks of Alzheimer’s disease and have been shown to contain large abundances of racemized amino acid residues in brain tissue1, 5, 6. In fact, Tau and Aβ have multiple Asp residues that are particularly susceptible to modification and truncation, especially at the N-terminus. Specifically, both racemization and isomerization of Asp7 have been observed in Aβ extracted from senile plaques, altering the Aβ42 conformation and enhancing its aggregation characteristics7, 8. Previous studies have also found that 55.7% of Aβ proteins isolated from the brain existed in the L-isoAsp form and 19.2% in the D-isoAsp form9, 10. In addition, both D-Asp1 and D-Asp7 were also detected in the isolated Aβ proteins10, suggesting the presence of β-linked and D-form amino acids in Alzheimer’s peptides may have potential roles in other LLPs where protein turnover is slow or protein degradation is biased against D-form amino acids.
Despite the important roles of D- and L-form Asp and isoAsp amino acid residues, their structural elucidation and separation has been analytically challenging, limiting a full understanding of their presence and biological roles. Characterization of these isomers is further complicated by their low abundances, requiring very sensitive techniques in addition to highly efficient separations. Chromatographic methods such as GC and LC are often used in chiral analyses of individual amino acid 11, 12. Thus, identification of which protein the chiral amino acids originated from is impossible unless only one isolated protein is studied 13. Mass spectrometry (MS)-based techniques are also used, but are problematic since isomerization does not change the mass of the resulting peptides. However, fragmentation methods such as collision induced dissociation (CID)14, 15, electron capture/transfer dissociated (ExD)16–18, and radical-directed dissociation (RDD)19, in conjunction with separations, have enabled the successful differentiation of diastereomers and made tandem MS a powerful tool for D/L-amino acid and isoAsp analyses. In this regard, IMS has also been of great interest in the structural separation of L- and D-amino acids and Asp and isoAsp residues, but its low resolution to date has hindered its full potential.
In the past decade, advances in IMS have made it an increasingly powerful tool for analyzing biological molecules20–22. IMS coupled with MS (IMS-MS) has been used to characterize peptide isomers23, 24, study D-amino acid containing peptides25–29, localize D-amino acid sites29, and understand oligomerization of the D-amino acid containing peptides30. However, D/L-peptide isomers normally cannot be resolved with currently available IMS instruments due to their low resolution. In this work, we applied a platform utilizing the recently developed structure for lossless ion manipulation technology (SLIM) to perform the IMS analyses. This platform enables long IMS path lengths, and efficient ion selection, trapping and accumulation capabilities to achieve both enhanced separations and sensitivity31–36. To evaluate its capabilities for separating β-linked and D-form amino acids, Aβ(6–16) isoforms were chosen for study. Aβ(6–16) is of great interest since its Asp7 residue is known to racemize and isomerize in senile plaques (His6-Asp-Ser-Gly-Tyr-Glu-Val-His-His-Gln-Lys16) and therefore is of great importance in Alzheimer’s studies. Further it is cleaved during the tryptic digestion of Aβ so it is often detected in bottom-up proteomic studies. Four Aβ(6–16) isoforms with different variants of the Asp7 residue were chosen for this study (the naturally existing L-Asp, and the three potential modifications D-Asp, L-isoAsp, and D-isoAsp) and were first investigated with a conventional 90-cm drift tube IMS-MS (DTIMS-MS) platform37, 38 and then the ultrahigh resolution SLIM IMS-MS platform39, 40. The IMS results for the doubly protonated peptides ([M + 2H]2+, m/z = 672.81) are shown in Figure 2.
Figure 2.
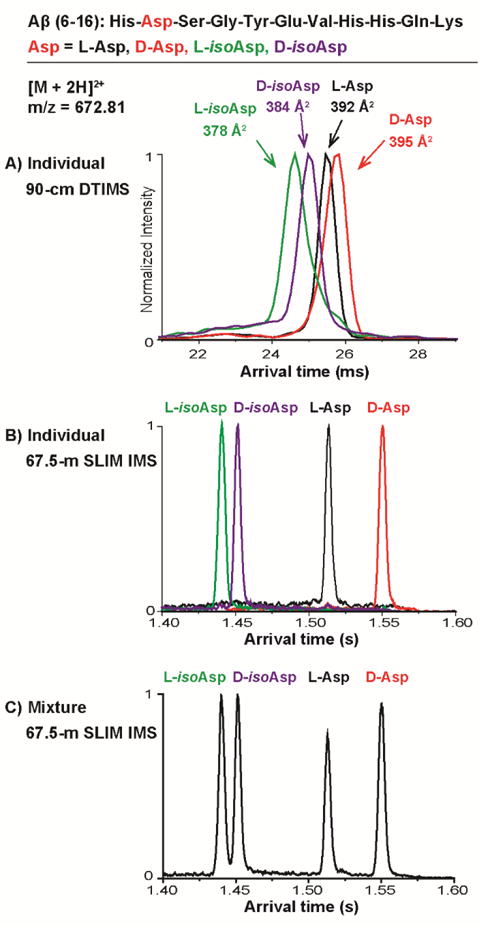
IMS profiling of the Aβ(6–16) isomer variants. Individual Aβ(6–16) peptide isomer standards containing L-aspartic acid (L-Asp), D-aspartic acid (D-Asp), L-isoaspartic acid (L-isoAsp) and D-isoaspartic acid (D-isoAsp) were analyzed using a A) 90-cm DTIMS-MS platform and B) 67.5-m SLIM IMS-MS platform. The SLIM IMS-MS platform was also able to baseline separate the isomer mixture allowing accurate quantitation of each peptide (C). The sequence of Aβ(6–16) is provided on the top panel.
The arrival time distributions (ATDs) obtained from the 90-cm DTIMS-MS platform (Figure 2A) illustrated a single peak for each peptide, suggesting only one conformation was present for each variant. When each conformer was analysed, two main trends were noted. First, the isoAsp residues, regardless of L- or D-form, traveled faster than the Asp containing peptides, indicating that isoAsp caused the peptides to fold into a more compact form than Asp. Second, the peptides with L-form amino acid traveled faster than the D-form (arrival time for L-isoAsp < D-isoAsp < L-Asp < D-Asp). Thus, the L to D conversion also increases the conformational size of the Aβ peptides. This was also noted in the measured DTIMS collision cross section (CCS) for each peptide: 378, 384, 392 and 395 Å2 for L-isoAsp, D-isoAsp, L-Asp and D-Asp (each measured with <1% error), where the L-isoAsp peptide has the smallest size while the D-Asp peptide is the largest. While these separations illustrate the promise of IMS for structural elucidation of D/L-form amino acids and isoAsp-containing peptide isomers, the 90-cm DTIMS-MS platform could only partially separate these isomers, limiting its isomeric quantification capabilities in complex samples. Thus, the new SLIM IMS-MS platform, which offers much greater IMS resolution, was explored39, 40. As shown in Figure 2B, after traversing 67.5-m in the SLIM IMS-MS platform, baseline separation of each isoform (IMS resolving power of ~350) was achieved in less than 2 seconds. Furthermore, the mixture of all four variants was also baseline resolved by the SLIM IMS-MS platform (Figure 2C). The ability to fully separate all of the variants is extremely beneficial for relative quantification of these specific peptides in a complex sample. This area is being actively explored at present and we expect important biological findings to result.
In conclusion, the ultrahigh resolution SLIM IMS analyses enabled baseline separation of challenging D- and L-form Asp and isoAsp variants in Aβ peptides of interest for Alzheimer’s studies. The development of this rapid technique also supports the identification and quantification of D-form Asp and isoAsp residues in other proteins. We are currently investigating these separations in complex biological samples (i.e. cerebral spinal fluid, plasma, tissue, etc.) and expect this capability will promote studies evaluating the effects of amino acid isomerization and racemization, along with other non-enzymatic post-translational modifications such as deamidation, oxidation and glycation that also result in numerous isomeric species. We expect that characterization of these isomers will enable a way of determining the age of the molecules and a better understanding the etiology of diseases. In addition to these advances, these new capabilities will greatly facilitate studies of D-amino acid incorporation in peptides and allow a better appreciation of how often such modifications are occurring.
Supplementary Material
Acknowledgments
Portions of this research were supported by grants from the National Institute of Environmental Health Sciences of the NIH (R01 ES022190), National Institute of General Medical Sciences (P41 GM103493), and the Laboratory Directed Research and Development Program and Microbes in Transition (MinT) Initiative at Pacific Northwest National Laboratory. This research utilized capabilities developed by the Pan-omics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program) and by the National Institute of Allergy and Infectious Diseases under grant U19 AI106772. This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL0 1830.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and References
- 1.Truscott RJW, Schey KL, Friedrich MG. Trends Biochem Sci. 2016;41:654–664. doi: 10.1016/j.tibs.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooi MYS, Truscott RJW. Age. 2011;33:131–141. doi: 10.1007/s11357-010-9171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seebach D, Matthews JL. Chem Commun. 1997;2015–2022 [Google Scholar]
- 4.Seebach D, Beck AK, Bierbaum DJ. Chem Biodivers. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 5.Lyons B, Friedrich M, Raftery M, Truscott R. Anal Chem. 2016;88:2675–2684. doi: 10.1021/acs.analchem.5b03891. [DOI] [PubMed] [Google Scholar]
- 6.Moro Maria L, Collins Matthew J, Cappellini E. Biochem Soc Trans. 2010;38:539–544. doi: 10.1042/BST0380539. [DOI] [PubMed] [Google Scholar]
- 7.Sugiki T, Utsunomiya-Tate N. Biochem Biophys Res Commun. 2013;441:493–498. [PubMed] [Google Scholar]
- 8.Fukuda H, Shimizu T, Nakajima M, Mori H, Shirasawa T. Biorg Med Chem Lett. 1999;9:953–956. doi: 10.1016/s0960-894x(99)00121-3. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T, Watanabe A, Ogawara M, Mori H, Shirasawa T. Arch Biochem Biophys. 2000;381:225–234. doi: 10.1006/abbi.2000.1955. [DOI] [PubMed] [Google Scholar]
- 10.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zürcher-Neely HA, Heinrikson RL, Ball MJ. J Biol Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 11.Ilisz I, Aranyi A, Pataj Z, Péter A. J Pharm Biomed Anal. 2012;69:28–41. doi: 10.1016/j.jpba.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Szökő É, Vincze I, Tábi T. J Pharm Biomed Anal. 2016;130:100–109. doi: 10.1016/j.jpba.2016.06.054. [DOI] [PubMed] [Google Scholar]
- 13.Stabler TV, Byers SS, Zura RD, Kraus VB. Arthrit Res Ther. 2009;11:R34. doi: 10.1186/ar2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai L, Romanova EV, Sweedler JV. Anal Chem. 2011;83:2794–2800. doi: 10.1021/ac200142m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livnat I, Tai H-C, Jansson ET, Bai L, Romanova EV, Chen T-t, Yu K, Chen S-a, Zhang Y, Wang Z-y, Liu D-d, Weiss KR, Jing J, Sweedler JV. Anal Chem. 2016;88:11868–11876. doi: 10.1021/acs.analchem.6b03658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cournoyer JJ, Lin C, O’Connor PB. Anal Chem. 2006;78:1264–1271. doi: 10.1021/ac051691q. [DOI] [PubMed] [Google Scholar]
- 17.Ni W, Dai S, Karger BL, Zhou ZS. Anal Chem. 2010;82:7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGraan-Weber N, Zhang J, Reilly JP. J Am Soc Mass Spectrom. 2016;27:2041–2053. doi: 10.1007/s13361-016-1487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao Y, Julian RR. Anal Chem. 2014;86:9733–9741. doi: 10.1021/ac502296c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu Rev Anal Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapthorn C, Pullen F, Chowdhry BZ. Mass Spectrom Rev. 2013;32:43–71. doi: 10.1002/mas.21349. [DOI] [PubMed] [Google Scholar]
- 22.Lanucara F, Holman SW, Gray CJ, Eyers CE. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 23.Shvartsburg AA, Creese AJ, Smith RD, Cooper HJ. Anal Chem. 2011;83:6918–6923. doi: 10.1021/ac201640d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim YM, Shvartsburg AA, Smith RD, Belov ME. Anal Chem. 2011;83:5617–5623. doi: 10.1021/ac200719n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srebalus CA, Li J, Marshall WS, Clemmer DE. Anal Chem. 1999;71:3918–3927. doi: 10.1021/ac9903757. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Siems WF, Klasmeier J, Hill HH. Anal Chem. 2000;72:391–395. doi: 10.1021/ac990601c. [DOI] [PubMed] [Google Scholar]
- 27.Kemper PR, Dupuis NF, Bowers MT. Int J Mass Spectrom. 2009;287:46–57. [Google Scholar]
- 28.Magalhães MT Q de, Barbosa EA, Prates MV, Verly RM, Munhoz VHO, Araújo IE de, Bloch C., Jr PLOS ONE. 2013;8:e59255. doi: 10.1371/journal.pone.0059255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia C, Lietz CB, Yu Q, Li L. Anal Chem. 2014;86:2972–2981. doi: 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang X, Jia C, Chen Z, Li L. J Am Soc Mass Spectrom. 2016;28:1–9. doi: 10.1007/s13361-016-1523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamid AM, Garimella SV, Ibrahim YM, Deng L, Zheng X, Webb IK, Anderson GA, Prost SA, Norheim RV, Tolmachev AV, Baker ES, Smith RD. Anal Chem. 2016;88:8949–8956. doi: 10.1021/acs.analchem.6b01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng L, Ibrahim YM, Hamid AM, Garimella SVB, Webb IK, Zheng X, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Baker ES, Smith RD. Anal Chem. 2016;88:8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng L, Ibrahim YM, Garimella SVB, Webb IK, Hamid AM, Norheim RV, Prost SA, Sandoval JA, Baker ES, Smith RD. Anal Chem. 2016;88:10143–10150. doi: 10.1021/acs.analchem.6b02678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng L, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Zhang X, Zheng X, Garimella SVB, Webb IK, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Smith RD. ChemistrySelect. 2016;1:2396–2399. doi: 10.1002/slct.201600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen TC, Ibrahim YM, Webb IK, Garimella SVB, Zhang X, Hamid AM, Deng L, Karnesky WE, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Baker ES, Smith RD. Anal Chem. 2016;88:1728–1733. doi: 10.1021/acs.analchem.5b03910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Garimella SVB, Prost SA, Webb IK, Chen TC, Tang K, Tolmachev AV, Norheim RV, Baker ES, Anderson GA, Ibrahim YM, Smith RD. Anal Chem. 2015;87:6010–6016. doi: 10.1021/acs.analchem.5b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Anal Chem. 2014;86:2107–2116. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibrahim YM, Baker ES, Danielson WF Iii, Norheim RV, Prior DC, Anderson GA, Belov ME, Smith RD. Int J Mass Spectrom. 2015;377:655–662. doi: 10.1016/j.ijms.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L, Webb IK, Garimella SVB, Hamid AM, Zheng X, Norheim RV, Prost SA, Anderson GA, Sandoval JA, Baker ES, Ibrahim YM, Smith RD. Anal Chem. 2017;89:4628–4634. doi: 10.1021/acs.analchem.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X, Renslow RS, Makola MM, Webb IK, Deng L, Thomas DG, Govind N, Ibrahim YM, Kabanda MM, Dubery IA, Heyman HM, Smith RD, Madala NE, Baker ES. J Phys Chem Lett. 2017;8:1381–1388. doi: 10.1021/acs.jpclett.6b03015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


