Abstract
BACKGROUND
Endometriosis, defined as the presence of ectopic endometrial stroma and epithelium, affects approximately 10% of reproductive-age women and can cause pelvic pain and infertility. Endometriotic lesions are considered to be benign inflammatory lesions but have cancerlike features such as local invasion and resistance to apoptosis.
METHODS
We analyzed deeply infiltrating endometriotic lesions from 27 patients by means of exomewide sequencing (24 patients) or cancer-driver targeted sequencing (3 patients). Mutations were validated with the use of digital genomic methods in micro-dissected epithelium and stroma. Epithelial and stromal components of lesions from an additional 12 patients were analyzed by means of a droplet digital polymerase-chain-reaction (PCR) assay for recurrent activating KRAS mutations.
RESULTS
Exome sequencing revealed somatic mutations in 19 of 24 patients (79%). Five patients harbored known cancer driver mutations in ARID1A, PIK3CA, KRAS, or PPP2R1A, which were validated by Safe-Sequencing System or immunohistochemical analysis. The likelihood of driver genes being affected at this rate in the absence of selection was estimated at P = 0.001 (binomial test). Targeted sequencing and a droplet digital PCR assay identified KRAS mutations in 2 of 3 patients and 3 of 12 patients, respectively, with mutations in the epithelium but not the stroma. One patient harbored two different KRAS mutations, c.35G→T and c.35G→C, and another carried identical KRAS c.35G→A mutations in three distinct lesions.
CONCLUSIONS
We found that lesions in deep infiltrating endometriosis, which are associated with virtually no risk of malignant transformation, harbor somatic cancer driver mutations. Ten of 39 deep infiltrating lesions (26%) carried driver mutations; all the tested somatic mutations appeared to be confined to the epithelial compartment of endometriotic lesions.
Endometriosis is a relatively common disease, affecting up to 10% of women of reproductive age.1 Its incidence is as high as 50% among adolescents with pelvic pain.1,2 Clinical symptoms include dysmenorrhea, pelvic pain, and infertility.1,3,4 Endometriotic lesions are considered to be benign (nonmalignant or non-neoplastic), inflammatory, estrogen-dependent lesions that are characterized by the ectopic presence of normal-appearing, functional endometrial tissue composed of glands and stroma outside of the uterus.1,3 The disease is often associated with multiple lesions that can be distributed throughout the abdominal–pelvic peritoneum and visceral organs.
There are three anatomical subtypes of endometriosis: superficial peritoneal endometriosis, ovarian endometriosis, and deep infiltrating endometriosis. Deep infiltrating endometriosis is characterized by nodules that locally invade pelvic structures, producing symptoms such as painful intercourse (deep dyspareunia) and painful bowel movements (dyschezia).5 Progestin-based hormonal therapy and gonadotropin-releasing hormone analogues have become the standard treatments; however, many patients have unacceptable systemic adverse effects in association with either or both treatments.1,6 Moreover, not all women with endometriosis have a response to hormonal therapy, particularly those who have deeply infiltrating disease.7 Surgical resection is an option for women who do not have a response to hormonal therapy or for those who desire pregnancy, but complete excision of deep infiltrating nodules requires surgical expertise and is not without risk.4
Despite its benign clinical behavior and normal-appearing histologic features, endometriosis can recapitulate some features of malignant neoplasms, including local invasion and resistance to apoptosis. The etiologic factors underlying endometriosis are controversial, and the disorder has been proposed to originate from several different processes: the migration of endometrial fragments from the uterus through the fallopian tubes during retrograde menstruation, the dissemination of these fragments to the peritoneal cavity, and their implantation on the serosal surface; the dissemination of endometrial (progenitor) cells through the lymphatic or blood circulation; the development of endometrial tissue through metaplasia of coelomic epithelium (i.e., mesothelium that lines the surface of the peritoneal cavity and organs); or differentiation from bone marrow–derived stem cells or fetal remnants of müllerian cells shed into the peritoneal cavity during retrograde menstruation.3,8–12
Genomewide association studies have identified genetic markers that are potentially related to an increased risk of endometriosis.13 Endometriosis, particularly ovarian endometriosis, is widely accepted as the direct precursor of clear-cell and endometrioid ovarian carcinomas,14,15 so-called endometriosis-related ovarian neoplasms. However, the study of somatic mutations in endometriosis has been restricted largely to endometriosis with concurrent cancer. A small number of candidate-gene studies have examined benign (non–cancer-associated) ovarian endometriosis lesions; one study identified a KRAS mutation (p.G12C) in a single lesion,16 and another identified PTEN mutations in 7 of 34 lesions (21%).17 Immunohistochemical studies have also shown rare partial or complete immunohistochemical loss of ARID1A (as a proxy for ARID1A loss-of-function mutations) in lesions from ovarian and nonovarian endometriosis without concurrent cancer.18–20 All of the above studies have been hampered by low-resolution genomic methods, failure to isolate specific endometrial stromal or epithelial cells from within a dominantly fibrotic endometriosis nodule (e.g., by laser-capture micro-dissection), and lack of orthogonal validation. Thus, these studies have yielded ambiguous answers to the fundamental question: do benign endometriosis lesions harbor somatic mutations in cancer driver genes outside of the process of transformation to endometriosis-related ovarian neoplasms, and if so, what is their role in the pathology of these lesions?
In this study, we performed exomewide sequencing and targeted amplicon sequencing and used two digital genomic methods to determine whether benign, but invasive, deep infiltrating endometriosis lesions harbor somatic mutations, including those frequently detected in human cancers.
METHODS
TISSUE SAMPLES AND PATIENT SELECTION
Tissue samples were obtained from three independent cohorts of patients with deep infiltrating endometriosis. All tissues were fixed archival specimens, either formalin-fixed and paraffin-embedded or Molecular-fixed (Sakura Finetek) and paraffin-embedded. Tissue samples from 17 patients were obtained from the Departments of Obstetrics and Gynecology and Pathology at Lenox Hill Hospital–Northwell Health (Hofstra University) in New York. Tissue samples from 7 patients were obtained from the Department of Pathology at Seirei Mikatahara Hospital in Hamamatsu, Japan. Tissue samples from 15 patients were obtained from the BC Women’s Centre for Pelvic Pain and Endometriosis in Vancouver, BC, Canada, and collected through the OVCARE Tissue Bank (part of the World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project).21 The institutional review boards at the respective hospitals approved all tissue collection. Inclusion criteria were pathologist-confirmed deep infiltrating endometriosis (lesions with a depth >5 mm, such as in the bowel or peritoneal wall) containing both epithelial and stromal components, the absence of cancer or dysplasia, and a lesion size sufficient for tissue coring, macrodissection, or laser-capture microdissection.
EXOME SEQUENCING
Endometriotic lesions and corresponding normal tissues from the same blocks were cored (2 mm for smaller lesions and 3 mm for larger lesions) from formalin-fixed and paraffin-embedded blocks. Sections that were stained with hematoxylin and eosin were prepared before and after coring to confirm the precision of the sampling and to ensure that at least 60% of the core area contained lesional tissue (or that 100% contained normal tissue). Paired-end libraries were generated with the use of standard Illumina procedures that involved DNA from core endometriosis and normal (matched control) samples. Coding regions were captured with the Agilent SureSelect Enrichment System and sequenced on Illumina sequencers. Data were processed as described previously.22
PROBABILISTIC EVALUATION OF OBSERVED DRIVER MUTATION FREQUENCIES
The probability that by pure chance, 5 mutations of the 80 that were found would be among the 125 driver genes considered was P = 0.001, calculated by the binomial test (Table S1 in Supplementary Appendix 2, available with the full text of this article at NEJM.org). Specifically, 1 – pbinom (4, 80, 276,824/27,000,000) = 0.001, in which the total number of positions sequenced is approximately 27 million bp (whole exome), and 276,824 is the estimated total number of positions, among all the driver genes considered, that may yield a driver mutation. The number of driver positions was derived by combining all positions found to be (somatically) mutated, across all cancer types, in the Cancer Genome Atlas database that are considered to be located within driver oncogenes. We conservatively assumed that only targeted positions in tumor suppressor genes were targets of driver mutations.
TARGETED SEQUENCING
The TruSeq Amplicon Cancer Panel (Illumina) and a previously described TruSeq Custom Amplicon Panel23 were applied, followed by Illumina MiSeq sequencing (Table S5 in Supplementary Appendix 2). Variants were identified with the use of the MiSeq Reporter Somatic Variant Caller tool. Only variants absent from germline DNA and present in both libraries (technical replicates) from both panels (i.e., quadruplicate libraries) were validated orthogonally by a droplet digital polymerase-chain-reaction (PCR) assay (Fig. S1 in Supplementary Appendix 1, available at NEJM.org).
DROPLET DIGITAL PCR ASSAYS
Dual-labeled 5′ exonuclease assays were used in droplet digital PCR assays (Table S7 in Supplementary Appendix 2). Droplets were generated on RainDrop Source (RainDance Technologies) or QX200 Droplet Generator (Bio-Rad Laboratories). After thermal cycling (see the Methods section in Supplementary Appendix 1), droplets were quantitated with RainDrop Sense (Rain-Dance Technologies) or QX200 Droplet Reader (Bio-Rad Laboratories).
SAFE-SEQUENCING SYSTEM
We used the single-molecule barcoding system Safe-Sequencing System (Safe-SeqS) as an error-reduction technology for fixed tissues and low-frequency mutations.24 (See the Methods section in Supplementary Appendix 1.) Purified products were sequenced on Illumina MiSeq and analyzed for mutations.
ARID1A IMMUNOHISTOCHEMISTRY
ARID1A immunoreactivity was used as a surrogate for ARID1A inactivating mutations.25–27 Formalin-fixed and paraffin-embedded tissue sections were stained manually with the use of a 1:2000 dilution of ARID1A antibody (Sigma-Aldrich HPA005456), as described previously.28 Additional details are provided in the Methods section in Supplementary Appendix 1.
RESULTS
ANALYSIS DESCRIPTION
We studied nonovarian, deep infiltrating endometriosis lesions from 39 women with a mean age of 37 years. Independent molecular genetic analyses were conducted in parallel and included unbiased whole-exome sequencing in one set of lesions, with validation of driver mutations by Safe-SeqS and immunohistochemical analysis; quadruplicate targeted panel-based sequencing confirmed by a droplet digital PCR assay in a second set of lesions; and droplet digital PCR analysis alone in a third set of lesions (Table 1, and Fig. S1 in Supplementary Appendix 1). The analyses carried out in the first two sets were designed and undertaken without knowledge of the results of the concurrent parallel experiments.
Table 1.
Cohort Description, Assay Pipeline, and Driver Gene Mutations in All Patients.*
| Patient | Age | Location Sampled |
Clinical Stage |
Whole-Exome Sequencing |
Safe-Sequencing System |
Targeted Amplicon Sequencing |
Droplet Digital PCR Assay |
Somatic Mutations Detected |
Driver Gene Mutations |
|---|---|---|---|---|---|---|---|---|---|
| Yr | no. | ||||||||
| 1 | 41 | Paraovarian tissue | III | Yes | No | No | No | 11 | None |
|
| |||||||||
| 2 | 36 | Sigmoid colon | III | Yes | No | No | No | 0 | None |
|
| |||||||||
| 3 | 25 | Appendix | IV | Yes | Yes | No | No | 5 | PIK3CA |
|
| |||||||||
| 4 | 26 | Right paraureteral serosa | III | Yes | No | No | No | 0 | None |
|
| |||||||||
| 5 | 29 | Right bladder peritoneum | IV | Yes | No | No | No | 0 | None |
|
| |||||||||
| 6 | 34 | Uterosacral peritoneum | IV | Yes | No | No | No | 17 | PPP2R1A |
|
| |||||||||
| 7 | 38 | Bladder peritoneum | III | Yes | No | No | No | 1 | None |
|
| |||||||||
| 8 | 33 | Appendix | IV | Yes | No | No | No | 2 | None |
|
| |||||||||
| 9 | 40 | Colonic wall | IV | Yes | No | No | No | 6 | ARID1A |
|
| |||||||||
| 10 | 23 | Sigmoid colon | IV | Yes | No | No | No | 1 | None |
|
| |||||||||
| 11 | 41 | Rectum | IV | Yes | No | No | No | 10 | None |
|
| |||||||||
| 12 | 39 | Appendix | IV | Yes | No | No | No | 4 | None |
|
| |||||||||
| 13 | 37 | Ileum | IV | Yes | No | No | No | 7 | None |
|
| |||||||||
| 14 | 38 | Paratubal soft tissue | III | Yes | No | No | No | 1 | None |
|
| |||||||||
| 15 | 42 | Hypogastric peritoneum | IV | Yes | Yes | No | No | 2 | KRAS |
|
| |||||||||
| 16 | 39 | Uterosacral ligament | III | Yes | No | No | No | 0 | None |
|
| |||||||||
| 17 | 49 | Paraovarian tissue | III | Yes | No | No | No | 1 | None |
|
| |||||||||
| 18 | 47 | Paratubal soft tissue | III | Yes | No | No | No | 4 | None |
|
| |||||||||
| 19 | 46 | Paratubal soft tissue | III | Yes | No | No | No | 2 | None |
|
| |||||||||
| 20 | 44 | Paratubal soft tissue | III | Yes | No | No | No | 2 | ARID1A |
|
| |||||||||
| 21 | 35 | Abdominal wall | IV | Yes | No | No | No | 1 | None |
|
| |||||||||
| 22 | 51 | Paratubal soft tissue | I | Yes | No | No | No | 2 | None |
|
| |||||||||
| 23 | 30 | Paratubal soft tissue | II | Yes | No | No | No | 1 | None |
|
| |||||||||
| 24 | 48 | Paratubal soft tissue | III | Yes | No | No | No | 0 | None |
|
| |||||||||
| 25 | 40 | Rectovaginal tissue | II | No | No | Yes | Yes | 1 | KRAS |
|
| |||||||||
| 26 | 36 | Rectovaginal tissue | III | No | No | Yes | Yes | 1 | KRAS |
|
| |||||||||
| 27 | 36 | Ureter | II | No | No | Yes | No | 0 | None |
|
| |||||||||
| 28 | 50 | Rectouterine pouch | IV | No | No | No | Yes | 1 | KRAS |
|
| |||||||||
| 29 | 27 | Posterior cul-de-sac | III | No | No | No | Yes | 1 | KRAS |
|
| |||||||||
| 30 | 25 | Uterosacral ligament | III | No | No | No | Yes | 1 | KRAS |
|
| |||||||||
| 31 | 33 | Sigmoid colon | III | No | No | No | Yes | 0 | None |
|
| |||||||||
| 32 | 40 | Uterosacral ligament | II | No | No | No | Yes | 0 | None |
|
| |||||||||
| 33 | 29 | Uterosacral ligament | II | No | No | No | Yes | 0 | None |
|
| |||||||||
| 34 | 41 | Rectovaginal tissue | IV | No | No | No | Yes | 0 | None |
|
| |||||||||
| 35 | 28 | Sigmoid colon | IV | No | No | No | Yes | 0 | None |
|
| |||||||||
| 36 | 41 | Peri ureter | IV | No | No | No | Yes | 0 | None |
|
| |||||||||
| 37 | 31 | Uterosacral ligament | IV | No | No | No | Yes | 0 | None |
|
| |||||||||
| 38 | 34 | Rectovaginal tissue | IV | No | No | No | Yes | 0 | None |
|
| |||||||||
| 39 | 29 | Rectovaginal tissue | III | No | No | No | Yes | 0 | None |
Tissue samples from Patients 1 through 17 were obtained from the Departments of Obstetrics and Gynecology and Pathology at Lenox Hill Hospital-Northwell Health (Hofstra University) in New York. Tissue samples from Patients 18 through 24 were obtained from the Department of Pathology at Seirei Mikatahara Hospital in Hamamatsu, Japan. Tissue samples from Patients 25 through 39 were obtained from the BC Women’s Centre for Pelvic Pain and Endometriosis in Vancouver, BC, Canada. PCR denotes polymerase chain reaction.
EXOMEWIDE ANALYSIS OF ENDOMETRIOTIC LESIONS
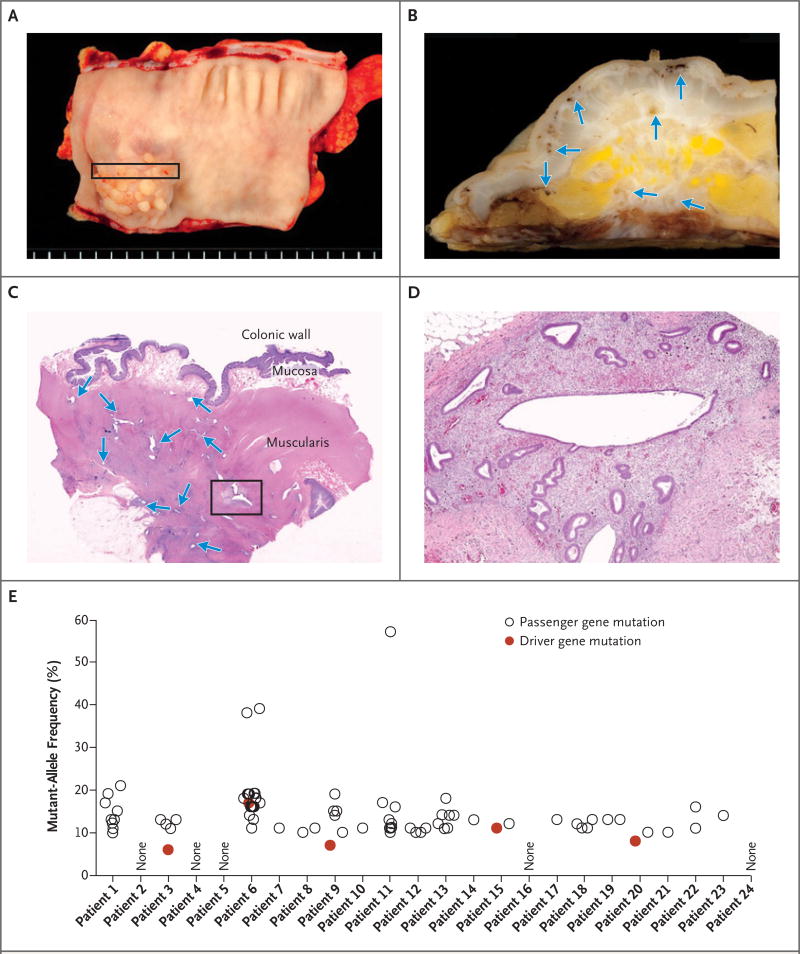
Whole-exome sequencing was performed on samples from 24 patients (Patients 1 through 24) with the use of DNA isolated from endometriosis tissue cores and matched normal tissue (Table 1 and Fig. 1). Coverage was on average 104× (range, 30 to 161) in endometriosis tissue cores and 53× (range, 26 to 97) in normal tissues (Table S1 in Supplementary Appendix 2). We identified 80 nonsynonymous, somatic mutations, including 61 missense and 5 nonsense mutations, 5 insertion–deletion mutations (indels) producing frameshifts, 7 indels that were in-frame, and 2 mutations at canonical splice sites (Table S2 in Supplementary Appendix 2). The number of mutations per lesion varied widely (from 0 to 17), with a mean of 3.3 mutations per lesion (Table 1); five lesions had no detectable mutations. The mutant-allele frequencies of somatic mutations were generally low (<20%) (Fig. 1E, and Table S2 in Supplementary Appendix 2), suggesting that only a subset of cells harbored mutations.
Figure 1. An Example of Deeply Infiltrative Endometriosis in the Colon.
In Panel A, a segment of involved colon shows a papillary lesion projecting into the lumen. Panel B shows the cross section of the colonic wall that is indicated by the rectangle in Panel A; arrows indicate endometriotic lesions. In Panel C, a section stained with hematoxylin and eosin shows multiple, discrete endometriotic foci (arrows) infiltrating into the muscle layer of the colon. The box indicates the approximate region that is core-targeted for molecular analysis. In Panel D, a higher magnification shows the characteristic morphologic features of endometriosis, with both glandular and stromal components. Results for all 24 exome-sequenced patients are shown in Panel E; not all patients’ samples yielded detectable somatic mutations. Both “driver” and “passenger” mutations are indicated (Tables S1 and S2 in Supplementary Appendix 2). As opposed to driver mutations, passenger mutations are defined as somatic mutations that are not known or presumed to directly contribute to cancer initiation or progression.
Five lesions harbored somatic mutations in cancer driver genes. Two lesions harbored frame-shift inactivating mutations in the tumor suppressor ARID1A (p.L2253Cfs*14 in Patient 9 and p.G276Efs*87 in Patient 20), and hotspot activating mutations in PIK3CA (c.3127A→G; p.M1043V), KRAS (c.35G→T; p.G12V), and PPP2R1A (c.767C→T;p.S256F) were each found in one lesion. The remaining mutations were in genes that are not suspected to be driver genes in cancer.
The mutant-allele frequencies of alterations that were identified in cancer driver genes such as ARID1A, PIK3CA, KRAS, and PPP2R1A ranged from 6% to 17% and did not differ significantly from those of non–cancer driver genes. Exome data were also used to estimate DNA copy number; however, small copy-number gains were detected only in Patient 10 (Table S3 in Supplementary Appendix 2).
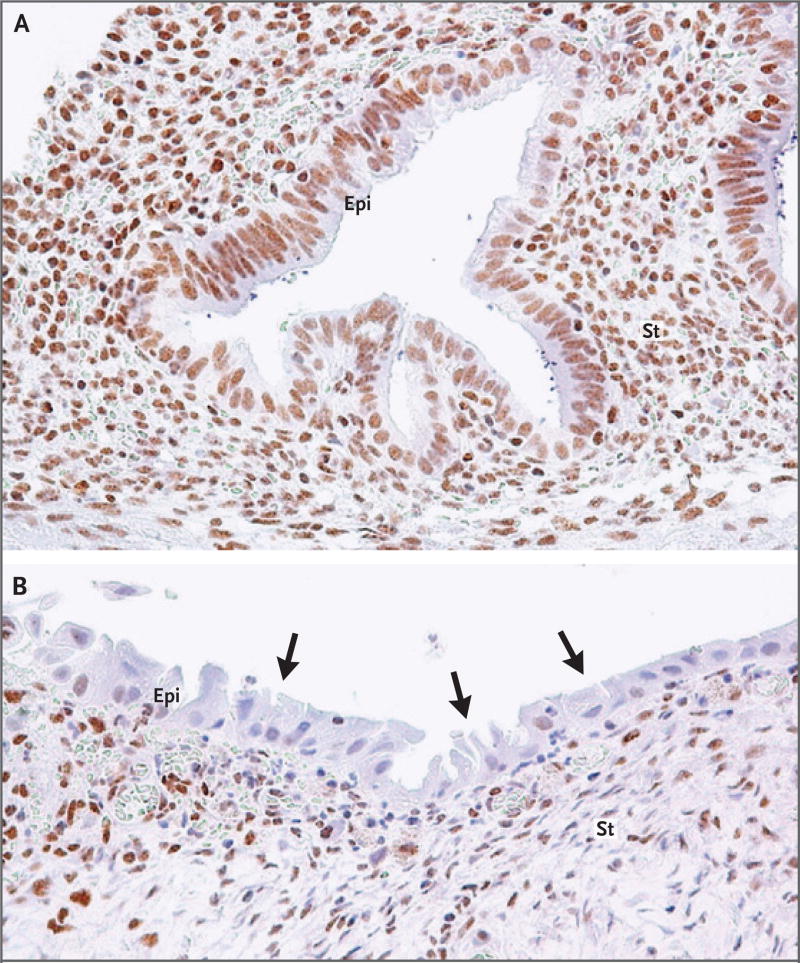
Sufficient DNA was available to validate a subset of these mutations from the same samples with the use of an orthogonal digital genomic method, Safe-SeqS.24 Safe-SeqS confirmed the existence of the somatic mutations and confirmed the allele frequencies from the original exome data (Table S4 in Supplementary Appendix 2). The hotspot KRAS mutation (in Patient 15) was further characterized by laser-capture micro-dissection of endometriotic lesions that separated epithelial and stromal compartments. Herein, the mutation was present in the epithelium but not in the endometrial-type stroma, a finding consistent with the low allelic fraction when both compartments were analyzed together (Table S4 in Supplementary Appendix 2). Loss of ARID1A immunoreactivity served as a surrogate for an inactivating mutation in Patient 20: ARID1A immunoreactivity was undetectable in a subset of the epithelial cells but was present in stromal cells (Fig. 2). In addition, the PPP2R1A mutation in Patient 6 was validated in the epithelial compartment by a droplet digital PCR assay (Table S7 in Supplementary Appendix 2); however, we were unable to recover a pure stromal sample from the remaining lesion.
Figure 2. Immunohistochemistry of ARID1A (BAF250A) in Deep Infiltrating Endometriosis.
ARID1A immunoreactivity was detected in all stromal (St) cells and epithelial (Epi) cells within an endometriotic lesion containing wild-type ARID1A (Panel A). In Patient 20, harboring an ARID1A inactivating mutation, loss of ARID1A immunoreactivity was observed in a subset of epithelial cells (arrows indicate examples), but immunoreactivity was preserved in a much larger fraction of the adjacent stromal cells within the same lesion (Panel B). The mutant-allele fraction of the ARID1A mutation in this patient was 8%.
CANCER DRIVER DEEP-SEQUENCING ANALYSIS OF ENDOMETRIOTIC LESIONS
In an independent set of experiments, we analyzed tissue samples from three patients (Patients 25, 26, and 27) using two targeted panels with overlapping coverage of the hotspot regions of five genes (NRAS, BRAF, FGFFR2, HRAS, and KRAS) and the full coding sequence of six genes (PIK3CA, PTEN, TP53 , CDKN2A , CTNNB1, and FBXW7) (Table S5 in Supplementary Appendix 2). Biphasic endometriotic lesions were isolated by laser-capture microdissection, and mutations were considered to be confirmed if they were present in four panel libraries for each endometriosis sample and absent from (patient-matched) germline DNA (Table S6 in Supplementary Appendix 2 and Fig. S1 in Supplementary Appendix 1). Of the three patients, two harbored activating mutations in KRAS; however, no other somatic alterations passed our filter (i.e., were present in all four libraries). Patient 25 harbored the c.35G→T (p.G12V) and c.35G→C (p.G12A) KRAS mutations, which were present at mean allele frequencies of 11.2% and 8.6%, respectively. Patient 26 had a single c.35G→A (p.G12D) KRAS mutation present at a mean allele frequency of 10.4% (Fig. S2 in Supplementary Appendix 1).
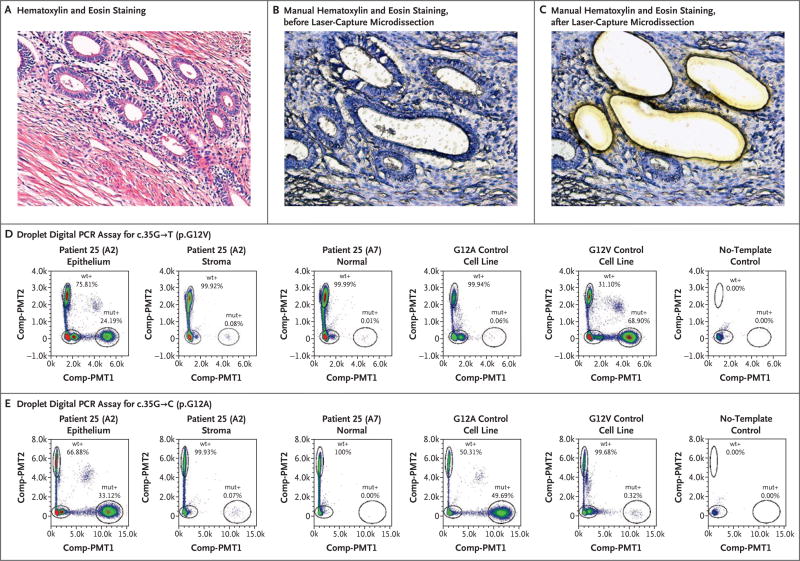
KRAS variants were validated orthogonally by droplet digital PCR assay. Two adjacent blocks from a single lesion in Patient 25 were assayed for both c.35G→T (p.G12V) and c.35G→C (p.G12A); the assay confirmed the presence of the two distinct mutations in the same endometriotic epithelial compartment: the c.35G→T (p.G12V) mutation was present in index and adjacent blocks at frequencies of 24% and 37%, respectively, and the c.35G→C (p.G12A) mutation was present at frequencies of 33% and 16%, respectively (Table 2 and Fig. 3). We examined two blocks in Patient 26, each from a distinct and anatomically separated endometriotic lesion. The KRAS c.35G→A (p.G12D) mutation was confirmed in the epithelial compartment of only the index tissue block (originally subjected to panel sequencing) at a frequency of 31% (Table 2, and Fig. S2 in Supplementary Appendix 1); it was not detected in the other lesion from this patient.
Table 2.
Droplet Digital PCR Assay from Laser-Capture Enriched Specimens.*
| Patient and Block |
Descriptor | Component | Assay | Mutant Droplets |
Wild-Type Droplets |
Variant Frequency |
Result |
|---|---|---|---|---|---|---|---|
| no. | % | ||||||
| 25 | |||||||
|
| |||||||
| A2 | Index | Epithelium | G12A | 21,399 | 43,210 | 33.121 | Mutant |
|
| |||||||
| A3 | Adjacent block | Epithelium | G12A | 16,903 | 86,580 | 16.334 | Mutant |
|
| |||||||
| A2 | Index | Stroma | G12A | 78 | 119,819 | 0.065 | Wild type |
|
| |||||||
| A3 | Adjacent block | Stroma | G12A | 118 | 41,096 | 0.286 | Wild type |
|
| |||||||
| A7 | Normal tissue | LCM: normal | G12A | 2 | 348,307 | 0.001 | Wild type |
|
| |||||||
| A2 | Index | Epithelium | G12V | 17,799 | 55,782 | 24.190 | Mutant |
|
| |||||||
| A3 | Adjacent block | Epithelium | G12V | 50,183 | 85,335 | 37.031 | Mutant |
|
| |||||||
| A2 | Index | Stroma | G12V | 96 | 116,715 | 0.082 | Wild type |
|
| |||||||
| A3 | Adjacent block | Stroma | G12V | 3 | 39,427 | 0.008 | Wild type |
|
| |||||||
| A7 | Normal tissue | LCM: normal | G12V | 25 | 342,829 | 0.007 | Wild type |
|
| |||||||
| 26 | |||||||
|
| |||||||
| B4 | Index | Epithelium | G12D | 13,051 | 29,148 | 30.927 | Mutant |
|
| |||||||
| F2 | Distant lesion | Epithelium | G12D | 5 | 13,138 | 0.038 | Wild type |
|
| |||||||
| B4 | Index | Stroma | G12D | 448 | 89,085 | 0.500 | Wild type |
|
| |||||||
| F2 | Distant lesion | Stroma | G12D | 80 | 60,865 | 0.131 | Wild type |
|
| |||||||
| B4 | Normal tissue | LCM: normal | G12D | 74 | 42,133 | 0.175 | Wild type |
|
| |||||||
| 28 | |||||||
|
| |||||||
| C2 | Index | Epithelium | G12D | 40,767 | 66,080 | 38.155 | Mutant |
|
| |||||||
| A8 | Distant lesion 1 | Epithelium | G12D | 12,643 | 27,849 | 31.223 | Mutant |
|
| |||||||
| B3 | Distant lesion 2 | Epithelium | G12D | 13,067 | 126,391 | 9.370 | Mutant |
|
| |||||||
| C2 | Index | Stroma | G12D | 4 | 170,166 | 0.002 | Wild type |
|
| |||||||
| A8 | Distant lesion 1 | Stroma | G12D | 109 | 23,854 | 0.455 | Wild type |
|
| |||||||
| B3 | Distant lesion 2 | Stroma | G12D | 4 | 147,595 | 0.003 | Wild type |
|
| |||||||
| A10 | Normal endometrium | Endometrium | G12D | 36 | 12,630 | 0.284 | Wild type |
|
| |||||||
| A5 | Normal endocervix | Endocervix | G12D | 6 | 11,253 | 0.053 | Wild type |
|
| |||||||
| A8 | Normal tissue | LCM: normal | G12D | 380 | 121,408 | 0.312 | Wild type |
|
| |||||||
| 29 | |||||||
|
| |||||||
| D1 | Index | LCM: stroma and epithelium | G12V | 2,698 | 72,473 | 3.589 | Mutant |
|
| |||||||
| D1 | Normal tissue | LCM: normal | G12V | 3 | 12,164 | 0.025 | Wild type |
|
| |||||||
| 30 | |||||||
|
| |||||||
| F1 | Index | LCM: stroma and epithelium | G12D | 2,444 | 115,916 | 2.065 | Mutant |
|
| |||||||
| F1 | Normal tissue | LCM: normal | G12D | 164 | 97,118 | 0.169 | Wild type |
LCM denotes laser-capture microdissection.
Figure 3. Confirmation of Activating, Somatic KRAS Mutations in the Glandular Epithelium but not Stromal Compartments of Deep Endometriotic Lesions.
Panel A shows a photomicrograph of endometriotic tissue from Patient 25, with standard hematoxylin and eosin staining. Panels B and C show manually stained, non–cover-slipped sections, also from Patient 25, that were prepared for laser-capture microdissection. Panels D and E show droplet digital polymerase-chain- reaction (PCR) plots illustrating the presence of both c.35G→T (p.G12V) and c.35G→C (p.G12A) KRAS mutations at different allelic frequencies but exclusive to the glandular epithelium in Patient 25. The allelic frequencies represent the percentage of droplets that were positive for the mutant allele (mut+) or positive for the wild-type allele (wt+). Control cell lines and no-template controls (including all reaction components except a DNA template) are also shown. Comp-PMT1 denotes spectrally compensated photomultiplier tube 1 (the dye channel used for the mutant assay), and Comp-PMT2 spectrally compensated photo-multiplier tube 2 (the dye channel used for the wild-type assay).
RECURRENT KRAS G12 VARIANTS IN DEEP INFILTRATING ENDOMETRIOSIS
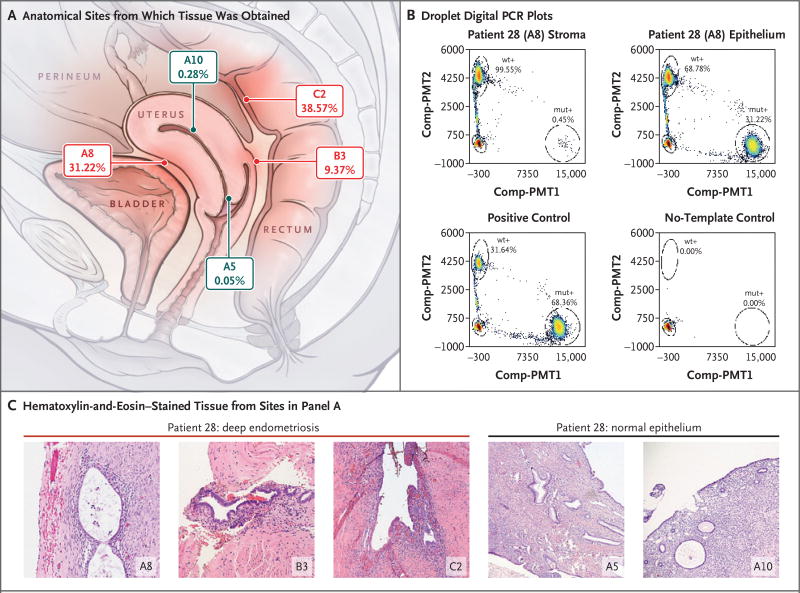
Because KRAS alterations were detected in the above two cohorts (3 of 27 patients), we analyzed deep infiltrating endometriosis lesions from an additional 12 patients (Patients 28 through 39) (Table 1) using droplet digital PCR assays for five KRAS codon 12 variants (c.35G→A [p.G12D], c.35G→T [p.G12V], c.35G→C [p.G12A], c.34G→C [p.G12R], and C.34G→T [p.G12C]). Droplet digital PCR assays were initially performed in manually macrodissected lesions (Table S7 in Supplementary Appendix 2), and lesions that showed positivity were then laser-capture microdissected, with stromal and epithelial compartments separated when possible. We detected KRAS mutations in 3 of 12 patients (25%) (Table 2). For Patient 28 with a c.35G→A (p.G12D) mutation, three independent deep lesions and normal endometrial epithelium were available for laser-capture microdissection and high-resolution droplet digital PCR analysis (Fig. 4). All three anatomically distinct endometriosis lesions harbored the same KRAS c.35G→A alteration in the glandular epithelium; however, we were not able to detect the mutation in endometriosis stroma or normal eutopic endometrial epithelium from the same patient.
Figure 4. Co-Occurring and Anatomically Distinct Deep Infiltrating Endometriosis Lesions That Harbor Identical KRAS Mutations.
Panel A shows an anatomical diagram outlining the position of three distinct deep infiltrating endometriosis lesions (in red) and normal sampling of eutopic endometrial and endocervical epithelium (in gray), all from Patient 28. The allelic frequency of the epithelial-restricted c.35G→A (p.G12D) KRAS mutation from droplet digital PCR experiments is also shown below each block identifier. Panel B shows droplet digital PCR plots confirming the KRAS mutation in the epithelial, but not stromal, component of the A8 endometriotic lesion, with the mutant-droplet-positive fraction (allelic frequency). Controls are also shown. Panel C shows hematoxylin-and-eosin photomicrographs of each location shown in Panel A, including endometriotic tissue taken from the three distinct lesions: in the anterior serosal surface of the uterus (A8) as well as the vaginal (B3) and rectal (C2) surfaces of the rectouterine pouch.
DISCUSSION
Despite the high prevalence of endometriosis and its effect on women’s health-related quality of life,29 very little is known about the biologic processes that underpin it, although more than a century has elapsed since this disease was first described.30,31 There is a socioeconomic imperative for endometriosis research; in the United States alone, the economic burdens are more than $12,000 per hospital stay,32 more than $11,000 per year per woman with the disorder,32,33 and more than $54 billion per year in total.34 In this study, we used a combination of next-generation sequencing and validation through highly sensitive digital genomic assays and found that the majority of benign deep infiltrating endometriosis lesions harbored somatic mutations, including mutations in the well-known cancer driver genes ARID1A, PIK3CA, KRAS, and PPP2R1A.35–37
Our reporting of these driver gene mutations in the affected patients should be considered conservative, because methods for sequencing ultra-low-input and formalin-fixed materials are still developing. Should a more complete genomic or epigenomic analysis be applied, additional driver mutations may be uncovered. Nonetheless, the driver genes that we observed to be altered are frequently mutated in endometriosis-related ovarian neoplasms and in (clonally related) adjacent and distant endometriotic lesions from patients with endometriosis-related ovarian neoplasms.14,38 Yet, it is critical to note that endometriosis is generally regarded to be a nonmalignant inflammatory condition.3 The finding of cancer-associated mutations in endometriotic lesions without concurrent cancer and, in particular, in nonovarian deep infiltrating lesions that rarely (if ever) transform into cancer was surprising. These mutations may be an intrinsic characteristic of deep infiltrating endometriosis and raise interesting questions about the pathobiology of endometriosis.
In the past, studies that examined endometriosis-related ovarian neoplasms with concurrent endometriosis suggested that driver mutations shared by both lesions were the mutations responsible for the progression of the endometriosis to cancer.16,17,39,40 However, given an estimated rate of malignant transformation for endometriosis close to 1%,41 our results suggest that the presence of driver mutations alone is neither sufficient to drive the transformation of endometriosis nor indicative of likely progression to cancer. At least one study of a mouse model of endometriosis has already provided evidence that (subclonal) activating Kras mutations can sustain endometriosis but are not sufficient for malignant transformation.42 Our data also agree with studies of other organ systems that have shown cancer driver mutations in benign lesions and normal tissues.43–46 Beyond this, because only a minority of our patients harbored detectable driver mutations, the current study may suggest that driver mutations are not required for the development of (deep infiltrating) endometriosis. Perhaps more important, our findings challenge past research, in which endometriosis was typically studied in conjunction with endometriosis-related ovarian neoplasms and in which candidate single-gene approaches were used; these types of studies are unlikely to be sufficient in addressing the risk of transformation.
The probability that 5 mutations, out of the 80 mutations found, were in a pool of 125 driver genes was calculated to be P = 0.001 (binomial test), which suggests that these mutations were highly unlikely to have arisen by chance alone. Because cancer driver mutations were present only in the epithelium but not the stroma of the same endometriosis lesion, we can assume that the observed mutations provide some key selective advantage to endometriotic epithelial cells. This apparent selective pressure in the epithelial compartment could result in the emergence of distinct clonal populations within the same lesion. For example, Patient 25 harbored two different KRAS codon 12 mutations. Again, not all lesions harbored detectable driver gene mutations; future studies are necessary to more accurately determine the prevalence of specific driver events, as well as to address whether cases of endometriosis without driver mutations have functionally equivalent genomic or epigenomic events that were not assessed in this study and whether mutation-harboring endometriosis represents a distinct pathology. Finally, not all lesions in a given patient were found to harbor a detectable (driver) mutation (e.g., Patient 26), a trend that has also been observed in the context of endometriotic lesions co-occurring with cancer 14; these findings potentially support the coexistence of multiple endometriosis lineages within the same patient.
Deep infiltrating endometriosis can invade visceral organs and distort local anatomy, whereas in the superficial subtype of endometriosis, lesions are noninvasive and anatomical relationships are maintained. The biologic processes underpinning these distinct phenotypes is unknown. Our findings challenge the current understanding of this invasive subtype of endometriosis and open the discussion on whether deep infiltrating endometriosis can be considered as a benign neoplasm. One patient (Patient 28) had the same KRAS mutation in three spatially distinct lesions, raising the possibility that some sites of benign deep infiltrating endometriosis arise through the neoplastic process of metastasis. However, several mechanisms should be considered as explanations for this observation. First, mutations may have arisen independently of each other. However, it is unlikely that exactly the same mutation occurred independently, by chance, in all three lesions. Second, the mutation may have arisen in a single lesion and subsequently disseminated to other sites. This possibility supports the “metastasis” of somatically altered endometriosis epithelial cells but does not specifically address the contentious issue of endometriosis originating through metaplasia or dissemination from the endometrium. Nonetheless, if we choose to accept this explanation, there is also the nonmutant stromal component of the endometriosis to consider; either this component must be required to maintain fitness of the epithelial cells and therefore co-migrates (is recruited) with the epithelium during the pathogenesis of endometriosis, or it develops after the establishment of the (mutant) endometrial epithelial cell through metaplasia of non-endometrial stromal cells. Lastly, a bipotent (uterine) stem cell origin leading to both stromal and epithelial components of endometriosis has been proposed.47,48 A stem-cell–related theory may provide a plausible route for single-cell metastasis; however, this would require a specific pressure for only the epithelial progeny to gain (or retain) the cancer driver mutations that we have observed thus far. Clearly, this hypothesis requires additional validation, but it may be more compatible with rare and distant sites of endometriosis (e.g., in the lung, spine, and brain).49
A spectrum of molecular analyses are needed to address key questions about the pathogenesis of endometriosis and its clinical behavior (Table S8 in Supplementary Appendix 2). The current clinical classification of endometriosis is neither biologically based nor strongly predictive of clinical behavior50; herein lies an opportunity to improve or redesign a biologically informed classification scheme. There is a broad range of challenges faced by women with endometriosis and their caregivers; this study provides a strong rationale and the molecular foundations for the extensive characterization of all endometriosis subtypes. Given the wide range and often subjective nature (e.g., self-reported pain) of clinical characteristics across patients with endometriosis, large and well-annotated cohorts will be needed.
In summary, although endometriosis is considered to be a benign disorder from both a clinical and a histopathological perspective, well-known cancer-associated somatic mutations were found in the glandular epithelium of some deep infiltrating endometriosis lesions. These findings create new opportunities for a more detailed examination of all forms of endometriosis with the use of research approaches that are common in the study of cancer.
Supplementary Material
Acknowledgments
Supported in part by the Richard W. TeLinde Gynecologic Pathology Research Program at Johns Hopkins University, the Virginia and D.K. Ludwig Fund for Cancer Research, the Ephraim and Wilma Shaw Roseman Foundation, the Endometriosis Foundation of America, the National Institutes of Health and National Cancer Institute (grants P50-CA62924, CA06973, GM07184, GM07309, CA09243, CA57345, P30-CA006973, CA215483, and UO1-CA200469), the Gray Family Ovarian Clear Cell Carcinoma Research Resource, the Canadian Cancer Society (grant 701603), the Canadian Institutes of Health Research (IHD-137431 and MOP-142273), the Canadian Foundation for Innovation (John R. Evans Leaders Fund) and British Columbia Knowledge Development Fund, the Women’s Health Research Institute (Nelly Auersperg Grant), and the Canadian Foundation for Women’s Health (General Research Grant). The BC Women’s Hospital and Health Centre Foundation provided support to the BC Women’s Centre for Pelvic Pain and Endometriosis. The BC Cancer Foundation and the VGH and UBC Hospital Foundation provided funding to OVCARE: BC’s Ovarian Cancer Research Team (Dr. Anglesio, Dr. Nazeran, Dr. Horlings, Ms. Lum, Ms. Senz, Ms. Ho, Ms. Lac, Dr. Tessier-Cloutier, Ms. Wang, Dr. Boyd, Dr. Gilks, Dr. Yong, and Dr. Huntsman), including donations from David and Darrell Mindell, Peter and Shelley O’Sullivan, and the Jemini Foundation to directly support this research. The Mentored Clinician-Scientist Award of the Vancouver Coastal Health Research Institute provided support to Dr. Yong. The Dr. Chew Wei Memorial Professorship in Gynecologic Oncology and the Canada Research Chairs program (Research Chair in Molecular and Genomic Pathology) provided support to Dr. Huntsman. The Dutch Cancer Society translational research fellowship (KWF 2013-5869) provided support to Dr. Horlings.
We thank all the women who have allowed their tissue to be used in our research studies and without whom this work would not have been possible; staff at Illumina who provided early-access reagents for targeted sequencing, including beta versions of the TruSeq Amplicon Cancer Panel; and staff at the Genetic Pathology Evaluation Centre in Vancouver and Sol Goldman Sequencing Facility at Johns Hopkins, both of which provided technical support.
Appendix
The authors’ full names and academic degrees are as follows: Michael S. Anglesio, Ph.D., Nickolas Papadopoulos, Ph.D., Ayse Ayhan, M.D., Ph.D., Tayyebeh M. Nazeran, M.D., Michaël Noë, M.D., Hugo M. Horlings, M.D., Ph.D., Amy Lum, B.Sc., Siân Jones, Ph.D., Janine Senz, B.Sc., Tamer Seckin, M.D., Julie Ho, M.H.A., Ren-Chin Wu, M.D., Ph.D., Vivian Lac, B.Sc., Hiroshi Ogawa, M.D., Basile Tessier-Cloutier, M.D., Rami Alhassan, M.D., Amy Wang, Yuxuan Wang, B.Sc., Joshua D. Cohen, B.Sc., Fontayne Wong, B.Sc., Adnan Hasanovic, M.D., Natasha Orr, B.Sc., Ming Zhang, Ph.D., Maria Popoli, B.Sc., Wyatt McMahon, Ph.D., Laura D. Wood, M.D., Ph.D., Austin Mattox, B.Sc., Catherine Allaire, M.D., James Segars, M.D., Christina Williams, M.D., Cristian Tomasetti, Ph.D., Niki Boyd, Ph.D., Kenneth W. Kinzler, Ph.D., C. Blake Gilks, M.D., Luis Diaz, M.D., Tian-Li Wang, Ph.D., Bert Vogelstein, M.D., Paul J. Yong, M.D., Ph.D., David G. Huntsman, M.D., and Ie-Ming Shih, M.D., Ph.D.
The authors’ affiliations are as follows: the Departments of Obstetrics and Gynaecology (M.S.A., C.A., C.W., P.J.Y., D.G.H.) and Pathology and Laboratory Medicine (M.S.A., T.M.N., J. Senz, B.T.-C., C.B.G., D.G.H.), University of British Columbia, the Department of Anatomical Pathology, Vancouver General Hospital (T.M.N., H.M.H., J.H., V.L., B.T.-C., A.W., C.B.G., D.G.H.), the Department of Molecular Oncology, British Columbia Cancer Agency (H.M.H., A.L., V.L., N.B., D.G.H.), and the BC Women’s Centre for Pelvic Pain and Endometriosis, BC Women’s Hospital and Health Centre (F.W., N.O., C.A., C.W., P.J.Y.) — all in Vancouver, BC, Canada; the Department of Oncology (N.P., C.T., K.W.K., L.D., T.-L.W., B.V., I.-M.S.) and Ludwig Center (N.P., Y.W., J.D.C., M.Z., M.P., W.M., A.M., K.W.K., L.D., B.V.), Sidney Kimmel Comprehensive Cancer Center, the Departments of Pathology (N.P., A.A., M.N., L.D.W., T.-L.W., B.V., I.-M.S.) and Gynecology and Obstetrics (J. Segars, I.-M.S.), Johns Hopkins Medical Institutions, Personal Genome Diagnostics (S.J.), and Johns Hopkins University Howard Hughes Medical Institute (B.V.) — all in Baltimore; the Department of Pathology, Seirei Mikatahara Hospital (A.A., H.O.), and the Department of Tumor Pathology, Hamamatsu University School of Medicine (A.A.), Hamamatsu, and the Department of Molecular Pathology, Hiroshima University School of Medicine, Hiroshima (A.A.) — all in Japan; the Department of Pathology, University Medical Center Utrecht, Utrecht, the Netherlands (M.N.); the Departments of Obstetrics and Gynecology (T.S.) and Pathology (R.A., A.H.), Lenox Hill Hospital–Northwell Health (Hofstra University), New York; and the Department of Pathology, Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Tao-Yuan City, Taiwan (R.-C.W.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Giudice LC. Endometriosis. N Engl J Med. 2010;362:2389–98. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reese KA, Reddy S, Rock JA. Endometriosis in an adolescent population: the Emory experience. J Pediatr Adolesc Gynecol. 1996;9:125–8. doi: 10.1016/s1083-3188(96)70021-9. [DOI] [PubMed] [Google Scholar]
- 3.Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–79. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 4.Leyland N, Casper R, Laberge P, Singh SS. Endometriosis: diagnosis and management. J Obstet Gynaecol Can. 2010;32(Suppl 2):S1–S32. [PubMed] [Google Scholar]
- 5.Tosti C, Pinzauti S, Santulli P, Chapron C, Petraglia F. Pathogenetic mechanisms of deep infiltrating endometriosis. Reprod Sci. 2015;22:1053–9. doi: 10.1177/1933719115592713. [DOI] [PubMed] [Google Scholar]
- 6.Ferrero S, Alessandri F, Racca A, Leone Roberti Maggiore U. Treatment of pain associated with deep endometriosis: alternatives and evidence. Fertil Steril. 2015;104:771–92. doi: 10.1016/j.fertnstert.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 7.Donnez J, Squifflet J, Pirard C, Jadoul P, Wyns C, Smets M. The efficacy of medical and surgical treatment of endometriosis-associated infertility and pelvic pain. Gynecol Obstet Invest. 2002;54(Suppl 1):2–7. doi: 10.1159/000066288. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson BR, Bennington JL, Haber SL. Histochemistry of mucosubstances and histology of mixed Müllerian pelvic lymph node glandular inclusions: evidence for histogenesis by Müllerian metaplasia of coelomic epithelium. Obstet Gynecol. 1969;33:617–25. [PubMed] [Google Scholar]
- 9.Sampson JA. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am J Pathol. 1927;3 93-110.43. [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson JA. Peritoneal endometriosis due to menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:422–69. [Google Scholar]
- 11.Sasson IE, Taylor HS. Stem cells and the pathogenesis of endometriosis. Ann N Y Acad Sci. 2008;1127:106–15. doi: 10.1196/annals.1434.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueira PG, Abrão MS, Krikun G, Taylor HS. Stem cells in endometrium and their role in the pathogenesis of endometriosis. Ann N Y Acad Sci. 2011;1221:10–7. doi: 10.1111/j.1749-6632.2011.05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung JN, Rogers PA, Montgomery GW. Identifying the biological basis of GWAS hits for endometriosis. Biol Reprod. 2015;92:87. doi: 10.1095/biolreprod.114.126458. [DOI] [PubMed] [Google Scholar]
- 14.Anglesio MS, Bashashati A, Wang YK, et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. J Pathol. 2015;236:201–9. doi: 10.1002/path.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurman RJ, Shih IM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186:733–47. doi: 10.1016/j.ajpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vestergaard AL, Thorup K, Knudsen UB, et al. Oncogenic events associated with endometrial and ovarian cancers are rare in endometriosis. Mol Hum Reprod. 2011;17:758–61. doi: 10.1093/molehr/gar049. [DOI] [PubMed] [Google Scholar]
- 17.Sato N, Tsunoda H, Nishida M, et al. Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 2000;60:7052–6. [PubMed] [Google Scholar]
- 18.Samartzis EP, Samartzis N, Noske A, et al. Loss of ARID1A/BAF250a–expression in endometriosis: a biomarker for risk of carcinogenic transformation? Mod Pathol. 2012;25:885–92. doi: 10.1038/modpathol.2011.217. [DOI] [PubMed] [Google Scholar]
- 19.Borrelli GM, Abrão MS, Taube ET, et al. (Partial) loss of BAF250a (ARID1A) in recto-vaginal deep-infiltrating endometriosis, endometriomas and involved pelvic sentinel lymph nodes. Mol Hum Reprod. 2016;22:329–37. doi: 10.1093/molehr/gaw009. [DOI] [PubMed] [Google Scholar]
- 20.Chene G, Ouellet V, Rahimi K, Barres V, Provencher D, Mes-Masson AM. The ARID1A pathway in ovarian clear cell and endometrioid carcinoma, contiguous endometriosis, and benign endometriosis. Int J Gynaecol Obstet. 2015;130:27–30. doi: 10.1016/j.ijgo.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Fassbender A, Rahmioglu N, Vitonis AF, et al. World Endometriosis Research Foundation Endometriosis Phenome, Biobanking Harmonisation Project: IV. Tissue collection, processing, and storage in endometriosis research. Fertil Steril. 2014;102:1244–53. doi: 10.1016/j.fertnstert.2014.07.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robles AI, Traverso G, Zhang M, et al. Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology. 2016;150:931–43. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anglesio MS, Wang YK, Maassen M, et al. Synchronous endometrial and ovarian carcinomas: evidence of clonality. J Natl Cancer Inst. 2016;108(6):djv428. doi: 10.1093/jnci/djv428. [DOI] [PubMed] [Google Scholar]
- 24.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayhan A, Mao TL, Seckin T, et al. Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int J Gynecol Cancer. 2012;22:1310–5. doi: 10.1097/IGC.0b013e31826b5dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan B, Mao TL, Panuganti PK, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–32. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu RC, Ayhan A, Maeda D, et al. Frequent somatic mutations of the telomerease reverse transcriptase promoter in ovarian clear cell carcinoma but not in other major types of gynaecological malignancy. J Pathol. 2014;232:473–81. doi: 10.1002/path.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao TL, Ardighieri L, Ayhan A, et al. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–8. doi: 10.1097/PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Graaff AA, D’Hooghe TM, Dunselman GA, Dirksen CD, Hummelshoj L, Simoens S. The significant effect of Endometriosis on physical, mental and social wellbeing: results from an international cross-sectional survey. Hum Reprod. 2013;28:2677–85. doi: 10.1093/humrep/det284. [DOI] [PubMed] [Google Scholar]
- 30.Cullen TS. Adenoma-myoma uteri diffusum benignum. Johns Hopkins Hosp Bull. 1896;6:133–7. [Google Scholar]
- 31.Russell WW. Aberrant portions of the Mullerian duct found in an ovary. Johns Hopkins Hosp Bull. 1899;10:8–9. [Google Scholar]
- 32.Gao X, Outley J, Botteman M, Spalding J, Simon JA, Pashos CL. Economic burden of endometriosis. Fertil Steril. 2006;86:1561–72. doi: 10.1016/j.fertnstert.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Klein S, D’Hooghe T, Meuleman C, Dirksen C, Dunselman G, Simoens S. What is the societal burden of endometriosis-associated symptoms? A prospective Belgian study. Reprod Biomed Online. 2014;28:116–24. doi: 10.1016/j.rbmo.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Simoens S, Dunselman G, Dirksen C, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27:1292–9. doi: 10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- 35.McConechy MK, Anglesio MS, Kalloger SE, et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol. 2011;223:567–73. doi: 10.1002/path.2848. [DOI] [PubMed] [Google Scholar]
- 36.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas EJ, Campbell IG. Molecular genetic defects in endometriosis. Gynecol Obstet Invest. 2000;50(Suppl 1):44–50. doi: 10.1159/000052878. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol. 2012;25:615–24. doi: 10.1038/modpathol.2011.189. [DOI] [PubMed] [Google Scholar]
- 41.Wei JJ, William J, Bulun S. Endometriosis and ovarian cancer: a review of clinical, pathologic, and molecular aspects. Int J Gynecol Pathol. 2011;30:553–68. doi: 10.1097/PGP.0b013e31821f4b85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng CW, Licence D, Cook E, et al. Activation of mutated Kras in donor endometrial epithelium and stroma promotes lesion growth in an intact immunocompetent murine model of endometriosis. J Pathol. 2011;224:261–9. doi: 10.1002/path.2852. [DOI] [PubMed] [Google Scholar]
- 43.Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution: high burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–6. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McConnell MJ, Lindberg MR, Brennand KJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–7. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in non-cancerous tissues. Proc Natl Acad Sci U S A. 2016;113:6005–10. doi: 10.1073/pnas.1601311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The conundrum of genetic “drivers” in benign conditions. J Natl Cancer Inst. 2016;108:108. doi: 10.1093/jnci/djw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda H, Matsuzaki Y, Hiratsu E, et al. Stem cell-like properties of the endometrial side population: implication in endometrial regeneration. PLoS One. 2010;5(4):e10387. doi: 10.1371/journal.pone.0010387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cervelló I, Mas A, Gil-Sanchis C, et al. Reconstruction of endometrium from human endometrial side population cell lines. PLoS One. 2011;6(6):e21221. doi: 10.1371/journal.pone.0021221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jubanyik KJ, Comite F. Extrapelvic endometriosis. Obstet Gynecol Clin North Am. 1997;24:411–40. doi: 10.1016/s0889-8545(05)70311-9. [DOI] [PubMed] [Google Scholar]
- 50.Vercellini P, Fedele L, Aimi G, Pietropaolo G, Consonni D, Crosignani PG. Association between endometriosis stage, lesion type, patient characteristics and severity of pelvic pain symptoms: a multivariate analysis of over 1000 patients. Hum Reprod. 2007;22:266–71. doi: 10.1093/humrep/del339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.