Abstract
Despite its limited immediate reinforcement value, alcohol has a potent ability to induce neuroadaptations that promote its incentive salience, escalation of voluntary alcohol intake and aversion-resistant alcohol seeking. A constellation of these traits, collectively called ‘post-dependent’, emerges following brain exposure to repeated cycles of intoxication and withdrawal. The medial prefrontal cortex (mPFC) and its subdivisions exert top-down regulation of approach and avoidance behaviors, including those that lead to alcohol intake. Here, we review an emerging literature which indicates that a reprogramming of mPFC function occurs with prolonged exposure of the brain to cycles of alcohol intoxication and withdrawal. This reprogramming results in molecular dysregulations that contribute to the post-dependent syndrome. Convergent evidence has identified neuroadaptations resulting in altered glutamatergic and BDNF-mediated signaling, and for these pathways, direct evidence for a mechanistic role has been obtained. Additional evidence points to a dysregulation of pathways involving calcium homeostasis and neurotransmitter release. Recent findings indicate that global DNA hypermethylation is a key factor in reprogramming the mPFC genome after a history of dependence. As one of the results of this epigenetic remodeling, several histone modifying epigenetic enzymes are repressed. Among these, PR-domain zinc-finger protein 2, a methyltransferase that selectively mono-methylates histone H3 at lysine 9 has been functionally validated to drive several of the molecular and behavioral long-term consequences of alcohol dependence. Information processing within the mPFC involves formation of dynamic neuronal networks, or functional ensembles that are shaped by transcriptional responses. The epigenetic dysregulations identified by our molecular studies are likely to alter this dynamic processing in multiple ways. In summary, epigenetic molecular switches in the mPFC appear to be turned on as alcoholism develops. Strategies to reverse these processes may offer targets for disease-modifying treatments.
Keywords: Alcohol use disorder, animal model, BDNF, DNA methylation, inhibitory control, mGlur2, miRNA, neuronal ensemble, post-dependent, transcriptome, viral vector
Alcohol addiction: emergence of the syndrome
Alcohol addiction [hereafter equated with alcohol dependence, moderate – severe alcohol use disorder (AUD) or simply ‘alcoholism’, the term preferred by Alcoholics Anonymous] develops over years to decades. Most attempts to capture this syndrome by different diagnostic systems are based on symptoms observed or reported here-and-now. However, because of its chronic, progressive nature, it is also informative to consider the development of this condition over time. A prolonged, repeated brain alcohol exposure at considerable levels (above 150–250 mg/dl), interspersed with alcohol-free intervals, is necessary albeit not sufficient for alcoholism to develop (Heilig et al. 2010). The transition into the clinical disorder is slow and gradual, as illustrated by the fact that the time from meeting diagnostic criteria for alcohol dependence to seeking treatment is on average about a decade (Hasin et al. 2007).
Epidemiological studies have long indicated that only a minority of people exposed to addictive drugs develop addiction (Anthony et al. 1994). While 87.6% of people aged 18 or older in the U.S. have consumed alcohol in their lifetime, only 6.8% become affected by an alcohol use disorder (SAMHSA 2014). The way alcohol addiction evolves in a significant minority of people who expose their brains to alcohol has some important implications. It suggests that neuroadaptive processes are triggered in these individuals, and that these processes do not occur in the majority of exposed subjects who do not develop a clinical syndrome. Alcohol-induced neuroadaptations appear to persistently change brain functions that affect the ability to control alcohol drinking. A biological understanding of these changes requires them to be modeled in experimental animals, where they can be studied at a molecular level. This is a challenging task, because commonly used laboratory mice and rats do not voluntarily consume amounts of alcohol sufficient to cause brain alcohol exposure levels approaching those that occur clinically. An additional challenge is the long duration of brain exposure required for alcoholism to develop in patients. In rats, following several months of voluntary consumption, some behavioral traits believed to be relevant for alcoholism do emerge in a significant minority of animals (Wolffgramm & Heyne 1995). However, exposure levels in this model are still modest, and the procedure is impractical because of its duration. Models based on genetic selection can result in voluntary consumption approaching adequate levels of exposure, but do so at the expense of other challenges. Perhaps most importantly, genetic selection leads to random allelic fixation throughout the genome, resulting in a segregation of multiple behavioral and biological traits of uncertain relation to alcohol seeking and taking (Sommer et al. 2006; Zhou et al. 2013).
In patients who ultimately develop alcohol addiction, brain alcohol exposure occurs with a pattern that alternates between cycles of intoxication and abstinence. This type of intermittent exposure is at the core of two animal models that have gained considerable popularity, every-second-day intermittent access (Hopf et al. 2010; Simms et al. 2008; Wise 1973, 1975), and long-term drinking with repeated deprivation phases (Spanagel & Holter 1999; Vengeliene et al. 2014). These models produce several behavioral traits of considerable interest, most importantly an escalation of voluntary alcohol consumption, and drinking that is resistant to aversive consequences such as quinine adulteration (‘aversion resistant’, or ‘compulsive’ drinking). However, unless exposure is for extended periods of time [3–4 months; (Hopf et al. 2010)], the consequences of alcohol exposure in both these models are limited, or of limited duration. This indicates that, to the extent they trigger neuroadaptive processes important for the evolution of alcohol addiction, these models do so in an incomplete manner.
The use of an intermittent brain exposure pattern can be pushed a step further through the use of an approach pioneered in its basic form by Dora Goldstein, in her classic studies of alcohol withdrawal (Goldstein & Pal 1971). Using this method, an intermittent pattern of brain alcohol exposure can be imposed on rats through vapor chambers, inducing blood-alcohol levels relevant for alcoholism (about 150–250 mg/dl) that can be maintained for an extended period of time [4–7 weeks; (O’Dell et al. 2004; Rimondini et al. 2002, 2003, 2008)]. This results in the emergence of a cluster of traits that persist for a long time after exposure is terminated, and make up what we have collectively called ‘the post-dependent state’ (Heilig & Koob 2007; Heilig et al. 2010). Key characteristics of this syndrome, and its utility as a tool for drug discovery have recently been reviewed (Meinhardt & Sommer 2015). In brief, exposure using this approach results in escalation of subsequent alcohol intake, both measured as home cage consumption on two-bottle free-choice access, and as rates of operant self-administration. Self-administration on a progressive ratio schedule is also increased, supporting an increased motivation to obtain alcohol. Furthermore, post-dependent rats continue to consume alcohol following adulteration with quinine to a higher extent than do non-dependent rats, showing evidence of ‘compulsivity’. These behavioral consequences of intermittent vapor exposure typically been observed 1–3 weeks after exposure has been terminated, in the absence of physical withdrawal. They have furthermore been studied up to 6 weeks following termination of exposure, and have appeared stable over this time (Meinhardt & Sommer 2015).
The use of chronic intermittent vapor exposure has allowed us to study post-dependent neuroadaptations at a molecular level. Our initial studies used a candidate-based approach, and focused on the role of neuroadaptations involving corticotropin-releasing hormone and its receptors within the amygdala complex (Gehlert et al. 2007; Sommer et al. 2008). This work, which converged with that of other laboratories, has been reviewed previously (Heilig & Koob 2007). In parallel, we have pursued transcriptome-wide approaches as an unbiased discovery strategy, with the objective of identifying novel mechanisms that might ultimately offer treatment targets. A series of these studies have focused on the medial prefrontal cortex (mPFC), a brain area that exerts important top-down control over structures that mediate approach and positively reinforced drug seeking, such as the Nucleus Accumbens (NAc), as well as structures that mediate aversion and negatively reinforced drug seeking (Koob & Volkow 2010; Peters et al. 2009).
In the following, we first briefly review important structural features of the rodent mPFC, and their potential role in the control of behaviors implicated in the control of alcohol seeking and drinking. We then review studies that have shown a wide-ranging and persistent reprogramming of the transcriptome within the mPFC in the post-dependent state, and the molecular mechanisms we have been able to identify in the course of these studies. An implication suggested by these findings is that molecular switches with broad and lasting functional consequences are turned on by a history of dependence, and that strategies to turn these switches back off may offer targets for novel, disease-modifying treatments.
Functional neuroanatomy of the PFC and control of behaviors
Drug use despite adverse consequences is a core feature of addiction. It is increasingly recognized that this behavior does not simply reflect increased motivation to seek and take drugs, but rather a breakdown of the balance between sub-cortical motivational processes and top-down executive control, exerted in large part by the prefrontal cortex (Goldstein & Volkow 2002, 2011). The past decade has therefore seen an increased interest in the prefrontal cortex and its role in addiction. This rise has coincided with seminal work that has established the structural and functional diversity of rodent prefrontal areas, their relation to the primate brain, and their role in attentional and emotional processing (Seamans et al. 2008; Uylings et al. 2003).
The prefrontal cortex can be separated into medial, lateral and orbital divisions. In rodents, the orbital and lateral divisions are formed by the orbital frontal cortex and agranular insular cortex, while the medial division encompasses the anterior cingulate cortex (ACC), the prelimbic and the infralimbic subdivisions of the mPFC (Uylings et al. 2003). The mPFC receives dopaminergic input from the ventral tegmental area (VTA), as well as other inputs from limbic and sensory areas, including amygdala and hippocampus (Hoover & Vertes 2007). Subdivisions of the PFC, including the agranular insular cortex, are highly interconnected. This allows them to evaluate external and internal states, integrate these with the motivational salience of specific stimuli – including those associated with alcohol or other drugs – and exert executive control in selecting approach or avoidance behaviors via projections to the NAc, amygdala and other targets. Although a matter of relative density rather than clear-cut boundaries, the projection patterns of the prelimbic and infralimbic mPFC subdivisions differ, with the former predominantly projecting to the NAc core and the latter predominantly to the shell (Vertes 2004).
Anatomical, cellular and functional differences between mPFC subdivisions have given rise to a widely held dichotomous view of pre- and infralimbic mPFC function in the control of both fear and drug responses (Moorman et al. 2015). According to this view, dorsal (largely prelimbic) mPFC promotes conditioned approach to both drug and natural rewards, whereas the ventral (infralimbic) mPFC suppresses it. A similar dichotomy has been thought to exist in controlling the expression and suppression of conditioned fear responses, respectively. Some support for a ‘prelimbic – go/infralimbic – no-go’ dichotomy in the control of conditioned appetitive and aversive behaviors has also been provided by human neuroimaging findings. These have shown that the activity of dorsal ACC and ventromedial PFC correlates with initiation and inhibition of drug and fear related behaviors, respectively (Peters et al. 2009). However, because the human PFC has undergone a vast expansion during evolution, it is likely that functional homologs of rodent mPFC subdivisions are more intermixed within the larger human mPFC volume.
Evidence has emerged in recent years to challenge the simple ‘go/no-go’ model for the functional contributions of pre-vs. infralimbic mPFC to drug seeking, respectively. This has in particular been the case outside the context of cocaine or fear conditioning with simple discrete cues (Gourley & Taylor 2016). A striking example was provided for heroin seeking, where selective deletion of a neuronal ensemble activated during context-induced reinstatement resulted in suppressed, rather than increased reinstatement responding on subsequent testing (Bossert et al. 2011, 2012). In contrast, it was found that ablation of a neuronal ensemble activated within the infralimbic cortex by alcohol cues promoted alcohol seeking. The effect of inactivating the tagged ensemble was specific for the infralimbic cortex, while manipulations of the prelimbic cortex were ineffective. The effect was also selective for cue-induced reinstatement, while stress-induced reinstatement was unaffected. Importantly, non-selective inactivation of infralimbic neurons was also without consequence for reinstatement behavior (Pfarr et al. 2015). The latter finding is in agreement with the observation that a reversible non-selective inactivation of the infralimbic cortex using a combination of baclofen and muscimol did not affect context-induced reinstatement of alcohol seeking (Willcocks & McNally 2013).
The functional ensemble that is activated within the infralimbic mPFC upon recall of an alcohol memory comprises about 10% of infralimbic neurons (Pfarr et al. 2015). This is a larger population than that identified by their c-Fos response after similar reinstatement experiments for cocaine or heroin seeking (Bossert et al. 2011; Koya et al. 2009). It can be speculated that alcohol cues engage a wider network than other drugs, because the more complex actions of alcohol generate more extensive internal representations. Because ablation of the ensemble activated by alcohol cues resulted in increased alcohol seeking, the function of this ensemble is likely to inhibit relapse-like behavior. However, because global inactivation of the infralimbic cortex did not affect reinstatement of alcohol seeking, neither when reversible (Willcocks & McNally 2013) nor permanent (Pfarr et al. 2015), this region does not seem to generally inhibit reinstatement of alcohol seeking. Furthermore, the stress-responsive subset of neurons within the infralimbic cortex must be different from that activated by alcohol cues, even though both promote the same behavioral output, i.e. reinstatement of lever pressing.
Our findings provide further evidence that neural mechanisms underlying drug seeking differ between drug classes, and rely on distinct interactions of mPFC circuits that are not easily accounted for by a simple ‘prelimbic – go/infralimbic – no-go’ model. This view is also supported by recent in vivo multiarray recording experiments. The use of this approach has shown that prelimbic and infralimbic neurons signal contextual information that promotes sucrose seeking in response to discriminative stimuli, irrespective of whether this involves initiation or suppression of behavioral responses (Moorman & Aston-Jones 2015). A more complex model of functional compartmentalization of the mPFC proposes that the prelimbic subdivision is mainly involved in the learning of rules, whereas the infralimbic supports flexibility of this response and the ability to shift toward new strategies when contingencies change (Heidbreder & Groenewegen 2003; Seamans et al. 2008).
A reprogrammed mPFC transcriptome in the post-dependent state
For some time, our work has been guided by the hypothesis that long-term changes in gene expression within key brain structures are a likely mechanism underlying the persistent behavioral changes observed in the post-dependent state. To examine this hypothesis, we carried out transcriptome profiling of several brain structures from post-dependent and control rats, and found the most pronounced differences within the mPFC (Meinhardt et al. 2013). Low magnitudes of differential gene expression are typically found in models of psychiatric disorders, largely due to the heterogeneous composition of the brain tissue samples (Reimers et al. 2005). As a result, the specificity achieved through commonly used statistical corrections for false discovery rates is at the expense of a low sensitivity, i.e. a low probability that true positives will be detected. To facilitate discovery, we therefore chose a different strategy, in which we have initially screened for transcripts with nominally (i.e. uncorrected) significant differential expression, and have then sought to confirm candidates of interest through independent, wet analysis. This is a strategy that is standard in clinical analytics, where optimal results are commonly achieved through the combined use of separate screening and validation steps, allowing these to prioritize sensitivity and specificity, respectively.
Our initial mPFC screen identified 165 transcripts with nominally differential expression that persisted 3 weeks following a history of alcohol dependence (Tapocik et al. 2013). Among the differentially expressed transcripts detected, 105 were significantly upregulated and 60 significantly downregulated, arguing against a global repression or cell death due to non-specific toxicity as major factors underlying the findings. As expected, the fold-changes in gene expression were relatively small, consistent with what has typically been found for brain expression in psychiatric disorders such as alcohol dependence (Reimers et al. 2005; Sommer et al. 2005). Among the differentially expressed transcripts identified in this initial screen, 37 had previously been associated with alcohol use, abuse and dependence (Tapocik et al. 2013).
Because a known effect of alcohol is to influence the excitation – inhibition balance controlled by gamma aminobutyric acid (GABA) and glutamate, a first, focused knowledge-based follow-up analysis used Gene-Set Enriched Analysis (Subramanian et al. 2005) to examine the potential presence of persistent neuroadaptations in GABAergic or glutamatergic mPFC neurons. For this purpose, two marker gene sets were used that have previously been described as highly divergent between GABAergic and glutamatergic neurons (Sugino et al. 2006). The results of this analysis indicated a marked enrichment of downregulated glutamatergic marker genes in the mPFC of post-dependent rats. Several transcripts corresponding to these genes were confirmed by independent quantitative polymerase chain reaction (qPCR) or in situ hybridization analysis (Meinhardt et al. 2013). Among them, Grm2, which encodes the metabotropic glutamate receptor 2 (mGluR2), was markedly downregulated in post-dependent rats, as well as in postmortem tissue from mPFC of human alcoholics.
The mGluR2 has recently been implicated in control of alcohol consumption in some Wistar rat populations, including the genetically selected Indiana alcohol preferring P-rat line (Wood et al. 2016; Zhou et al. 2013), and is considered a promising candidate medication target (Augier et al. 2016; Holmes et al. 2013). In situ hybridization was used to localize the downregulation of mGluR2 expression in greater anatomical detail. This analysis once again confirmed a marked downregulation of mGluR2 expression, but also showed that this downregulation was restricted to the infralimbic mPFC; expression in the prelimbic area was unaffected. The specificity of infralimbic mGluR2 repression in post-dependent rats was further highlighted by the observation that the expression of a closely related metabotropic glutamate receptor subtype, mGluR3, was unaffected by a history of dependence, both in the pre- and the infralimbic areas. These findings suggested that the infralimbic mPFC is a key structure affected by alcohol-induced neuroadaptations, and that presynaptic control of glutamate release by mGluR2s is a candidate mechanism underlying some of these neuroadaptations. Based on these observations, an extensive functional validation effort was undertaken, which is described in detail in the next section.
As a second knowledge-based strategy, a broader, unbiased Gene Ontology (GO) analysis was carried out (Tapocik et al. 2013). Overall, 90 of the 165 differentially regulated genes belonged to significantly overrepresented GO categories. Among the top categories of genes with differential expression in the mPFC identified as significant by the GO analysis were two related categories, associated with ion channels and synaptic transmission, respectively. These categories are functionally related in that they both have as members genes that influence transmitter release. Specifically, several synaptic proteins involved in vesicle docking and transmitter release were downregulated, including synapsin II (Syn2) and synaptotagmin 7 (Syt7). This was also the case for several ion channels that influence transmitter release through effects on membrane potentials and Ca++ flux, such as, e.g. the pore-forming alpha 1B subunit of the N-type voltage-dependent calcium channel. We have subsequently discovered differential expression of genes encoding additional calcium channel and synaptic protein transcripts in an RNA sequencing screen of mPFC gene expression in the post-dependent model. These findings were confirmed by qPCR, and subjected to functional validation (Barbier et al. 2015), in work that is discussed in a subsequent section.
A third category of differentially expressed genes identified by the GO analysis was associated with neuronal plasticity. Within this category, several members of the Early Growth Response gene family (Egr1, 2 and 4) were downregulated together with other transcription factors of the immediate early gene class (e.g. of the Fos, Jun or Nr4a families). Downregulation of these transcripts was in each case independently confirmed by qPCR of bulk mPFC tissue extracts or by in situ hybridization; the downregulation of Egr2 and Egr4 was also confirmed in identified projection neurons from the infralimbic mPFC to the NAc, isolated using laser capture microscopy (Meinhardt et al. 2013). This experiment showed that modest expression changes detected in bulk extracts of neuronal tissue may reflect much more robust expression differences, 10- to 500-fold, within specific neuronal populations. A further transcript in the category of plasticity-associated genes that was of functional interest was brain-derived growth factor (Bdnf), which was robustly downregulated in the array analysis, and confirmed by independent qPCR analysis of bulk mPFC tissue as well as in situ hybridization (Tapocik et al. 2013). The Bdnf and several of the plasticity-associated transcripts were significantly linked when the array dataset was evaluated using the Ingenuity Pathway Analysis software (IPA, QIAGEN; Methodological details on IPA can be found at http://www.ingenuity.com/science/knowledge_base.html.). Collectively, the expression data, the bioinformatics analysis, and prior data indicating a role for Bdnf within corticostriatal projections in regulating excessive alcohol intake (Jeanblanc et al. 2006, 2009; Logrip et al. 2009; McGough et al. 2004) made Bdnf a high priority candidate for a functional validation effort, described in a subsequent section.
The expression changes within the mPFC of post-dependent rats have been replicated in multiple batches of animals in different laboratory settings. Downregulation of Egr1 and Bdnf was also observed in the PFC of mice immediately following chronic intermittent alcohol exposure, but was not detected 8 h into withdrawal (Melendez et al. 2012). When the expression analysis in this model was extended 7 days into abstinence, only a limited overlap with the rat studies was found within the mPFC (Smith et al. 2016). Differences between species, procedures and time points analyzed could each contribute to the differences in findings. The time-course is a particularly important factor to assess, given recent findings of highly dynamic changes in dopamine D1 receptor and dopamine transporter expression in the striatum of post-dependent rats across different phases following initiation of abstinences (Hirth et al. 2016).
Limited human data are available to assess the extent to which our rat expression data translate to the human situation. Early microarray studies from prefrontal cortex of alcoholics found robust downregulation of the immediate early genes (IEG) JunB and Nr4a1 (Iwamoto et al. 2004). A subsequent report used expression data from rodent and human PFC to enrich the results of a genome-wide association study (GWAS) of alcoholism, and identified Jun, Fosl2, Nr4a3 and Bdnf as significant hits (Zhao et al. 2012). Recently, a more comprehensive study found that the expression of multiple ion channels within the PFC was associated with lifetime alcohol consumption. Glutamatergic and GABAergic receptors were among the most central elements of these altered gene networks (Farris et al. 2015). Another report that integrated a GWAS of alcohol dependence with human and animal gene expression data found members of the synaptotagmin and metabotropic glutamate receptor families among its top hits (Levey et al. 2014). A brain bank that provides carefully phenotyped postmortem brain tissue samples matched for age and postmortem interval, the Tissue Resource Center of the University of South Wales, was used for the following work (Sheedy et al. 2008). The Grm2 expression was determined in the anterior cingulate cortex (ACC), a brain region that is anatomically and functionally related to the rodent mPFC (Uylings et al. 2003). Within the ACC, a significant, 2.6-fold decrease in Grm2 transcript levels was found in alcoholics compared with controls (Meinhardt et al. 2013).
In summary, there is convergent evidence for long-lasting reprogramming of gene expression networks in the pre-frontal cortex by a history of alcohol dependence, with some candidate genes receiving support from both human and rodent studies. This holds particularly true for the metabotropic glutamate receptor mGluR2 and for the neurotrophin Bdnf. Both these potentially offer molecular targets for novel alcoholism therapeutics, and therefore merit a detailed functional analysis.
Functional role of infralimbic mGluR2 receptor repression
In a series of experiments, the functional consequences of the suppressed mGluR2 expression observed following a history of dependence was assessed in infralimbic mPFC neurons that project to the NAc shell (NAcSh). Extracellular glutamate levels in the target area of this projection were measured using in vivo microdialysis in freely moving rats. Given its role as a presynaptic autoreceptor, stimulation of mGluR2 was expected to reduce glutamate release. As expected, systemic administration of the mGluR2/3 agonist LY379268 decreased extracellular glutamate levels in dialysate obtained from the NAcSh of control rats. This effect of the agonist was absent in post-dependent rats. Thus, downregulation of mGluR2 expression in the mPFC-NAcSh pathway resulted in a loss of mGluR2 function (Meinhardt et al. 2013).
Next, behavioral consequences of suppressed mGluR2 expression within this circuit were assessed. Post-dependent rats showed markedly higher response rates following cue-induced reinstatement of extinguished alcohol seeking, a widely used model of relapse (Sanchis-Segura & Spanagel 2006; Shaham et al. 2003). A rescue of infralimbic mGluR2 expression in the mPFC of post-dependent rats using local injections of a lentiviral construct, was found to result in a rescue of cue-induced alcohol seeking. Following viral injections, reinstatement response rates of post-dependent animals declined by about 40%, bringing them into the range of non-dependent controls (Meinhardt et al. 2013). Together, these experiments provided consistent evidence that a profound, anatomically restricted mGluR2 pathology in the mPFC causally contributes to excessive alcohol seeking in post-dependent rats; and that restoring mGluR2 function in this region is sufficient for regaining control over alcohol-seeking behavior.
It will be important to examine possible functional consequences of a post-dependent mGluR2 repression for other behaviors controlled by the mPFC. One useful test for this purpose is the attentional set shifting task (ASST), a rodent equivalent to the human Wisconsin Card Sorting Test (WCST); both assess an aspect of executive function commonly referred to as cognitive flexibility, and their performance depends largely on mPFC in both primates and rodents (Berg 1948; Brown & Tait 2016). In alcohol-dependent patients, impaired performance on the WCST is a predictor of relapse (Wicks et al. 2001). In mice, experiments have shown performance deficits in the ASST after 1 week of abstinence from chronic intermittent alcohol exposure (Kroener et al. 2012). Patch clamp recordings in brain slices from these mice showed increased presynaptic transmitter release, consistent with impaired mGluR2 autoreceptor function.
The mechanism underlying reduced mGluR2 expression in the mPFC of post-dependent rats is presently not known. The Grm2 was not detected by a DNA methylation screen (see below), but could be a target for other types of direct epigenetic regulation that have not been assessed in those studies. For instance, treatment with L-acetylcarnitine, a drug that promotes histone acetylation and is approved as a medication for the treatment of neuropathic pain, leads to increased K27-acetylation of H3 histones bound to the Grm2 promoter as well as Bdnf promoter in the PFC, and this is associated with respective increased expression of the respective gene in this region, as well as rapid antidepressant actions in rodent models of depression (Nasca et al. 2013). Also, mice with a deletion of the 5-HT2A receptor show repressive histone marks at the Grm2 promoter, and this is associated with a pronounced downregulation of prefrontal Grm2 expression (Kurita et al. 2013). Regulation of mGluR2 expression by alcohol exposure could also be indirect, resulting from the downregulated expression of transcription factors belonging to the Egr-family (Meinhardt et al. 2013). The Grm2 promoter contains binding sites for this family of transcription factors, and viral mediated Egr1 overexpression can induce mGluR2 expression (Kurita et al. 2013). Furthermore, Egr2, which has a specific binding site within the Grm2 promoter, can itself be subject to epigenetic regulation via hypermethylation of intron 1 (Swanberg et al. 2009; Unoki & Nakamura 2003). It is thus possible that a network of IEG is dysregulated by epigenetic mechanisms, ultimately leading to reduced Grm2 in the mPFC of post-dependent rats.
Small molecule mGluR2/3 agonists are available, and are able to reduce self-administration and drug seeking for several drugs of abuse, including alcohol, nicotine, cocaine, methamphetamine and heroin (Baptista et al. 2004; Bossert et al. 2006; Cannella et al. 2013; Crawford et al. 2013; Liechti et al. 2007; Peters & Kalivas 2006; Zhao et al. 2006). These studies indicate that presynaptic group II mGluRs act as gatekeepers that regulate glutamate release at synapses relevant for addictive behaviors. Available orthosteric agonists are unable to discriminate between mGluR2 and mGluR3, but positive allosteric modulators (PAMs) selective for mGluR2 have recently been developed (Augier et al. 2016; Holmes et al. 2013; Johnson & Lovinger 2015; Nickols & Conn 2014). Using these PAMs, several investigators have been able to demonstrate that mGluR2 activation can reduce drug intake and drug-seeking behaviors (Caprioli et al. 2015; Justinova et al. 2015; Nikiforuk et al. 2010). Clinical trials are currently ongoing to evaluate whether these promising data can be translated to humans (Salih et al. 2015).
microRNA reprogramming of the mPFC
Expression regulation by microRNAs (miRNAs) represents an appealing candidate mechanism for post-dependent neuroadaptation of mPFC function. The miRNAs are small (∼22–30 nucleotide in length), highly conserved noncoding RNAs that modulate gene expression at the posttranscriptional level. After being processed into a mature form, they bind to miRNA recognition elements in the 3′-untranslated region (UTR) of their target transcripts and cause translational degradation or repression (Guo et al. 2010). The miRNA regulation of mRNA expression is inherently divergent, because recognition elements are shared between multiple transcripts. A limited set of miRNAs could therefore account for a substantial proportion of differential gene expression observed in the post-dependent state at the mRNA level.
The miRNAs may play an important role in neuroadaptive processes triggered by chronic drug exposure (Pietrzykowski 2010), including tolerance to alcohol (Pietrzykowski et al. 2008). Postmortem analysis has also identified upregulation of multiple miRNAs in the frontal cortex of alcohol-dependent patients (Lewohl et al. 2011), but the functional role of these dysregulations is unknown. Human studies offer limited opportunities to isolate the consequences of brain alcohol exposure from those of preexisting vulnerability factors, or of environmental influences associated with alcoholism such as head trauma or nutritional deficiency. Animal models offer an important complementary tool for this type of analysis.
The smaller number of miRNAs (about 1800 in the rat genome) and their lower complexity make the analysis of their expression more tractable than that of mRNA transcripts. Microarray-based screening for differential miRNA expression in the mPFC of individual post-dependent rats resulted in 41 candidates (Tapocik et al. 2013). Among these, 17 were upregulated and 24 were decreased, again suggesting that non-specific global processes such as cell death were unlikely to account for the observations. Principal component analysis indicated that a history of dependence accounted for a higher proportion of the variance in miRNA than in mRNA expression. Accordingly, hierarchical clustering of miRNA expression segregated post-dependent and control animals more robustly than that of mRNA expression. Thus, the effect size of post-dependent neuroadaptations is higher at the miRNA than at the mRNA level.
The bioinformatic analysis identified 89 mPFC mRNA transcripts as being potentially regulated by miRNAs. These were subjected to a pathway analysis using the Ingenuity software tool. A major pathway that emerged from this analysis was associated with neuronal plasticity (Fig. 1). This network contained multiple genes thought to be involved in neuropsychiatric disorders and addiction, including Bdnf, Fos, Per2 and Arc, as well as the nuclear receptors/transcription factors Nr4a1 and Nr4a3. The 15 differentially expressed mRNAs within this network were targeted by 26 of the miRNAs found in the miRNA microarray analysis, with several of the miRNAs converging on the same mRNA targets (Tapocik et al. 2013). This observation is in agreement with prior observations that multiple miRNAs can act synergistically to repress gene expression (Nachmani et al. 2010). Several candidate miRNA–mRNAs pairs identified by these analyses above were chosen for confirmation by qPCR. For the miRNAs selected for confirmation, results were highly consistent with the microarray data; for the selected mRNAs, confirmation was obtained in most, but not all cases. Notably, an experimentally robust and functionally compelling expression pair that emerged from this analysis was that of Bdnf and miR-206.
Figure 1. Ingenuity pathway analysis showing one of the top gene networks that is downregulated in the mPFC by history of alcohol dependence.
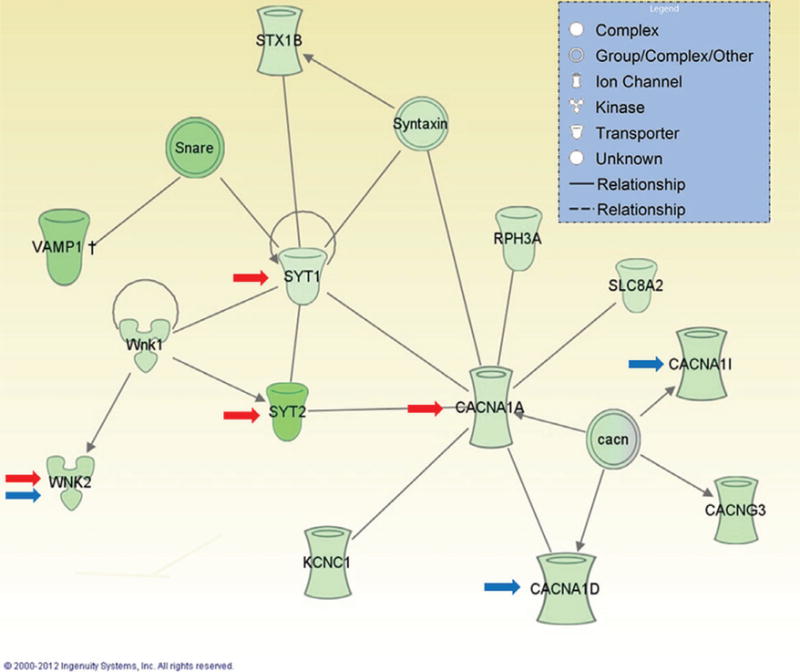
Green color indicates decreased expression in post-dependent compared with control rats. Red arrows indicate genes regulated by DNA methylation. Blue arrows indicate genes regulated by Prdm2.
A functional role for miRNA-mediated BDNF repression in the mPFC
Dysregulation of BDNF expression within the mPFC offers a potential mechanism for the persistent neuroadaptations observed in the post-dependent state. The BDNF signaling leads to phosphorylation of the cAMP response element-binding protein (CREB) and upregulation of CREB target genes, including the activity-regulated cytoskeleton-associated (ARC) protein. The ARC is of particular interest for long-term neuroadaptive processes, because it is directly involved in remodeling of neuronal connectivity via dendritic spine formation. For instance, physical modulation of spine density by ARC is required for consolidation of long-term synaptic potentiation (reviewed in Bramham 2008). Previous research has also shown that ARC mediates alcohol-induced neuroadaptations within the amygdala (Pandey et al. 2008).
The Bdnf gene has a unique, highly conserved organization. In rodents, it is composed of at least eight 5′-exons, each with its own promoter, that are spliced to a common 3′-terminal exon containing the coding frame and two alternative 3′-UTRs (Aid et al. 2007; Timmusk et al. 1993). The promoters for exons I and II mediate constitutive expression, while the promoters for exons IV and VI have been shown to share properties with IEG and seem to mediate much of the activity-dependent gene regulation of Bdnf (Hansson et al. 2006, 2011; Jiang et al. 2008). The short and long 3′-UTR Bdnf transcripts are involved in different cellular functions. Short 3′-UTR mRNAs are restricted to cell bodies, while long 3′-UTR mRNAs are also localized in dendrites, where local synthesis of BDNF occurs (An et al. 2008). The 3′-UTR of the long Bdnf transcript contains three predicted target sites for miR-206, and is repressed by this miRNA in the course of muscle cell differentiation (Miura et al. 2012).
Under normal circumstances, miR-206 does not seem to be expressed in the brain at significant levels, but its expression is induced by accumulation of amyloid in a transgenic mouse model of Alzheimers disease, where inhibition of miR-206 activity resulted in a rescue of low BDNF levels and spine density numbers (Lee et al. 2012). The miR-206 induction may therefore be a response to neuronal insult. Based on these findings, a chain of events can be hypothesized to occur as post-dependent neuroadaptations develop. According to this hypothesis, miR-206 is induced in mPFC neurons by cycles of intoxication and withdrawal, represses BDNF expression, and results in decreased expression and activity of ARC. Decreased activity of ARC then contributes to lasting changes in the connectivity of mPFC neurons with their projection targets, and impaired top-down regulation of motivated behaviors.
A series of studies was carried out to evaluate this hypothesis (Tapocik et al. 2014). In situ hybridization confirmed a persistent induction of miR-206 in the mPFC of post-dependent rats. This induction was region specific, as no increase was seen in the VTA, NAc or the amygdala. When the activity of miR-206 was examined in vitro using a luciferase reporter assay, a 60% repression of the Bdnf reporter construct was found. The three miR-206 target sites in the Bdnf 3′-UTR contributed synergistically to this effect, whereby the presence of two of the sites, site 2 (located at bp 392–398) and 3 (bp 1294–1300) is necessary. Functional in vivo validation of these results was obtained by adeno-associated virus (AAV)-mediated overexpression of miR-206 in the mPFC of rats that did not have any history of alcohol dependence. Vector-mediated miR-206 expression resulted in a significant reduction of BDNF levels within the mPFC. This was accompanied by a significant escalation of alcohol self-administration rates in these animals, mimicking what is observed following a history of dependence.
Together, these findings provide consistent support for the hypothesis that miR-206 expression is induced in the mPFC of post-dependent rats, and causally contributes to one of the core elements of the resulting behavioral syndrome, escalation of alcohol self-administration, through repression of BDNF. This notion has subsequently received independent support by data from the laboratory of Dorit Ron. Using a mouse model, this group showed that another miRNA, miR-30a-5p, mediates a transition to escalated drinking through repression of BDNF expression within the mPFC (Darcq et al. 2015).
A role for BDNF in addiction-related behaviors and neu-roadaptations has previously been reported in studies with cocaine, but these effects have been highly dependent on the brain region examined (reviewed in McGinty et al. 2010). Specifically, BDNF injections into the mPFC have been found to result in decreased cocaine self-administration and suppression of both cue- and priming-induced relapse to cocaine seeking. In contrast, BDNF administration into the VTA or the NAc has largely produced the opposite effect. The mPFC findings on alcohol intake are generally in agreement with the cocaine studies in which BDNF has been administered into this brain area. The cocaine studies suggest the possibility that BDNF may simply be a general mediator of drug-induced plasticity, with different functional effects depending on the circuitry in which it acts. Based on the presented mPFC data alone, it is therefore difficult to predict the net effect of interventions with global effects on BDNF activity for alcohol addiction. Indeed, opposite effects of escalating alcohol intake have been reported on cortical and striatal BDNF expression, i.e. decreased expression in the mPFC and increase in the striatum (Jeanblanc et al. 2006, 2009; Logrip et al. 2009; McGough et al. 2004). It is unclear whether this reflects the actions of locally produced BDNF within this structure, BDNF provided by inputs to the striatum such as those originating from the prefrontal cortex, or both. At a minimum, findings to date support the notion that remodeling of mPFC output connectivity is an important mechanism in the transition to excessive alcohol drinking, and that BDNF is an important mediator of this remodeling.
However, some data additionally suggest that a global potentiation of BDNF activity may in fact have a therapeutic potential. Specifically, a hemizygous deletion of BDNF was reported to result in increased alcohol consumption in mice (Hensler et al. 2003), while systemic injections of a fusion protein that delivers Rack1 to the brain and promotes BDNF activity resulted in decreased drinking. These findings provide a rationale for developing small molecule BDNF mimics as therapeutics for alcohol addiction.
Global hypermethylation of the mPFC and synaptic transmission
Similar to miRNAs, epigenetic mechanisms (i.e. DNA methylation and histone modification) represent an attractive candidate mechanism to drive alcohol-induced long-term neuroadaptation. Like miRNAs, DNA methylation and his-tone modification can regulate multiple genes and therefore be one of the main upstream regulators of alcohol-induced transcriptomic reprogramming. Moreover, DNA methylation has the ability to change dynamically in response to external factors. For instance, stress (Murgatroyd et al. 2009; Weaver et al. 2002) and exposure to drugs of abuse (Tian et al. 2012; Wong et al. 2011) regulate methylation patterns in the brain. Both human and animal studies suggest that alcohol can also modify methylation patterns in the brain and more specifically in the mPFC. Manzardo and colleagues found no global methylation differences between alcoholics and control subjects. However, they found different methylation patterns at the gene promoter level between control and alcoholic subjects, suggesting a ‘gene specific effect’ (Manzardo et al. 2012). In accordance with this study, Wang and his collaborators recently observed different methylation patterns between mPFC tissue of controls and subjects with alcoholism (Wang et al. 2016). Contrary to Manzardo’s study, which measured methylation level at the promoter region only, Wang et al., extended their analysis to the full gene sequence and measured methylation at individual CpGs. They found that the majority of the differentially methylated CpGs identified in male alcoholics were hypermethylated and were dispersed over different gene regions. These results suggest that alcohol-induced hypermethylation in non-promoter regions that comprise, for example, enhancer and insulator regions, may regulate splicing and gene expression by influencing the binding of regulatory proteins (Jones 2012). More experiments are needed in order to better understand this mechanism.
Although studies of DNA methylation in human postmortem brain tissues are informative, this approach does not allow for determination of the functional consequences of differential DNA methylation. Animal models offer an important complement to these studies, as they allow assessment of dependence-induced DNA methylation changes to be carried out in relevant brain regions. More importantly, they allow the evaluation of the functional impact of DNA methylation changes, as it is possible to manipulate the enzymes that regulate DNA methylation levels and assess the behavioral consequences. For example, manipulation of DNA methyltransferase in a rat model suggests that DNA methylation is involved in alcohol addiction. Systemic inhibition of DNA methyltransferase activity (DNMT) activity decreased excessive alcohol drinking and seeking behaviors in rodents (Warnault et al. 2013). Similarly, intraventricular infusion of the DNMT inhibitor RG-108 significantly decreased alcohol self-administration in post-dependent but not in control rats (Barbier et al. 2015). Interestingly, micro-infusion of RG-108 into the mPFC was sufficient to inhibit escalation in alcohol intake in post-dependent rats, suggesting that alcohol-induced hypermethylation in this region is involved in increased alcohol intake.
In order to identify methylation-dependent gene expression changes that occur after a history of alcohol dependence, whole transcriptome analysis was performed using RNA-seq (Barbier et al. 2015). Seven hundred and eighty-four genes with significant expression changes in the mPFC of post-dependent compared with control rats were identified. Similar to what was previously found (Meinhardt et al. 2013; Tapocik et al. 2013), bioinformatics analysis identified within this list two overrepresented categories of biological functions related to gene expression and neurotransmis-sion. Specifically, a history of alcohol dependence decreased expression of a gene network that includes proteins regulating synaptic vesicle formation and function (Fig. 1). A subset of seven genes coding for synaptic proteins (synap-totagmin 1: Syt1, synaptotagmin 2: Syt2, P/Q type channel: Cacna1a, T-type calcium channel: Cacan1i and the serine/threonine-protein kinase 1 and 2: Wnk1 and Wnk2, and Potassium Channel, voltage-gated Shaw related subfamily C, member 1: Kcnc1) was selected for subsequent analysis. Expression of these genes was not influenced by alcohol exposure in other brain regions known to play a role in addiction (Nac, amygdala and hippocampus). These data suggest a persistent dysregulation of synaptic transmission in the mPFC after a history of alcohol dependence, which is consistent with the hypothesis of mPFC hypoac-tivity associated with drug addiction (Goldstein & Volkow 2011).
Importantly, we found that micro-infusion of RG-108 into the mPFC restored the expression of four out of the seven transcripts (Syt1, Syt2, Cacna1a and Wnk2), indicating a regulatory effect of alcohol-induced DNA hypermethylation on those genes. Pyrosequencing analysis did not show altered methylation on the promoter region of Cacna1a, suggesting an indirect regulation of this transcript’s expression by DNA methylation. In contrast, chronic intermittent alcohol exposure did induce hypermethylation on exon 1 of Syt2. There is now evidence suggesting that, similar to promoter hypermethylation, DNA methylation on exon 1 is associated with gene silencing (Brenet et al. 2011). Together, these results suggest a direct regulation of Syt2 by alcohol-induced DNA hypermethylation. Interestingly, it was also found that although Syt2 knockdown in non-dependent rats does not alter alcohol consumption, it increases tolerance to quinine adulteration. Aversion-resistant alcohol intake has been previously interpreted to indicate compulsive motivation to consume alcohol (Wolffgramm et al. 2000). Therefore, decreased Syt2 may be part of the molecular events that drive compulsive-like behavior observed in alcohol post-dependent rat (Vendruscolo et al. 2012).
Fine-tuning histone function: PRDM2
In addition to direct methylation of DNA, environmentally sensitive epigenetic mechanisms can also manifest as post-translational modifications to unstructured histone tails (Feil & Fraga 2011). Histone tail modifications include acetylation, methylation and phosphorylation, among others. While there are three enzymes that catalyze DNA methylation (DNMT1, DNMT3A, DNMT3B), there are numerous epigenetic enzymes that oversee the addition, interpretation and removal of various histone modifications (Borrelli et al. 2008). For example, there are over 50 human epigenetic enzymes that catalyze the addition of methyl groups to histones (his-tone methyltransferases), and approximately 30 enzymes that remove these methyl groups (histone demethylases) (Copeland et al. 2009). Histone methylation can either facilitate or repress transcription depending on the particular amino acid residue that is modified. For example, methylation at histone H3 lysine 4 (H3K4) is typically associated with transcriptional activation, while methylation at H3K27 condenses chromatin and inhibits gene expression. This ‘histone code’ is further complicated by the fact that lysine can be mono-, di- or tri-methylated, and the degree of methylation can differentially influence gene expression. Each epigenetic enzyme specifically catalyzes modification of a particular amino acid on a particular histone, and further exhibits specificity toward the degree of methylation at that site. Despite the fact that many of these epigenetic enzymes are expressed in the brain, little is known about their neurological function.
RNA-seq analysis showed that 22 epigenetic enzyme mRNAs were dysregulated in mPFC tissue from post-dependent rats (Barbier et al. 2015). One of these enzymes, histone methyltransferase PR-domain containing 2 (PRDM2), is strongly enriched in the brain compared with peripheral tissues and is also selectively expressed in neurons. Further, qPCR showed that downregulation of Prdm2 in alcohol-dependent rats could be restored by treatment with the DNA methyltransferase inhibitor RG-108, suggesting that PRDM2 is part of an epigenetic pathway downstream of DNA methylation changes. While PRDM2 had previously been implicated in brain cancer, no studies had examined a role for PRDM2 in psychiatric conditions.
In order to test whether PRDM2 contributes to alcohol-seeking behavior, a lentiviral-mediated siRNA knockdown approach was used to downregulate Prdm2 expression in the mPFC of alcohol-naïve rats (Barbier et al. 2015). The Prdm2 knockdown was sufficient to cause escalated alcohol self-administration, aversion-resistant drinking in a quinine adulteration task, and increased sensitivity to stress-induced reinstatement, essentially resulting in a phenocopy of the post-dependent state. These effects were specific for alcohol intake, as Prdm2 knockdown did not affect sucrose-seeking behavior.
Target genes of PRDM2 that could underlie the observed alcohol-seeking behaviors, were identified using a tripartite approach. The PRDM2 is known to specifically catalyze the addition of a single methyl group at histone H3, lysine 9 (H3K9me1) resulting in transcriptional activation (Barski et al. 2007; Kim et al. 2003). The H3K9me1 is decreased in the mPFC of post-dependent rats, consistent with Prdm2 down-regulation. Therefore, we first used chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) to identify the genomic regions differentially regulated by H3K9me1 in control as compared to post-dependent mPFC. These results were overlaid with the corresponding RNA-seq data in order to narrow down the list of H3K9me1 target genes to those that are also transcriptionally dysregulated as a consequence of protracted alcohol exposure. Pathway analysis of these genes showed a significant enrichment of biological functions related to synaptic communication, specifically regulation of voltage-gated calcium signaling (Barbier et al. 2015).
Finally, laser capture microdissection was used to quantify the expression of known alcohol-associated genes in mPFC neurons of alcohol-naïve rats subjected to Prdm2 siRNA compared to scramble control. This third arm of our tripartite approach allowed us to narrow the list of H3K9me1 target genes to those most likely to be functionally involved in the ability of the Prdm2 knockdown to create a phenocopy of the post-dependent state. There were four convergent target genes identified by our ChIP-seq, RNA-seq and Prdm2 knockdown studies: Bsn, Cacna1i, Cacna1d and Wnk2. All four genes were downregulated, consistent with PRDM2 downregulation, decreased H3K9me1, and subsequent transcriptional repression in alcohol dependence. Cacna1i and Cacna1d encode subunits of voltage-gated calcium channels, while Bsn encodes Bassoon, a protein that supports calcium channel activity by anchoring the channels to the presynaptic terminal. The WNK2 is a kinase that regulates cation-chloride transporters, and along with Cacna1i and Cacna1d, was part of the gene network we found to be regulated by DNA methylation in dependent rats (Barbier et al. 2015); see Fig. 1). Calcium channels have previously been implicated in altered patterns of cortical excitability that may underlie excessive alcohol intake and increase the propensity for relapse. For instance, nonhuman primates that chronically self-administer alcohol display plasticity in T-type calcium channel currents that is sensitive to periods of intoxication and withdrawal (Carden et al. 2006; Welsh et al. 2011). Furthermore, our findings are in accordance with prior postmortem studies, which have shown changes in the expression pattern of genes involved in synaptic neurotransmission (Liu et al. 2006, 2007).
The fact that this same gene network was implicated in distinct experimental approaches emphasizes the validity and central importance of this epigenetically regulated signaling pathway in mediating dependence-associated neuroadaptive changes within the mPFC. Interestingly, functional enrichment analysis of miRNAs significantly enriched for target genes within the H3K9me1 ChIP-seq results showed miR-206 as the number one hit (WebGestalt adj. P = 0.0004), relating these recent findings to our earlier studies (Tapocik et al. 2014). Therefore, a plausible hypothesis to test in future experiments is that PRDM2 directly mediates miR-206 dysregulation in alcohol dependence, or that Prdm2 knockdown or miR-206 overexpression both cause escalation of alcohol self-administration by targeting overlapping gene networks. Altogether, this line of experimental inquiry showed that PRDM2-mediated histone modifications are an essential component of genomic reprogramming in the mPFC that contributes to neuroadaptive changes underlying a transition into alcohol addiction. Because epigenetic enzymes such as PRDM2 are emerging as novel drug-gable targets with potential to treat complex human disease (Copeland et al. 2009), this work underscores the potential for epigenetic-targeted therapies to impact substance abuse disorders.
Integrating molecular changes with mPFC function
Our work over the past 5 years has provided evidence that prolonged brain alcohol exposure causes a lasting reprogramming of mPFC transcriptional activity and function. It remains to be determined by what exact mechanisms that these molecular changes impact the control of alcohol seeking. Alcohol cues induce neural activity throughout the entire mPFC, as shown by our c-fos studies (Pfarr et al. 2015) and this process is altered in the post-dependent state (Meinhardt et al. 2013). Although only the deletion of Fos-positive neurons within the infralimbic subregion has affected cue-induced reinstatement, activated neurons in other mPFC subregions may serve other behavioral processes. The emerging picture is that mPFC neurons can form highly dynamic networks. These seem to cross anatomical boundaries, and differentially contribute to behavioral output according to contextual demands at the time.
Similar to the functional studies, the epigenetic dysregulation identified by our molecular studies affects both infralimbic and prelimbic subregions, and is likely to alter the dynamic communication within and between mPFC networks in multiple ways. A loss of autoreceptor function normally provided by mGluR2 in corticostriatal projection neurons impairs their capacity for synaptic plasticity, as showed by the loss of post-synaptic long-term depression in animals that do not express mGluR2 due to a stop-codon mutation (Meinhardt et al. 2013; Zhou et al. 2013). Loss of BDNF expression is likely to result in loss of structural plasticity that is required for long-term maintenance of synaptic plasticity (Tapocik et al. 2013, 2014). Repression of synaptic proteins and Ca++ channels is likely to result in impaired transmitter release from affected cell populations, leading to a loss of inhibitory control over alcohol intake despite adverse consequences (Barbier et al. 2015).
In conclusion, neuronal populations within the mPFC form dynamic ensembles that control alcohol-related behaviors in a stimulus-specific manner (Fig. 2). Formation of these ensembles and their output depends on rapid transcriptional responses mediated by immediate early gene networks, as well as more lasting epigenetic mechanisms that shape the transcriptional program of these cells. A prolonged exposure to cycles of alcohol intoxication and withdrawal results in persistent dysregulation of gene expression and function within these circuits. This dysregulation is in large part mediated through a broad range of epigenetic mechanisms that include induction of miR-expression, altered DNA methylation and altered activity of epigenetic enzymes that modify histone function. The resulting functional impairment interferes with fundamental mechanisms of top-down control of motivational and emotional processes. The breakdown of top-down control promotes the risk of relapse in alcoholism, but presently we have only an incomplete understanding of the exact circuit-level mechanisms through which this occurs. We hypothesize that future research leading to an improved understanding of mPFC function and its dys-regulation could contribute to the development of treatment responsive biomarkers, and the development of improved, personalized therapies (Heilig et al. 2016).
Figure 2. Post-dependent reprogramming of transcriptome responses in the mPFC affects local cue-responsive neuronal networks.
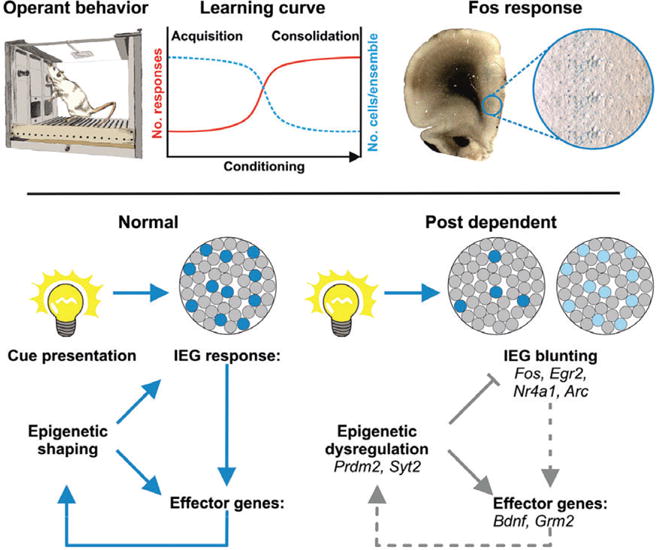
Local neuronal networks characterized by coherent activity play a critical role in mPFC function. We hypothesize that such functional ensembles are loosely formed by many neurons during early learning (task acquisition) and as performance consolidates these networks become more stable, comprised of discrete sets of neurons. A distinct feature associated with coherent neuronal activity is stimulus-induced expression of Fos (here represented by blue labeled neurons across most of the mPFC after presentation of a cue) and of other IEG, which in turn regulate downstream effector genes. These transcriptional responses are coordinated by epigenetic mechanisms and are crucial for shaping future outcomes of neural responses from the mPFC that ultimately determine the level of control over behavior. A history of alcohol dependence – such in the post-dependent state – causes epigenetic dysregulation and blunted IEG responses. This molecular reprogramming is likely impacting on neuronal communication and consequently on ensemble formation, which either could result in fewer neurons participating or lessened stimulus-driven transcriptional activity in the ensembles. This may underlie reduced behavioral flexibility and consequently increased risk of relapse.
Acknowledgments
Work in the Heilig laboratory reviewed here is supported by the Swedish Science Council (Dnr 2013-7434). The Sommer laboratory receives funding by Deutsche Forschungsgemeinschaft center grants (SFB636, SFB1134, subprojects D7 and B04, respectively). Work in the Wahlestedt laboratory was supported by US National Institute of Health NIAAA R01 no. 1R01AA023781-01A1. Related epigenomics and RNA work in the Wahlestedt laboratory has received funding by US National Institute of Health awards DA035592, MH084880 and NS071674.
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony JC, Warner LA, Kessler RC. Comparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: basic findings from the National Comorbidity Survey. Exp Clin Psychopharmacol. 1994;2:244–268. [Google Scholar]
- Augier E, Dulman RS, Rauffenbart C, Augier G, Cross AJ, Heilig M. The mGluR2 positive allosteric modulator, AZD8529 and cue-induced relapse to alcohol seeking in rats. Neuropsychopharmacology. 2016 doi: 10.1038/npp.2016.107. 2016 Jul 20. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista MA, Martin-Fardon R, Weiss F. Preferential effects of the metabotropic glutamate 2/3 receptor agonist LY379268 on conditioned reinstatement versus primary reinforcement: comparison between cocaine and a potent conventional reinforcer. J Neurosci. 2004;24:4723–4727. doi: 10.1523/JNEUROSCI.0176-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci. 2015;35:6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berg EA. A simple objective technique for measuring flexibility in thinking. J Gen Psychol. 1948;39:15–22. doi: 10.1080/00221309.1948.9918159. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Gray SM, Lu L, Shaham Y. Activation of group II metabotropic glutamate receptors in the nucleus accumbens shell attenuates context-induced relapse to heroin seeking. Neuropsychopharmacology. 2006;31:2197–2209. doi: 10.1038/sj.npp.1300977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FR, Cifani C, Koya E, Hope BT, Shaham Y. Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nat Neurosci. 2011;14:420–422. doi: 10.1038/nn.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FR, Marchant NJ, Wang HL, Morales M, Shaham Y. Role of projections from ventral medial prefrontal cortex to nucleus accumbens shell in context-induced reinstatement of heroin seeking. J Neurosci. 2012;32:4982–4991. doi: 10.1523/JNEUROSCI.0005-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown VJ, Tait DS. Attentional set-shifting across species. Curr Top Behav Neurosci. 2016;28:363–395. doi: 10.1007/7854_2015_5002. [DOI] [PubMed] [Google Scholar]
- Cannella N, Halbout B, Uhrig S, Evrard L, Corsi M, Corti C, Deroche-Gamonet V, Hansson AC, Spanagel R. The mGluR2/3 agonist LY379268 induced anti-reinstatement effects in rats exhibiting addiction-like behavior. Neuropsychopharmacology. 2013;38:2048–2056. doi: 10.1038/npp.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli D, Venniro M, Zeric T, Li X, Adhikary S, Madangopal R, Marchant NJ, Lucantonio F, Schoenbaum G, Bossert JM, Shaham Y. Effect of the novel positive allosteric modulator of metabotropic glutamate receptor 2 AZD8529 on incubation of methamphetamine craving after prolonged voluntary abstinence in a rat model. Biol Psychiatry. 2015;78:463–473. doi: 10.1016/j.biopsych.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carden WB, Alexander GM, Friedman DP, Daunais JB, Grant KA, Mu J, Godwin DW. Chronic ethanol drinking reduces native T-type calcium current in the thalamus of nonhuman primates. Brain Res. 2006;1089:92–100. doi: 10.1016/j.brainres.2006.02.135. [DOI] [PubMed] [Google Scholar]
- Copeland RA, Solomon ME, Richon VM. Protein methyl-transferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- Crawford JT, Roberts DC, Beveridge TJ. The group II metabotropic glutamate receptor agonist, LY379268, decreases methamphetamine self-administration in rats. Drug Alcohol Depend. 2013;132:414–419. doi: 10.1016/j.drugalcdep.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. 2015;20:1219–1231. doi: 10.1038/mp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2015;20:1438–1447. doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Gehlert DR, Cippitelli A, Thorsell A, Le AD, Hipskind PA, Hamdouchi C, Lu J, Hembre EJ, Cramer J, Song M, McKinzie D, Morin M, Ciccocioppo R, Heilig M. 3-(4-Chloro-2-morpholin-4-yl-thiazol-5-yl)-8-(1-ethylpropyl)-2,6-dimethyl-imidazo [1,2-b]pyridazine: a novel brain-penetrant, orally available corticotropin-releasing factor receptor 1 antagonist with efficacy in animal models of alcoholism. J Neurosci. 2007;27:2718–2726. doi: 10.1523/JNEUROSCI.4985-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Pal N. Alcohol dependence produced in mice by inhalation of ethanol: grading the withdrawal reaction. Science. 1971;172:288–290. doi: 10.1126/science.172.3980.288. [DOI] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND. Drug addiction and its underlying neurobiological basis: neuroimaging evidence for the involvement of the frontal cortex. Am J Psychiatry. 2002;159:1642–1652. doi: 10.1176/appi.ajp.159.10.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourley SL, Taylor JR. Going and stopping: dichotomies in behavioral control by the prefrontal cortex. Nat Neurosci. 2016;19:656–664. doi: 10.1038/nn.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson AC, Sommer WH, Metsis M, Stromberg I, Agnati LF, Fuxe K. Corticosterone actions on the hippocampal brain-derived neurotrophic factor expression are mediated by exon IV promoter. J Neuroendocrinol. 2006;18:104–114. doi: 10.1111/j.1365-2826.2005.01390.x. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Rimondini R, Heilig M, Mathe AA, Sommer WH. Dissociation of antidepressant-like activity of escitalopram and nortriptyline on behaviour and hippocampal BDNF expression in female rats. J Psychopharmacol. 2011;25:1378–1387. doi: 10.1177/0269881110393049. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heidbreder CA, Groenewegen HJ. The medial prefrontal cortex in the rat: evidence for a dorsoventral distinction based upon functional and anatomical characteristics. Neurosci Biobehav Rev. 2003;27:555–579. doi: 10.1016/j.neubiorev.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Heilig M, Koob GF. A key role for corticotropin-releasing factor in alcohol dependence. Trends Neurosci. 2007;30:399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, Becker HC. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict Biol. 2010;15:169–184. doi: 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Sommer WH, Spanagel R. The need for treatment responsive translational biomarkers in alcoholism research. Curr Top Behav Neurosci. 2016;28:151–171. doi: 10.1007/7854_2015_5006. [DOI] [PubMed] [Google Scholar]
- Hensler JG, Ladenheim EE, Lyons WE. Ethanol consumption and serotonin-1A (5-HT1A) receptor function in heterozygous BDNF (+/−) mice. J Neurochem. 2003;85:1139–1147. doi: 10.1046/j.1471-4159.2003.01748.x. [DOI] [PubMed] [Google Scholar]
- Hirth N, Meinhardt MW, Noori HR, Salgado H, Torres-Ramirez O, Uhrig S, Broccoli L, Vengeliene V, Rossmanith M, Perreau-Lenz S, Kohr G, Sommer WH, Spanagel R, Hansson AC. Convergent evidence from alcohol-dependent humans and rats for a hyperdopaminergic state in protracted abstinence. Proc Natl Acad Sci U S A. 2016;113(11):3024–3029. doi: 10.1073/pnas.1506012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Spanagel R, Krystal JH. Glutamatergic targets for new alcohol medications. Psychopharmacology. 2013;229:539–554. doi: 10.1007/s00213-013-3226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover WB, Vertes RP. Anatomical analysis of afferent projections to the medial prefrontal cortex in the rat. Brain Struct Funct. 2007;212:149–179. doi: 10.1007/s00429-007-0150-4. [DOI] [PubMed] [Google Scholar]
- Hopf FW, Chang SJ, Sparta DR, Bowers MS, Bonci A. Motivation for alcohol becomes resistant to quinine adulteration after 3 to 4 months of intermittent alcohol self-administration. Alcohol Clin Exp Res. 2010;34:1565–1573. doi: 10.1111/j.1530-0277.2010.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Washizuka S, Kakiuchi C, Kato T. Expression of HSPF1 and LIM in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. J Hum Genet. 2004;49:227–231. doi: 10.1007/s10038-004-0136-5. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, McGough NN, Logrip ML, Phamluong K, Janak PH, Ron D. The dopamine D3 receptor is part of a homeostatic pathway regulating ethanol consumption. J Neurosci. 2006;26:1457–1464. doi: 10.1523/JNEUROSCI.3786-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu XZ, Xu K, Kenney H, Egan SE, Turley H, Harris AL, Marini AM, Lipsky RH. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci. 2008;28:1118–1130. doi: 10.1523/JNEUROSCI.2262-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Lovinger DM. Metabotropic glutamate receptor 2 positive allosteric modulators: closing the gate on drug abuse? Biol Psychiatry. 2015;78:436–438. doi: 10.1016/j.biopsych.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Justinova Z, Panlilio LV, Secci ME, Redhi GH, Schindler CW, Cross AJ, Mrzljak L, Medd A, Shaham Y, Goldberg SR. The novel metabotropic glutamate receptor 2 positive allosteric modulator, AZD8529, decreases nicotine self-administration and relapse in squirrel monkeys. Biol Psychiatry. 2015;78:452–462. doi: 10.1016/j.biopsych.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, Bossert JM, Nair SG, Uejima JL, Marin MT, Mitchell TB, Farquhar D, Ghosh SC, Mattson BJ, Hope BT. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat Neurosci. 2009;12:1069–1073. doi: 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroener S, Mulholland PJ, New NN, Gass JT, Becker HC, Chandler LJ. Chronic alcohol exposure alters behavioral and synaptic plasticity of the rodent prefrontal cortex. PLoS One. 2012;7:e37541. doi: 10.1371/journal.pone.0037541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita M, Moreno JL, Holloway T, Kozlenkov A, Mocci G, Garcia-Bea A, Hanks JB, Neve R, Nestler EJ, Russo SJ, Gonzalez-Maeso J. Repressive epigenetic changes at the mGlu2 promoter in frontal cortex of 5-HT2A knockout mice. Mol Pharmacol. 2013;83:1166–1175. doi: 10.1124/mol.112.084582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Chu K, Jung KH, Kim JH, Huh JY, Yoon H, Park DK, Lim JY, Kim JM, Jeon D, Ryu H, Lee SK, Kim M, Roh JK. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72:269–277. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- Levey DF, et al. Genetic risk prediction and neurobiological understanding of alcoholism. Transl Psychiatry. 2014;4:e391. doi: 10.1038/tp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res. 2011;35:1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti ME, Lhuillier L, Kaupmann K, Markou A. Metabotropic glutamate 2/3 receptors in the ventral tegmental area and the nucleus accumbens shell are involved in behaviors relating to nicotine dependence. J Neurosci. 2007;27:9077–9085. doi: 10.1523/JNEUROSCI.1766-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Logrip ML, Janak PH, Ron D. Escalating ethanol intake is associated with altered corticostriatal BDNF expression. J Neurochem. 2009;109:1459–1468. doi: 10.1111/j.1471-4159.2009.06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Henkhaus RS, Butler MG. Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene. 2012;498:5–12. doi: 10.1016/j.gene.2012.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Whitfield TW, Jr, Berglind WJ. Brain-derived neurotrophic factor and cocaine addiction. Brain Res. 2010;1314:183–193. doi: 10.1016/j.brainres.2009.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough NN, He DY, Logrip ML, Jeanblanc J, Phamluong K, Luong K, Kharazia V, Janak PH, Ron D. RACK1 and brain-derived neurotrophic factor: a homeostatic pathway that regulates alcohol addiction. J Neurosci. 2004;24:10542–10552. doi: 10.1523/JNEUROSCI.3714-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt MW, Sommer WH. Postdependent state in rats as a model for medication development in alcoholism. Addict Biol. 2015;20:1–21. doi: 10.1111/adb.12187. [DOI] [PubMed] [Google Scholar]
- Meinhardt MW, Hansson AC, Perreau-Lenz S, Bauder-Wenz C, Stahlin O, Heilig M, Harper C, Drescher KU, Spanagel R, Sommer WH. Rescue of infralimbic mGluR2 deficit restores control over drug-seeking behavior in alcohol dependence. J Neurosci. 2013;33:2794–2806. doi: 10.1523/JNEUROSCI.4062-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez RI, McGinty JF, Kalivas PW, Becker HC. Brain region-specific gene expression changes after chronic intermittent ethanol exposure and early withdrawal in C57BL/6J mice. Addict Biol. 2012;17:351–364. doi: 10.1111/j.1369-1600.2011.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Amirouche A, Clow C, Belanger G, Jasmin BJ. Brain-derived neurotrophic factor expression is repressed during myogenic differentiation by miR-206. J Neurochem. 2012;120:230–238. doi: 10.1111/j.1471-4159.2011.07583.x. [DOI] [PubMed] [Google Scholar]
- Moorman DE, Aston-Jones G. Prefrontal neurons encode context-based response execution and inhibition in reward seeking and extinction. Proc Natl Acad Sci U S A. 2015;112:9472–9477. doi: 10.1073/pnas.1507611112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman DE, James MH, McGlinchey EM, Aston-Jones G. Differential roles of medial prefrontal subregions in the regulation of drug seeking. Brain Res. 2015;1628:130–146. doi: 10.1016/j.brainres.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. 2010;11:806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- Nasca C, Xenos D, Barone Y, Caruso A, Scaccianoce S, Matrisciano F, Battaglia G, Mathe AA, Pittaluga A, Lionetto L, Simmaco M, Nicoletti F. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci U S A. 2013;110:4804–4809. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols HH, Conn PJ. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis. 2014;61:55–71. doi: 10.1016/j.nbd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforuk A, Popik P, Drescher KU, van Gaalen M, Relo AL, Mezler M, Marek G, Schoemaker H, Gross G, Bespalov A. Effects of a positive allosteric modulator of group II metabotropic glutamate receptors, LY487379, on cognitive flexibility and impulsive-like responding in rats. J Pharmacol Exp Ther. 2010;335:665–673. doi: 10.1124/jpet.110.170506. [DOI] [PubMed] [Google Scholar]
- O’Dell LE, Roberts AJ, Smith RT, Koob GF. Enhanced alcohol self-administration after intermittent versus continuous alcohol vapor exposure. Alcohol Clin Exp Res. 2004;28:1676–1682. doi: 10.1097/01.alc.0000145781.11923.4e. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008;28:2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Kalivas PW. The group II metabotropic glutamate receptor agonist, LY379268, inhibits both cocaine- and food-seeking behavior in rats. Psychopharmacology. 2006;186:143–149. doi: 10.1007/s00213-006-0372-9. [DOI] [PubMed] [Google Scholar]
- Peters J, Kalivas PW, Quirk GJ. Extinction circuits for fear and addiction overlap in prefrontal cortex. Learn Mem. 2009;16:279–288. doi: 10.1101/lm.1041309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr S, Meinhardt MW, Klee ML, Hansson AC, Vengeliene V, Schonig K, Bartsch D, Hope BT, Spanagel R, Sommer WH. Losing control: excessive alcohol seeking after selective inactivation of cue-responsive neurons in the infralimbic cortex. J Neurosci. 2015;35:10750–10761. doi: 10.1523/JNEUROSCI.0684-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ. The role of microRNAs in drug addiction: a big lesson from tiny molecules. Int Rev Neurobiol. 2010;91:1–24. doi: 10.1016/S0074-7742(10)91001-5. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Post-transcriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers M, Heilig M, Sommer WH. Gene discovery in neuropharmacological and behavioral studies using Affymetrix microarray data. Methods. 2005;37:219–228. doi: 10.1016/j.ymeth.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16:27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- Rimondini R, Sommer W, Heilig M. A temporal threshold for induction of persistent alcohol preference: behavioral evidence in a rat model of intermittent intoxication. J Stud Alcohol. 2003;64:445–449. doi: 10.15288/jsa.2003.64.445. [DOI] [PubMed] [Google Scholar]
- Rimondini R, Sommer WH, Dall’Olio R, Heilig M. Long-lasting tolerance to alcohol following a history of dependence. Addict Biol. 2008;13:26–30. doi: 10.1111/j.1369-1600.2007.00079.x. [DOI] [PubMed] [Google Scholar]
- Salih H, Anghelescu I, Kezic I, Sinha V, Hoeben E, Van Nueten L, De Smedt H, De Boer P. Pharmacokinetic and pharmacodynamic characterisation of JNJ-40411813, a positive allosteric modulator of mGluR2, in two randomised, double-blind phase-I studies. J Psychopharmacol. 2015;29:414–425. doi: 10.1177/0269881115573403. [DOI] [PubMed] [Google Scholar]
- SAMHSA. National Survey on Drug Use and Health (NSDUH) Serious Mental Health Challenges among Older Adolescents and Young Adults. In: Lipari RN, Hedden SL, editors. The CBHSQ Report. Rockville (MD): Substance Abuse and Mental Health Services Administration (US); 2014. 2013–2014 May 6. [PubMed] [Google Scholar]
- Sanchis-Segura C, Spanagel R. Behavioural assessment of drug reinforcement and addictive features in rodents: an overview. Addict Biol. 2006;11:2–38. doi: 10.1111/j.1369-1600.2006.00012.x. [DOI] [PubMed] [Google Scholar]
- Seamans JK, Lapish CC, Durstewitz D. Comparing the prefrontal cortex of rats and primates: insights from electrophysiology. Neurotox Res. 2008;14:249–262. doi: 10.1007/BF03033814. [DOI] [PubMed] [Google Scholar]
- Shaham Y, Shalev U, Lu L, De Wit H, Stewart J. The reinstatement model of drug relapse: history, methodology and major findings. Psychopharmacology. 2003;168:3–20. doi: 10.1007/s00213-002-1224-x. [DOI] [PubMed] [Google Scholar]
- Sheedy D, Garrick T, Dedova I, Hunt C, Miller R, Sundqvist N, Harper C. An Australian Brain Bank: a critical investment with a high return! Cell Tissue Bank. 2008;9:205–216. doi: 10.1007/s10561-008-9076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms JA, Steensland P, Medina B, Abernathy KE, Chandler LJ, Wise R, Bartlett SE. Intermittent access to 20% ethanol induces high ethanol consumption in Long-Evans and Wistar rats. Alcohol Clin Exp Res. 2008;32:1816–1823. doi: 10.1111/j.1530-0277.2008.00753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Lopez MF, Archer KJ, Wolen AR, Becker HC, Miles MF. Time-course analysis of brain regional expression network responses to chronic intermittent ethanol and withdrawal: implications for mechanisms underlying excessive ethanol consumption. PLoS One. 2016;11:e0146257. doi: 10.1371/journal.pone.0146257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer W, Arlinde C, Heilig M. The search for candidate genes of alcoholism: evidence from expression profiling studies. Addict Biol. 2005;10:71–79. doi: 10.1080/13556210412331327821. [DOI] [PubMed] [Google Scholar]
- Sommer W, Hyytia P, Kiianmaa K. The alcohol-preferring AA and alcohol-avoiding ANA rats: neurobiology of the regulation of alcohol drinking. Addict Biol. 2006;11:289–309. doi: 10.1111/j.1369-1600.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- Sommer WH, Rimondini R, Hansson AC, Hipskind PA, Gehlert DR, Barr CS, Heilig MA. Upregulation of voluntary alcohol intake, behavioral sensitivity to stress, and amygdala crhr1 expression following a history of dependence. Biol Psychiatry. 2008;63:139–145. doi: 10.1016/j.biopsych.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Holter SM. Long-term alcohol self-administration with repeated alcohol deprivation phases: an animal model of alcoholism? Alcohol Alcohol. 1999;34:231–243. doi: 10.1093/alcalc/34.2.231. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum Mol Genet. 2009;18:525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR, Schwandt M, Sommer WH, Heilig M. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J. 2013;13:286–296. doi: 10.1038/tpj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, Sun H, Schank JR, King C, Heilig M. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34:4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Zhao M, Li M, Song T, Zhang M, Quan L, Li S, Sun ZS. Reversal of cocaine-conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex. PLoS One. 2012;7:e33435. doi: 10.1371/journal.pone.0033435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y. Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett. 2003;554:67–72. doi: 10.1016/s0014-5793(03)01092-5. [DOI] [PubMed] [Google Scholar]
- Uylings HBM, Groenewegen HJ, Kolb B. Do rats have a prefrontal cortex? Behav Brain Res. 2003;146:3–17. doi: 10.1016/j.bbr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Vendruscolo LF, Barbier E, Schlosburg JE, Misra KK, Whitfield TW, Jr, Logrip ML, Rivier C, Repunte-Canonigo V, Zorrilla EP, Sanna PP, Heilig M, Koob GF. Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J Neurosci. 2012;32:7563–7571. doi: 10.1523/JNEUROSCI.0069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengeliene V, Bilbao A, Spanagel R. The alcohol deprivation effect model for studying relapse behavior: a comparison between rats and mice. Alcohol. 2014;48:313–320. doi: 10.1016/j.alcohol.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Vertes RP. Differential projections of the infralimbic and prelimbic cortex in the rat. Synapse. 2004;51:32–58. doi: 10.1002/syn.10279. [DOI] [PubMed] [Google Scholar]
- Wang F, Xu H, Zhao H, Gelernter J, Zhang H. DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci Rep. 2016;6:19430. doi: 10.1038/srep19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling – a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Szyf M, Meaney MJ. From maternal care to gene expression: DNA methylation and the maternal programming of stress responses. Endocr Res. 2002;28:699. doi: 10.1081/erc-120016989. [DOI] [PubMed] [Google Scholar]
- Welsh JP, Han VZ, Rossi DJ, Mohr C, Odagiri M, Daunais JB, Grant KA. Bidirectional plasticity in the primate inferior olive induced by chronic ethanol intoxication and sustained abstinence. Proc Natl Acad Sci U S A. 2011;108:10314–10319. doi: 10.1073/pnas.1017079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S, Hammar J, Heilig M, Wisen O. Factors affecting the short-term prognosis of alcohol dependent patients undergoing inpatient detoxification. Subst Abus. 2001;22:235–245. doi: 10.1080/08897070109511465. [DOI] [PubMed] [Google Scholar]
- Willcocks AL, McNally GP. The role of medial prefrontal cortex in extinction and reinstatement of alcohol-seeking in rats. Eur J Neurosci. 2013;37:259–268. doi: 10.1111/ejn.12031. [DOI] [PubMed] [Google Scholar]
- Wise RA. Voluntary ethanol intake in rats following exposure to ethanol on various schedules. Psychopharmacologia. 1973;29:203–210. doi: 10.1007/BF00414034. [DOI] [PubMed] [Google Scholar]
- Wise RA. Maximization of ethanol intake in the rat. Adv Exp Med Biol. 1975;59:279–294. doi: 10.1007/978-1-4757-0632-1_19. [DOI] [PubMed] [Google Scholar]
- Wolffgramm J, Heyne A. From controlled drug intake to loss of control: the irreversible development of drug addiction in the rat. Behav Brain Res. 1995;70:77–94. doi: 10.1016/0166-4328(95)00131-c. [DOI] [PubMed] [Google Scholar]
- Wolffgramm J, Galli G, Thimm F, Heyne A. Animal models of addiction: models for therapeutic strategies? J Neural Transm. 2000;107:649–668. doi: 10.1007/s007020070067. [DOI] [PubMed] [Google Scholar]
- Wong CC, Mill J, Fernandes C. Drugs and addiction: an introduction to epigenetics. Addiction. 2011;106:480–489. doi: 10.1111/j.1360-0443.2010.03321.x. [DOI] [PubMed] [Google Scholar]
- Wood CM, Nicolas CS, Choi SL, Roman E, Nylander I, Fernandez-Teruel A, Kiianmaa K, Bienkowski P, de Jong TR, Colombo G, Chastagnier D, Wafford KA, Collingridge GL, Wildt SJ, Conway-Campbell BL, Robinson ES, Lodge D. Prevalence and influence of cys407* Grm2 mutation in Hannover-derived Wistar rats: mGlu2 receptor loss links to alcohol intake, risk taking and emotional behaviour. Neuropharmacology. 2016;14 doi: 10.1016/j.neuropharm.2016.03.020. pii: S0028-3908(16)30092-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dayas CV, Aujla H, Baptista MA, Martin-Fardon R, Weiss F. Activation of group II metabotropic glutamate receptors attenuates both stress and cue-induced ethanol-seeking and modulates c-fos expression in the hippocampus and amygdala. J Neurosci. 2006;26:9967–9974. doi: 10.1523/JNEUROSCI.2384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, Edenberg HJ, Riley BP, Dick DM, Bettinger JC, Davies AG, Grotewiel MS, Schuckit MA, Agrawal A, Kramer J, Nurnberger JI, Jr, Kendler KS, Webb BT, Miles MF. Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study. BMC Genomics. 2012;13(Suppl. 8):S16. doi: 10.1186/1471-2164-13-S8-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Karlsson C, Liang T, Xiong W, Kimura M, Tapocik JD, Yuan Q, Barbier E, Feng A, Flanigan M, Augier E, Enoch MA, Hodgkinson CA, Shen PH, Lovinger DM, Edenberg HJ, Heilig M, Goldman D. Loss of metabotropic glutamate receptor 2 escalates alcohol consumption. Proc Natl Acad Sci U S A. 2013;110:16963–16968. doi: 10.1073/pnas.1309839110. [DOI] [PMC free article] [PubMed] [Google Scholar]


