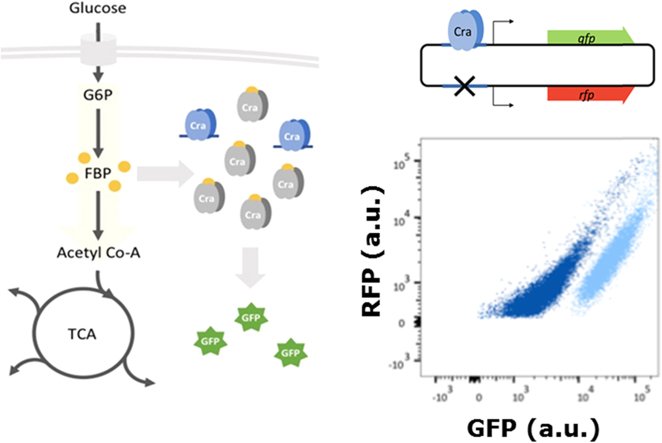
Abstract
We describe the development of an optimized glycolytic flux biosensor and its application in detecting altered flux in a production strain and in a mutant library. The glycolytic flux biosensor is based on the Cra-regulated ppsA promoter of E. coli controlling fluorescent protein synthesis. We validated the glycolytic flux dependency of the biosensor in a range of different carbon sources in six different E. coli strains and during mevalonate production. Furthermore, we studied the flux-altering effects of genome-wide single gene knock-outs in E. coli in a multiplex FlowSeq experiment. From a library consisting of 2126 knock-out mutants, we identified 3 mutants with high-flux and 95 mutants with low-flux phenotypes that did not have severe growth defects. This approach can improve our understanding of glycolytic flux regulation improving metabolic models and engineering efforts.
Keywords: Cra, Glycolytic flux, Escherichia coli, Transcription factor, Genome-wide screening, Biosensor
Graphical abstract
1. Introduction
Recent developments in synthetic biology allow the affordable construction of diverse, engineered cell libraries (Goodman et al., 2013, Kosuri and Church, 2014, Cavaleiro et al., 2015, Gibson, 2014, Bonde et al., 2015, Jiang et al., 2013). These capabilities enable a deeper understanding of biological processes and regulation (Bonde et al., 2016, Kosuri et al., 2013) and facilitate more rapid and efficient cell factory and protein engineering (Wang et al., 2009). Similarly, inexpensive deep sequencing simplifies the identification of beneficial genetic variants, often by multiplexing (Kosuri et al., 2013). Nevertheless, the development of new biotechnologically relevant production pathways is still not trivial.
A major challenge remains the identification of candidates for genetic modifications that have a desired characteristic out of the abundance of different variants. In certain cases, as, for example, the expression of a colored compound, the identification can be straightforward; however, in many cases, it is more challenging.
Genetically encoded biosensors enable the expression of a reporter molecule to be linked to the concentration of a certain ligand. If the intracellular concentration of a small molecule is coupled to the readout of fluorescent protein production, differences in intracellular concentrations can be easily identified at the single-cell level (Binder et al., 2012). Biosensors have been applied in several high-throughput screens, demonstrating their relevance to enzyme engineering and cell factory optimization (Binder et al., 2013, Mustafi et al., 2012, Michener et al., 2012, Schendzielorz et al., 2014, Siedler et al., 2014b, Raman et al., 2014, Taylor et al., 2015). There is a vast application range for biosensors. They have been established in a number of different areas, e.g., screening for improved enzymes (Siedler et al., 2014a, Tang et al., 2013, Binder et al., 2012, Schendzielorz et al., 2014), production pathways (Tang and Cirino, 2011, Dietrich et al., 2013, Yang et al., 2013) or evolutionarily adapted variants (Chou et al., 2013, Mahr et al., 2015). Biosensors can also be applied to identify novel enzymes with desired functions from metagenomic libraries (Uchiyama and Miyazaki, 2010, Genee et al., 2016).
Biosensors are either product-specific or responsive to an intermediate metabolite, thereby limiting the application range of each individual biosensor. In contrast, recent work has demonstrated that the E. coli transcription factor Cra (catabolite repressor/activator) can be used as a glycolytic flux biosensor, as it responds to the concentration of the glycolytic flux-dependent metabolite fructose-1,6-bisphosphate (FBP) (Kochanowski et al., 2013). Linking fluorescent protein production to a Cra-regulated promoter enables in vivo measurement of the glycolytic flux in a time-dependent manner, therefore limiting the need for otherwise laborious in vitro flux measurements (Kochanowski et al., 2013).
Metabolic flux analysis is a powerful tool to identify underlying mechanisms of perturbations to the cellular metabolic network (Nikel et al., 2009, Siedler et al., 2012, Mazumdar et al., 2013). However, it is time consuming and costly, and there are still limitations for high-throughput approaches (Heux et al., 2017). Furthermore, a flux-dependent biosensor can be useful for many different biotechnological applications, as it is not end product-specific.
We set out to construct an optimized glycolytic flux biosensor that enables single cell measurements for parallelized, high-throughput applications by characterizing different Cra-regulated promoters. Thus far, 164 binding sites have been identified in the genome of E. coli (Shimada et al., 2011), mainly related to central carbon metabolism, as Cra acts as a switch between glycolysis and gluconeogenesis (Ramseier et al., 1995, Ramseier, 1996). We characterized three promoters for their utility as biosensors and used a final construct (pFlux) with the ppsA promoter of E. coli controlling the expression of a green fluorescent protein (GFP) to identify flux-regulating genes and to achieve an improved understanding of the cellular networks affecting glycolytic flux.
2. Materials and methods
2.1. Bacterial strains, plasmids and oligonucleotides
E. coli Top10 and E. coli DH5a strains were used for the construction of pFlux. E. coli BW25113, and knockout strains from the KEIO collection (Baba et al., 2006) were used for the characterization and application of pFlux. For clarification, the Cra knockout strain E. coli BW25113 JW0078-1 is called ∆cra throughout this manuscript, even though it was originally labeled ∆fruR in the KEIO collection due to old terminology. Oligonucleotides were obtained from Integrated DNA Technologies (Leuven, Belgium). The adapters and primers for Illumina sequencing were additionally high-performance liquid chromatography (HPLC) purified and, in the case of the UAD_tail, contained a 3′-phosphorothioate bond and a 5′-phosphate for the barcoded sequencing adapters. The strains, plasmids and primers are listed in Tables SI, SII and SIII.
2.2. Cultivation and growth conditions
The growth media used in this study were Luria-Bertani (LB) complex medium, Super Optimal broth with Catabolite repression (SOC) medium and M9 minimal medium (Kochanowski et al., 2013) supplemented with filter-sterile trace element solution, resulting in final concentrations of 6.3 µM ZnSO4, 7.0 µM CuCl2, 7.1 µM MnSO4, 7.6 µM CaCl2 and 60 µM FeCl3. The M9 medium contained 5 g/l of the indicated carbon source (fructose, glucose, mannitol, sorbitol, galactose, glycerol, sodium pyruvate or sodium acetate). When solid medium was required, the bacteria were grown on LB-agar plates. When required, spectinomycin and kanamycin were added for final concentrations of 25 and 50 µg/ml, respectively. If not stated otherwise, the cells were cultured in 5 ml medium in 15-ml cultivation tubes or 2 ml medium in 24-deep-well plates. The cultivation tubes were incubated at 37 °C, 190 rpm in a common shaking incubator and the 24-deep-well plates at 37 °C and 900 rpm on a tabletop plate shaker (Titramax 1000 incubator, Heidolph Instruments GmbH, Germany). Strains were stored in 15% v/v glycerol at −80 °C.
2.3. Plasmid construction
All plasmids were assembled by USER cloning (Nour-Eldin et al., 2006). Phusion™ U Hot Start DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) or in-house-synthesized Pfu-X polymerase (Nørholm, 2010) was used for PCR amplification with a standard thermocycler program, matching the Tm values of the respective primers. Amplified PCR products were purified with the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co. KG) and digested with DpnI FastDigest (Thermo Fisher Scientific), and the plasmids were ligated with USER enzyme mix (New England BioLabs, Ipswich, MA, USA) according to the protocols. The resulting plasmids were transformed into the respective chemically competent E. coli strains.
The template for the pFlux backbones is pZA11MCS, a modular constructed plasmid backbone from EXPRESSYS with a p15A origin, ampicillin resistance, a tetracycline-inducible promoter (PLtetO-1) and a multiple cloning site (MCS) (Lutz, 1997). The gfp sequence was derived from (Calero et al., 2016), and the rfp gene was obtained from the Standard European Vector Architecture database (Silva-Rocha et al., 2013). As spectinomycin resistance was desired and the respective EXPRESSYS was not available, the resistance was amplified from another EXPRESSYS plasmid using the primer PC055/PC060 for the spectinomycin resistance and PC055/PC070 on pZA11MCS for the backbone, creating the pZA41MCS plasmid. For the assembly of the plasmids pGFPppsA, pGFPppc and pGFPpykF, pZA14MCS was amplified with the primers PC004/PC031, eliminating the promoter region but maintaining the p15A origin of replication and the spectinomycin resistance marker. The different natural promoter regions were obtained by colony PCR from E. coli BW25113 using the primer pairs PC001/PC002, PC003/PC004 and PC005/PC006; gfp was amplified with the primers PC019/PC021.
To generate the constitutive ppsA promoter with a scrambled Cra binding site, pGFPppsA was amplified with the primer pair PC063/PC064. This constitutive promoter was subsequently amplified with PC033/PC065, and rfp was amplified with PC069/PC070. The pGFPppsA backbone was amplified with PC071/PC072, resulting in an opening of the backbone downstream of the gfp gene. The PCR products were ligated and transformed as described to obtain the plasmid pFlux. The DNA sequences of the different promoters used in this study can be found in Table SIV and SV, and the sequence of pFlux is given in Fig. SI.
2.4. Flow cytometry
To measure the fluorescence signals of the pFlux plasmid, the different E. coli strains were initially grown overnight in LB medium at 37 °C and 190 rpm. Minimal medium with 5 g/l of the selected carbon source (fructose, glucose, mannitol, sorbitol, mannose, galactose, malate, glycerin, sodium pyruvate or sodium acetate) was inoculated 1:50 with the LB preculture. The cultures in minimal medium were incubated overnight at 37 °C and 190 rpm, after which they were used to inoculate fresh minimal medium (1:200) and grown under the same conditions for four hours. The fluorescence of the bacterial cells was always analyzed in the early exponential phase. The cells were diluted in FACSFlow (BD) to prepare them for screening on a FACSAria flow cytometer (BD, Franklin Lakes, New Jersey,USA), equipped with 488 and 561 nm lasers. To collect the GFP and RFP signals, fluorescein isothiocyanate (FITC, 530/30) and phycoerythrin (PE)-Texas Red (610/20) filters were used. To maintain comparability among multiple runs on different days, the E. coli strain BW25113 was aligned with the diagonal between the FITC and PE-Texas Red channels. The obtained data were analyzed with FlowJo software (FlowJo LLC, Oregon, US).
2.5. Validation in a mevalonate production strain
The E. coli BW25113 strain containing the pFlux plasmid was subsequently transformed with pMevT (Martin et al., 2003) or pZA1 and grown in medium containing 25 ng/ml spectinomycin and 25 ng/ml chloramphenicol. For mevalonate production, the cells were grown in M9 medium containing 5 g/l glucose. When the cultures were transferred from the preculture to fresh medium, they were induced with 0.05 mM IPTG. Fluorescence was determined 5 h after induction by flow cytometry.
2.6. KEIO library generation and plasmid transformation
For the library screen, the KEIO collection was pooled. To obtain the best possible coverage, the individual strains were plated from the glycerol stock on LB agar plates with 50 µg/ml kanamycin. The plates were incubated overnight at 37 °C, and the colonies were washed off with 1 ml liquid LB medium without antibiotics. One hundred microliters of each cell suspension was used to inoculate 150 ml LB medium containing 50 µg/ml kanamycin in one 500-ml shaking flask. The resulting inoculation volume for the 150 ml medium was 5.6 ml in total. The flasks were incubated at 37 °C, 190 rpm until OD 0.5 was reached. To prepare the cells for electroporation and to remove all traces of salts, the cells were prechilled on ice for 10 min and afterwards washed three times with ice-cold 10% (v/v) glycerol. Between each washing step, the cells were pelleted for 5 min at 4000 rpm and 0 °C, and the supernatant was discarded. The first two washing steps were performed in a 50-ml Falcon tube with 50- and 25-ml cell suspensions, respectively. The last washing step was performed in a 2-ml reaction tube, and the cells were pelleted in a prechilled tabletop centrifuge. The pellet was resuspended in 200 µl of ice-cold 10% (v/v) glycerol. Fifty microliters of this cell suspension was transferred to a prechilled 1-mm electroporation cuvette, and 100 ng of pFlux was added. The cells were electroporated at 1.8 kV and resuspended in 950 µl prewarmed SOC medium. After being transferred to a 1.5-ml reaction tube, the cells recovered for 1 h at 37 °C and 500 rpm. The cell suspension was used to inoculate 50 ml LB medium (with an additional 0.5 mM MgSO4 and 25 µg/ml kanamycin) in a 250-ml shaking flask. The cultures grew overnight at 37 °C and 190 rpm. Seven hundred microliters of the overnight culture was diluted to 15% glycerol stocks and stored at −80 °C until use.
2.7. Fluorescence-activated cell sorting (FACS)
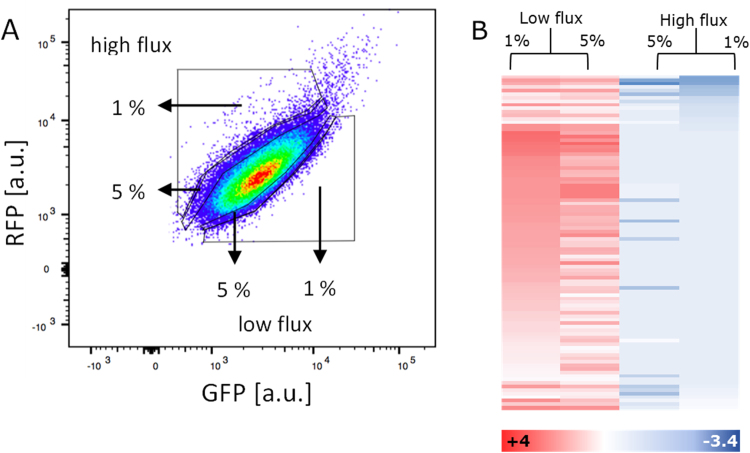
For the individual cell sorting rounds, 50 ml LB medium containing 25 ng/ml spectinomycin was inoculated with 1 ml of the KEIO cryo-stocks to maintain diversity. The cultures were incubated at 37 °C, 190 rpm overnight. Fifty milliliters of M9 medium containing 5 g/l glucose or galactose and the respective antibiotics was inoculated from the LB precultures to an OD600 of 0.01. The cultures were incubated at 37 °C, 190 rpm overnight. Fresh M9 medium was inoculated to an OD600 of 0.05. After 4 h of shaking incubation, samples were taken and diluted in FACSFlow (BD) to prepare them for sorting on the FACSAria (BD) with 488 and 561 nm lasers. To collect the GFP and RFP signals, the FITC (530/30) and PE-Texas Red (610/20) filters were used. The individual cells were sorted according to their signal in the FITC (GFP fluorescence) and PE-Texas Red (RFP fluorescence) channels. The top and bottom 1% of cells by FITC per PE-Texas Red signal ratio were collected (Fig. 4A). The cells were sorted into 12-cm FACS tubes with 1 ml LB medium (25 ng/ml spectinomycin) and grown at 37 °C, 190 rpm overnight. The cells were pelleted at 4500 rpm and stored at −20 °C.
Fig. 4.
Identification of gene deletions resulting in different glycolytic fluxes. (A) Dot plot of the RFP and GFP signals of the KEIO library grown in 5 g/l galactose. The four indicated gates were used to sort the cells by flux phenotype, sorting 100,000 cells into the 1% and 5% gates. Additionally, a sample of 1,000,000 cells was collected to determine the genetic composition of the total population at the point of sorting. (B) Heat map of the 95 genes enriched in the low-flux pools and depleted in the high-flux pools compared to the total knockout library. Enrichment is shown in red and depletion in blue. The genes are sorted according to their abundance in the 1% high-flux pool.
2.8. Genome purification, amplification and sequencing
Library preparation and validation closely followed the TnSeq protocol of Lennen et al. (Lennen and Herrgård, 2014). To adjust the protocol to the KEIO strains, the biotinylated PCR primer was designed to match the 19-base-pair flippase recognition target (FRT) scar (GAAGCAGCTCCAGCCTACA) that was left from the deletion process to generate the knockout library (Baba et al., 2006). To amplify the knockout regions, a biotinylated primer (/5BiotinTEG/AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGAAGCAGCTCCAGCCTACA) and a standardized UAD-tail primer (GATCTACACTCTTTCCCTACACGACG) were used. The barcoded adapter matched the Illumina Nextera platform. The sequencing was performed on an Illumina MiSeq, 150 bp, running 1 pM of DNA per sample.
2.9. Data analysis
To analyze the obtained sequence reads from the MiSeq, we ran a customized script (Table SVI), consisting of data preparation, quality checking, creating a database of quality reads, searching for E. coli genes using their bar codes and summarizing the results in a table.
The PCR amplification of the region following the FRT scar allows evaluation of the occurrence of a deletion mutant in the different pools without mapping it to the entire genome. Instead, it can be simply matched to the list of primers that Baba et al. (Baba et al., 2006) used to generate the knockout strains. This mapping results in less bias and a clear output list of each gene and the number of annotated reads.
In the data preparation process, the short-read FASTQ files were converted into FASTA files and a BLAST database build for each experiment, along with a tabular file with reads indexed by their read identification number. The quality check assured that only reads that began with the FRT-specific DNA sequence GAAGCAGCTCCAGCCTACA were taken into consideration and BLASTed (BLASTN) considering the parameters given in Supplementary Table SII to search for small sequences. We then extracted the reads that had at least 80% coverage of the primers with a maximum of two mismatches and discarded those that did not meet these criteria. The extracted reads form the BLAST database is used to search for the barcodes. The barcode list was based on the reverse primers that were used by Baba et al., (2006) to delete the respective genes. For each barcode, corresponding to one gene deletion, the number of reads in the BLAST database was counted, allowing a maximum of three mismatches.
We normalized the reads for every gene in each sequencing run to the overall number of reads in the run to make the results of the different sequencing runs comparable. The threshold for consideration of a gene was set to a minimum of 10 annotated reads in the library. Afterwards, the number of reads of the sorted population was compared to the library, which was grown under the same conditions, to identify enrichment and depletion in the different pools.
2.10. Clustering and Gene Ontology (GO) analysis
The enrichment and depletion of the individual genes in the different pools were analyzed and grouped based on their enrichment profiles. Genes enriched in the low-flux pools and depleted in the high-flux pools were grouped, as were all genes enriched in the high-flux pools and depleted in the low-flux pools. A GO analysis based on biological functions was performed with the genes of the group with low-flux phenotypes (Ashburner et al., 2000, The Gene Ontology Consortium, 2015). In detail, the corresponding data file for this GO analysis contains all genes of the E. coli K-12 genome, grouped based on their biological functions. An initial step uses the observed gene coverage to calculate how many genes would be expected per functional group if the distribution were entirely random. In a second step, these expected numbers are compared with the actual detected numbers of each group. The fold enrichment compared to the expected count is computed, and the statistical significance of the result is tested.
2.11. Growth rate characterization
The gene deletions that showed interesting phenotypes in the flux data analysis were tested individually for their growth rate. Two milliliters of LB medium (25 nm/ml kanamycin) in a 24-deep-well plate was inoculated with strains from a cryo-stock and grown at 37 °C and 1000 rpm in a tabletop plate shaker (Titramax 1000 incubator, Heidolph Instruments GmbH, Germany) until the exponential or stationary phase. The cells were subsequently diluted 1:50 in M9 medium containing 5 g/l galactose and grown at the same conditions overnight. The cells were diluted 1:200 in fresh M9 medium containing the selected carbon source. Two hundred microliters of the fresh culture was transferred to a microtiter plate. The plate was sealed with Breathe-Easy sealing membrane (Sigma-Aldrich). The OD630 was measured in a plate reader over a period of 16 h, and the growth rate was determined. Under those conditions, the cells might be oxygen limited and might not reach the optimal growth rate. We compared all strains under the same conditions.
3. Results and discussion
3.1. Design and characterization of an optimized glycolytic flux biosensor
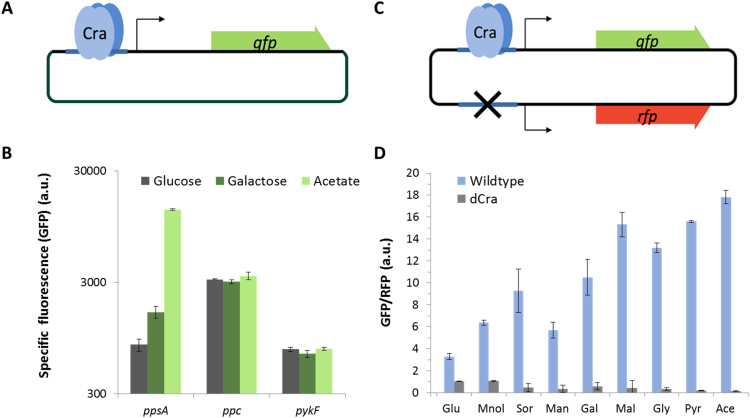
To generate an optimized glycolytic flux-dependent biosensor with a higher dynamic range and clear output signal, which will be applicable in high-throughput screening approaches, we compared the originally applied pykF promoter (Kochanowski et al., 2013) with the ppsA and ppc promoters of E. coli. All three promoters are sigma 70-dependent, and, to date, all are considered to be regulated exclusively by Cra (Keseler et al., 2013). By choosing these promoter regions, we reduced potential bias through cross-interactions via stress responses and other regulators. Gene expression from the ppsA promoter is activated by Cra, whereas the pykF and ppc promoters are repressed. The promoters were cloned in front of gfp to enable Cra-dependent regulation of GFP output fluorescence (Fig. 1A). The fluorescent signal was measured at the single-cell level during the exponential growth phase in various carbon sources using flow cytometry. The uptake of the used carbon sources and their entry point in the glycolysis process differ, which generates distinct alterations in intracellular FBP concentrations and glycolytic flux (Fig. 1B).
Fig. 1.
Glycolytic flux dependencies of different promoters (A) Different Cra-regulated promoter regions (ppsA, ppc and pykF) were cloned in front of gfp, generating three different reporter plasmids. (B) The fluorescence of E. coli KEIO wild-type strain BW25113 with the different flux sensor constructs was measured by flow cytometry in the exponential phase, 4 h after induction. The cells grew in minimal media containing carbon sources inducing high glycolytic flux (glucose/gray), medium flux (galactose/dark green) and low flux (acetate, light green). By applying a gate in the FSC/SSC dot plot, the bacterial cells could be separated from background noise. The bars represent the mean fluorescent signal of the bacterial population in the FITC channel. The standard deviations were calculated from three individual experiments. (C) Schematic map of pFlux. gfp transcription is controlled by a Cra-dependent ppsA promoter, whereas rfp transcription is controlled by a ppsA promoter with a scrambled Cra binding site. (D) GFP/RFP emission ratios for E. coli W25113 wild-type (blue bars) and ∆cra (gray bars) when grown on different carbon sources, inducing different glycolytic fluxes. The carbon sources are ordered according to previously defined glycolytic fluxes with the lowest flux on acetate and the highest flux on glucose (Kochanowski et al., 2013). Abbreviations: Glu, glucose; Mnol: mannitol; Sor: sorbitol; Man: mannose; Gal: galactose; Mal: maltose; Gly: glycerol; Pyr: pyruvate; Ace: acetate.
It is expected that the signal intensity of the activated promoter of ppsA increases with decreasing flux, whereas the signal intensities of the two repressed promoters, pykF and ppc, decrease with decreasing flux. In the case of the ppsA promoter, a 16-fold induction of the fluorescent signal was detected after growth on acetate (15318 ± 168 a.u.) compared to glucose (830 ± 103 a.u.). The differences in the signal intensities of the two repressing promoter regions were very low and not suitable for a high-throughput approach dependent on a single time point measurement (Fig. 1B). Furthermore, even though the expression levels of GFP followed the expected tendencies of a glycolytic flux reporter when grown on glucose compared to galactose, the growth on the gluconeogenic carbon source pyruvate resulted in the highest fluorescent signals for all three cases (Fig. 1B). There is experimental evidence for the modes of action of the different promoters (Bledig et al., 1996, Nègre et al., 1998, Shimada et al., 2011), indicating that the highest values of the pykF and ppc promoters in pyruvate must be due to other reasons. It can be assumed that this observed effect correlates with the highly different growth rates between glycolytic and gluconeogenic carbon sources (Klumpp et al., 2009). Indeed, if a bacterial culture is growing rapidly, the gfp expression and maturation is not fast enough to compensate for the expanding cell volume, and the signal is diluted. In contrast, if the cells are growing slowly, there is more time to accumulate gfp in the bacterial cells. This observed effect made comparison of the promoters more difficult, as there was a definite bias in the data. This effect was also observed by Kochanoskwi et al., and solved by measuring the promoter strength over time (dGFP/dt/OD) during exponential growth (Kochanowski et al., 2013). The relative promoter strength was calculated by the difference of the native pykF promoter strength, regulated by Cra, and the strength of a pykF promoter variant where Cra binding was omitted. Based on this data, we wanted to generate a versatile approach where end-point measurement in single cells can be obtained.
The ppsA promoter appeared to be the most interesting candidate for further optimization, as it showed the highest dynamic range in addition to comparably low expression in the OFF state (glucose) (Fig. 1B). In the exponential growth phase, the intracellular oxygen consumption competes with the oxygen needed for maturation of the GFP molecules. Interestingly, we identified relatively stable GFP per OD600 values during exponential growth in glucose and glycerol in a plate reader (Fig. SII).
To reduce the noise in the output signal of the glycolytic flux biosensor, we implemented an intrinsic protein production control by constitutive expression of a red fluorescent protein (RFP) (Kosuri et al., 2013, Kochanowski et al., 2013). The final biosensor construct contains the native ppsA promoter with a Cra binding site regulating the expression of GFP and a second ppsA promoter variant without a functional Cra binding site controlling the expression of RFP (Fig. 1C, Table SV). The expression levels of both GFP and RFP should be equally affected by the protein synthesis bias during growth on different carbon sources. As Cra controls only the promoter upstream of gfp, information regarding the glycolytic flux is solely conveyed into the intensity of the GFP signal. Therefore, relative glycolytic flux can be obtained by calculating the ratio of the fluorescence intensities of GFP and RFP. To confirm the assumption that this construct is actually capable of eliminating possible growth defects but also shows a Cra-dependent expression pattern, the construct was tested in a wild-type E. coli W25113 strain and an E. coli W25113 ∆cra deletion strain.
The two strains were grown in M9 medium with a range of carbon sources, resulting in different physiologically relevant fluxes. The highest flux was reached during growth on glucose, and the lowest was expected on pyruvate. The GFP/RFP ratios were analyzed by flow cytometry, and the ratio was normalized, with the ∆cra strain grown in glucose set to 1.0. The ratios in the wild-type strain followed the expected trend based on previously measured and estimated glycolytic fluxes (Kochanowski et al., 2013) (Fig. 1D and Fig. SIII). Furthermore, the flux sensor provided a large dynamic range with ratios of 3.3 ± 0.4 GFP/RFP fluorescence on glucose and 17.8 ± 0.6 on acetate. The ratios were not significantly changed in the ∆cra strain in glycolytic carbon sources, indicating the dependence on a functional Cra regulator. However, there was a small decrease in the GFP/RFP values in the gluconeogenic carbon sources, caused by a slight shift towards a higher red signal. This effect was potentially due to different maturation times and stabilities of the two fluorescent proteins.
3.2. Analyzing glycolytic fluxes in different E. coli strains
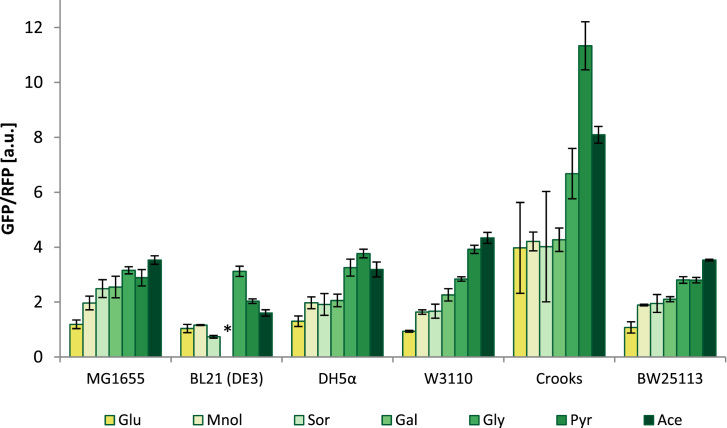
To validate the versatility of pFlux, we transformed the plasmid into different E. coli strains. The selected six E. coli strains are common laboratory and industrially relevant strains and include K-strains and one B-strain (BL21), enabling analysis of the biosensor signal in a variety of genetic backgrounds. The strains were grown in the presence of glycolytic and gluconeogenic carbon sources, and the GFP and RFP signals were measured by flow cytometry. MG1655, DH5α and W3110 followed the same pattern as previously observed for E. coli BW25113 (Fig. 2).
Fig. 2.
GFP/RFP ratios of six commonly used E. coli strains. The cells were grown in M9 minimal medium with different carbon sources. The fluorescence signals of GFP and RFP were measured by flow cytometry for 10,000 cells per run. The bars show the averages of the mean GFP per RFP values of three independent experiments. Abbreviations: Fru: Fructose, Glu: Glucose, Mnol: Mannitol; Sor: Sorbitol; Gal: Galactose; Gly: Glycerol; Pyr: Sodium pyruvate; Ace: Sodium acetate. * BL21(DE3) is unable to grow on galactose as a sole carbon source.
E. coli Crooks showed a generally higher GFP/RFP ratio, which correlated with lower fluorescent signals in the GFP and RFP channels compared with those for the other E. coli strains. Crooks has the highest growth rate and glucose uptake rate of the tested strains and should therefore have the highest flux values on glucose (Monk et al., 2016). It is known that different fluorescent proteins have different maturation times in different hosts, depending, for example, on growth rates and length of lag phases (Hebisch et al., 2013), which could explain the differences in this strain and the differential regulation of gene expression. Another reason might be different Cra activity or expression, resulting in higher relative expression of the GFP gene. BL21(DE3) did not grow in M9 medium containing galactose as the sole carbon source, as it contains the gal mutation in galactose metabolism, making it galactose non-utilizing. BL21(DE3) showed generally lower GFP/RFP ratios, especially on sorbitol, pyruvate and acetate. Compared with the other tested strains, it is known to have a higher flux through the citric acid cycle and a higher capacity for the glyoxylate shunt. Furthermore, it has a lower flux through the pentose phosphate pathway, resulting in a higher glycolytic flux (Monk et al., 2016). The higher capacity of the glyoxylate shunt, which is essential for growth on gluconeogenic carbon sources, might explain the higher FBP concentrations measured in this strain under growth on pyruvate and acetate. Metabolic flux analysis would be needed to confirm this hypothesis.
Our data suggest that even though the actual expression levels and subsequent ratios slightly differ in the tested strains, the differences should not have an impact on the usability of pFlux, as the glycolytic flux response was comparable in all tested E. coli strains.
3.3. Applying pFlux to measure altered glycolytic flux in a mevalonate production strain
After testing the sensitivity of the glycolytic flux biosensor to different fluxes and its function in a variety of E. coli backgrounds, its applicability to sense different fluxes in a production strain was assessed. We chose mevalonate as an example, as its production results in higher glycolytic flux due to higher demand for acetyl-CoA (Martin et al., 2003) (Fig. 3A).
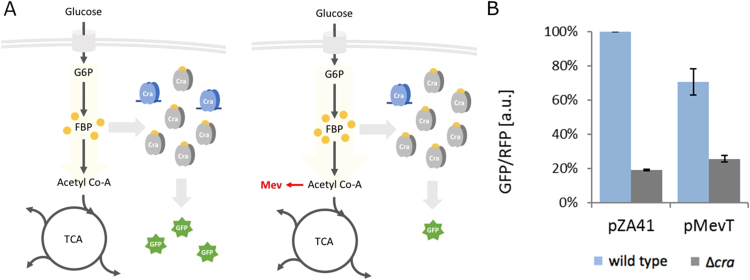
Fig. 3.
Analysis of different glycolytic fluxes in a production strain. (A) Schematic overview of increased flux during mevalonate production and the Cra-dependent response. During mevalonate production, a higher flux through glycolysis is expected, resulting in higher FBP concentrations, lower Cra activity and, consequently, lower GFP/RFP values. Abbreviations: G6P: glucose-6-phosphate, FBP: fructose-1,6-bisphosphate, TCA: citric acid cycle, Mev: mevalonate (B) The GFP per RFP ratios obtained by flow cytometry in the absence (pZA41) and presence of the mevalonate pathway (pMevT). The cells were grown in M9 medium containing 5 g/l glucose. The wild-type (gray) and ∆cra strains (blue) were compared (n=3).
Together with pFlux, the pMevT plasmid (Martin et al., 2003) was transformed into E. coli BW25113 wild-type and Δcra strains. As a control, we used the empty plasmid pZA1. The GFP per RFP ratio dropped to 70% in wild-type, glucose-grown cells expressing the genes of the mevalonate pathway, indicating a higher flux through glycolysis, while the GFP per RFP ratio did not significantly change in the Δcra mutant (Fig. 3B). This result demonstrates that pFlux can be applied for glycolytic flux measurements in production strains, where the glycolytic flux exceeds normally observed fluxes.
The integration of novel or enhanced biotechnologically relevant pathways often causes a change in the metabolic flux, as seen in this example of mevalonate production. However, lower glycolytic flux rates can also be found in production strains. For instance, during lysine production, the carbon flux is directed through the pentose phosphate pathway for improved NADPH supply, leading to reduced glycolytic flux (Kiefer et al., 2004). The fact that compound production alters glycolytic flux enables a broad range of possible applications for this glycolytic flux sensor, as monitoring flux changes might indicate higher production, in case no product sensor is available.
3.4. Applying the glycolytic flux biosensor in a genome-wide glycolytic flux screen
We wanted to use the glycolytic flux sensor to identify knockout mutants with altered glycolytic flux phenotypes from a genome-wide knockout library. The glycolytic flux biosensor plasmid pFlux was transformed into a library of the KEIO collection, a collection of non-lethal single gene deletions in E. coli (Baba et al., 2006). The library of knockout strains was grown in M9 minimal medium containing 5 g/l galactose. Galactose takes the same glycolytic route as glucose, but with a lower flux rate (Haverkorn van Rijsewijk et al., 2011). During growth on glucose, Cra is mostly inactive; thus, we assume that choosing galactose as the carbon source provides the possibility of better sensitivity for changes towards higher flux. Single cells were sorted by FACS into the top 1% and 5% GFP/RFP pools representing the low-flux phenotypes and the bottom 1% and 5% GFP/RFP pools representing the high-flux phenotypes (Fig. 4A). The terms low-flux and high-flux are defined as lower and higher glycolytic carbon flux from fructose-6-phosphate to pyruvate compared to the wild type, which is reflected by changes in the FBP concentrations. After recovery in LB medium, the gene regions downstream of the FRT sequence were amplified, sequenced and analyzed. The biological replicates showed good agreement (R2 range of 0.87–0.99) (Fig. SIV). The 1% and 5% pools of the high-flux pools were very similar (p-value 0.08), whereas the low-flux pools were more differentially distributed.
A total of 2126 individual deletion mutants were identified in the library after growth in minimal medium with galactose, which represents approximately 56% of the whole KEIO collection. We assume that knockout mutants, which do not appear in the library, had excessive fitness costs and were outcompeted by the other strains during the initial growth of the cell library (Cao et al., 2014). After coverage and quality filtering (see the Materials and Methods section), 504 genes remained (Table SVII and Fig. SV). The 504 genes were analyzed according to their enrichment and depletion patterns in the different pools compared to the total library.
3.4.1. Identification of gene deletions that result in higher glycolytic flux
Only 3 sequences, related to the gene deletions ∆ompC, ∆rpiA and ∆ynfH, were enriched at least two-fold in the high-flux pools and also depleted in the low-flux pools (Table SVIII). OmpC is a porin in the outer membrane and forms non-specific pores that allow the diffusion of small hydrophilic molecules across the outer membrane (Heller and Wilson, 1981), whereas YnfH is considered a subunit of a putative selenite reductase (Guymer et al., 2009). ompC and ynfH deletions have been shown to give E. coli a growth benefit compared to the wild-type strain in the presence of antibiotics. ∆ompC was tested with antibiotics of the ß-lactam family (Liu et al., 2012), and ∆ynfH was tested with spectinomycin directly (Vlasblom et al., 2015). We tested the growth of the deletion mutants harboring the plasmid pFlux in minimal medium containing 5 g/l galactose and 25 µg spectinomycin (Table SX). The ∆ynfH deletion mutant did not show a higher growth rate than that of the wild type, whereas the ∆ompC mutant had an increased growth rate (∆ompC 0.356 ± 0.006 h−1 and wt 0.223 ± 0.006 h−1) and OD600 after 16 h (∆ompC 0.51 ± 0.07 and wt 0.14 ± 0.044). The higher-flux phenotype of ∆ompC and possibly ∆ynfH mutants in the presence of antibiotics compared to those of the overall knockout library and the wild-type strain can be explained by their growth advantages in the presence of antibiotics. The remaining gene knockout strain with a significant high-flux phenotype is ∆rpiA, which encodes ribose-5-phosphate isomerase A. RpiA catalyzes the first step of the non-oxidative branch of the pentose phosphate pathway (PPP) and is therefore a step towards nucleotide and aromatic amino acid biosynthesis (Skinner and Cooper, 1971). Using a plate reader, we compared the growth rates of ∆rpiA and the wild-type strain on minimal medium with glucose or galactose supplemented. We detected a significant growth difference between ∆rpiA and the wild-type strain on galactose (∆rpiA 0.308 ± 0.005 h−1 and wt 0.223 ± 0.006 h−1), whereas no advantage was detected on glucose (∆rpiA 0.362 ± 0.007 h−1 and wt 0.416 ± 0.052 h−1). These findings are surprising, as a ∆rpiA mutant should not be able to grow on glucose (Sørensen and Hove-Jensen, 1996) and possibly not on galactose. It is known that the isoenzyme RpiB is capable of supplementing RpiA in a deletion strain, but the expression needs to be induced e.g., by ribose, and is not active on glucose (Sørensen and Hove-Jensen, 1996). RpiB was generally considered a substituting enzyme of minor function, but recent studies of ∆rpiB deletion strains have found surprisingly strong effects on biomass production. Kim and Reed found that the ∆rpiB mutant had a 30% decrease in biomass yield compared to the parental strain (Kim and Reed, 2012). In regard to our findings, this decrease could mean that the ∆rpiA mutant gained secondary mutations followed by up-regulation of RpiB expression, resulting in as-yet-uncharacterized positive effects on the glycolytic flux. This hypothesis will need to be further tested, and the isoenzymes RpiA and RpiB will serve as very interesting targets for further research on glycolytic flux control and optimization.
We compared our results to intracellular FBP concentrations identified in a knock-out library during growth on glucose (Fuhrer et al., 2017). Significant higher FBP concentrations were found in the ΔompC and ΔynfH mutants (2.43 a.u. and 0.77 a.u., respectively) compared to the wild type (0.27 a.u.), validating our screening results. Deletion of rpiA did not result in higher FBP concentrations (0.27 a.u.). Taking in mind, that these concentrations were obtained during growth on glucose, might explain the low FBP concentration in the ΔrpiA strain. We were only able to identify significant growth differences of the ΔrpiA strain during growth on galactose and not on glucose, which suggests that higher FBP concentrations would only be present during growth on galactose.
To conclude, we were able to identify three gene deletion strains that previously showed higher FBP concentration, higher growth rates or both.
3.4.2. Identification of gene deletions that lead to lower glycolytic flux
In total, 95 genes were associated with a low-flux phenotype; depleted in both high-flux GFP/RFP fractions and enriched in the low-flux GFP/RFP fractions with a threshold of > 0.5-fold enrichment and p-value < 0.05. This list showed a high similarity of the 1% and 5% low-flux pools (p-value 0.001 compared to the full list p-value 0.83) (Fig. 4B, Table SIX).
We identified the deletion mutant of the transcriptional regulator GalS in this fraction as a positive control. GalR was also > 2-fold enriched in the 5% low-flux pool but was depleted in the 1% low-flux pool. GalR and GalS take part in the regulation of operons involved in the transport and catabolism of d-galactose in the presence of high galactose concentrations and under glucose limitation (Semsey et al., 2007, Weickert and Adhya, 1992). It is expected that the deletion of these genes leads to reduced glycolytic flux in cells grown on galactose; therefore, our results for these genes can be seen as a validation of the applicability of the glycolytic flux biosensor in this experimental setup.
A GO analysis of the 95 genes with the low-flux phenotype was performed. A GO analysis is helpful to provide a global picture of which groups of genes with biologically related functions are significantly enriched or depleted in a dataset compared to the statistical expectation (Ashburner et al., 2000, The Gene Ontology Consortium, 2015).
This analysis identified the glyoxylate pathway, including all necessary genes (∆aceA, ∆aceB and ∆aceK), as significantly enriched in the low-flux phenotype. The genes of aceA and aceB encode isocitrate lyase and malate synthase, respectively, two enzymes that are essential for a functional glyoxylate pathway and are sufficient together with other genes of the TCA cycle. AceK controls the branch point between the TCA cycle and the glyoxylate cycle by phosphorylation of isocitrate dehydrogenase (ICD) and consequent modulation of ICD activity (LaPorte and Koshland, 1982, Cortay et al., 1988). A deletion of aceK results in constant activation of ICD and reduced glyoxylate pathway activity. As it was shown in previous 13C metabolic flux analysis (Haverkorn van Rijsewijk et al., 2011), the glyoxylate shunt is very active when E. coli is grown on galactose. Our data confirm the importance of the glyoxylate cycle on galactose, as deletion of the genes involved in this process leads to an overall reduction of glycolytic flux.
The second enriched GO pathway was the galactitol metabolic pathway (3 genes out of 7 in the E. coli genome, ρ = 1.11×10−02). Gene deletions include gatA and gatC, subunits of galactitol/sorbitol PTS permease. Either the transporter is also accepting galactose to some extent, increasing the intracellular galactose concentration, and subsequently a deletion reduces the uptake and hence the flux, or galactitol, an alcohol of galactose, is potentially present as a byproduct in the galactose solution taken up by the cells, and its co-utilization results in a higher overall flux.
Several other genes in the periphery of the central carbon metabolism were identified. Two genes (ulaE and ulaF) of the l-ascorbic acid metabolic process were identified (2 out of 4, ρ = 3.79×10−03), as well as the deletion of nanK, an N-acetylmannosamine kinase, which has low glucokinase activity in vitro. Their primary functions are not to utilize galactose, but they may potentially have some minor activity with metabolites of the galactose metabolic pathway that were not previously detected using other methods. Our data might be useful to help optimize metabolic models, as our sensor enables the detection of changes in the glycolytic flux in gene deletions that were not described before. Most genes identified in our study are not present in metabolic models (Literature), and several have unknown functions (Table SIX).
Another strength of our technology is its capacity for identifying gene deletions that result in a lower glycolytic flux but do not interfere with the cell fitness. Many groups have been studying the effects of gene deletions on fitness in different growth medium and stress environments (Lennen and Herrgård, 2014, Wetmore et al., 2015, Rau et al., 2016). It is well known that deletion of central carbon metabolism genes results in a lower glycolytic flux (e.g., pgi, pfkA, tpiA). These knockout strains do not appear in our library, as they result in a huge growth defect and are outcompeted by the better-growing mutants. To further validate that the identified strains in the low-flux pools do not have a growth defect, we analyzed the growth behaviors of the 10 mutants most strongly enriched in the 1% low-flux pools (Table SIX). No strain showed significant changes in growth rates or OD after 16 h compared to the wild-type strain (Table SX), demonstrating that these strains do not have a fitness defect.
We also compared the 10 strains with the highest enrichment in the 1% low flux pools to intracellular FBP concentration (Fuhrer et al., 2017) showing relatively lower FBP concentrations in those mutants compared to the wild type in seven strains (Table SX and Fig. SVI). Interestingly, deletion of aceK did not result in lower intracellular FBP concentrations during growth on glucose, pointing again to the relevance of the glyoxylate cycle during growth on galactose and not glucose.
Regarding biotechnological applications, the identified gene knockouts with low-flux phenotypes might be very interesting. These deletion mutants have lower maintenance flux but now growth defect under the tested conditions. Reduction of by-product formation and rerouting of the carbon flux have been shown to increase product and/or biomass formation (Vermuri et al., 2006, Balzer et al., 2013). Further experiments will aim at elucidating, whether the identified mutants provide additional flux capacity that could be redirected towards a production process.
Gene deletion is seldom correlated with a gain of function. Consequently, it was comparatively more difficult to identify gene deletions that caused higher flux, resulting in only three identified mutants. Additionally, we could demonstrate, by the example of the mevalonate pathway, that the glycolytic flux biosensor is capable of detecting changes towards even higher flux than observed during growth on glucose.SI
4. Conclusions
To conclude, we developed and validated a glycolytic flux biosensor that can be applied to screen for flux variants in libraries with hundreds of mutants. The developed biosensor, pFlux, has a wide application range, as it was able to detected alterations in glycolytic flux in six different industrially relevant E. coli strains during growth on various carbon sources. As pFlux enables the detection of glycolytic flux changes at the single cell level, the biosensor has great potential helping to reveal how different gene knock-outs affect glycolytic flux under diverse conditions. Furthermore, this biosensor can be applied in biotechnologically relevant production strains. We showed that production of mevalonate alters the glycolytic flux, which can be detected by pFlux. Our biosensor might be used to identify mutants with an altered production phenotype of diverse production strains, with the major advantage of endproduct independence. Accordingly, pFlux can be applied in a wide range of flux-altering production scenarios. Finally, the presented findings support the claims of Kochanowski et al. that Cra functions as a direct glycolytic flux sensor in E. coli, even though it has different functions in other organisms (Chavarría et al., 2014).
Author contributions
MOAS, CEL and SS designed the study; CEL performed all experimental work; MMHE processed the sequencing data; and MOAS, CEL and SS analyzed the results and wrote the paper.
Acknowledgments
This work was supported by The Novo Nordisk Foundation, a Ph.D. grant from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme [FP7-People-2012-ITN] under grant agreement no. 317058, “BACTORY”, and the European Union Seventh Framework Programme (FP7-KBBE-2013-7-single-stage) under grant agreement no. 613745, Promys. We acknowledge Stefano Cardinale for critical comments to the manuscript as well as early assistance in the computational analysis of sequencing data.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ymben.2017.07.002.
Appendix A. Supporting information
Supplementary material
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. (2006.0008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer G.J., Thaker C., Bennett G.N., San K.Y. Metabolic engineering of Escherichia coli to minimize byproduct formate and improving succinate productivity through increasing NADH availability by heterologous expression of NAD(+)-dependent formate dehydrogenase. Metab. Eng. 2013;20:1–8. doi: 10.1016/j.ymben.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Binder S., Schendzielorz G., Stäbler N., Krumbach K., Hoffmann K., Bott M., Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S., Siedler S., Marienhagen J., Bott M., Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41:6360–6369. doi: 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledig S.A., Ramseier T.M., Saier M.H. FruR mediates catabolite activation of pyruvate kinase (pykF) gene expression in Escherichia coli. J. Bacteriol. 1996;178:280–283. doi: 10.1128/jb.178.1.280-283.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M.T., Kosuri S., Genee H.J., Sarup-Lytzen K., Church G.M., Sommer M.O.A., Wang H.H. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth. Biol. 2015;4:17–22. doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde M.T., Pedersen M., Klausen M.S., Jensen S.I., Wulff T., Harrison S., Nielsen A.T., Herrgård M.J., Sommer M.O.A. Predictable tuning of protein expression in bacteria. Nat. Methods. 2016;13:233–236. doi: 10.1038/nmeth.3727. [DOI] [PubMed] [Google Scholar]
- Calero P., Jensen S.I., Nielsen A.T. Broad-host-range ProUSER vectors enable fast characterization of inducible promoters and optimization of p-coumaric acid production in Pseudomonas putida KT2440. ACS Synth. Biol. 2016;5:741–753. doi: 10.1021/acssynbio.6b00081. [DOI] [PubMed] [Google Scholar]
- Cao H., Butler K., Hossain M., Lewis J.D. Variation in the fitness effects of mutations with population density and size in Escherichia coli. PLoS One. 2014;9:e105369. doi: 10.1371/journal.pone.0105369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaleiro A.M., Kim S.H., Seppälä S., Nielsen M.T., Nørholm M.H.H. Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth. Biol. 2015;4:1042–1046. doi: 10.1021/acssynbio.5b00113. [DOI] [PubMed] [Google Scholar]
- Chavarría M., Durante-Rodríguez G., Krell T., Santiago C., Brezovsky J., Damborsky J., de Lorenzo V. Fructose 1-phosphate is the one and only physiological effector of the Cra (FruR) regulator of Pseudomonas putida. FEBS Open Bio. 2014;4:377–386. doi: 10.1016/j.fob.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.H., Keasling J.D., Elena S.F., Cooper V.S., Lenski R.E., Desai M.M., Fisher D.S., Sniegowski P.D., Gerrish P.J., Lenski R.E., Zhang Q., Stich M., Manrubia S.C., Lázaro E., Giruad A., Dietrich J.A., McKee A.E., Keasling J.D., Hibbert E.G., Greener A. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013;4:1802–1804. doi: 10.1038/ncomms3595. [DOI] [PubMed] [Google Scholar]
- Cortay J.C., Bleicher F., Rieul C., Reeves H.C., Cozzone A.J. Nucleotide sequence and expression of the aceK gene coding for isocitrate dehydrogenase kinase/phosphatase in Escherichia coli. J. Bacteriol. 1988;170:89–97. doi: 10.1128/jb.170.1.89-97.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J.A., Shis D.L., Alikhani A., Keasling J.D. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth. Biol. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T., Zampieri M., Sévin D., Sauer U., Zamboni N. Genomewide landscape of gene-metabolome associations in Escherichia coli. Mol. Syst. Biol. 2017;13(1):907. doi: 10.15252/msb.20167150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genee H.J., Bali A.P., Petersen S.D., Siedler S., Bonde M.T., Gronenberg L.S., Kristensen M., Harrison S.J., Sommer M.O.A. Functional mining of transporters using synthetic selections. Nat. Chem. Biol. 2016 doi: 10.1038/nchembio.2189. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. Programming biological operating systems: genome design, assembly and activation. Nat. Methods. 2014;11:521–526. doi: 10.1038/nmeth.2894. [DOI] [PubMed] [Google Scholar]
- Goodman D.B., Church G.M., Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Sci. (80-.) 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- Guymer D., Maillard J., Sargent F. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 2009;191:519–528. doi: 10.1007/s00203-009-0478-7. [DOI] [PubMed] [Google Scholar]
- Haverkorn van Rijsewijk B.R.B., Nanchen A., Nallet S., Kleijn R.J., Sauer U. Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol. Syst. Biol. 2011;7:477. doi: 10.1038/msb.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebisch E., Knebel J., Landsberg J., Frey E., Leisner M. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PLoS One. 2013;8:e75991. doi: 10.1371/journal.pone.0075991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K.B., Wilson T.H. Selectivity of the Escherichia coli outer membrane porins ompC and ompF. FEBS Lett. 1981;129:253–255. doi: 10.1016/0014-5793(81)80177-9. [DOI] [PubMed] [Google Scholar]
- Heux S., Bergès C., Millard P., Portais J.-C., Létisse F. Recent advances in high-throughput 13C-fluxomics. Curr. Opin. Biotechnol. 2017;43:104–109. doi: 10.1016/j.copbio.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martínez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., Latendresse M., Muñiz-Rascado L., Ong Q., Paley S., Schröder I., Shearer A.G., Subhraveti P., Travers M., Weerasinghe D., Weiss V. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer P., Heinzle E., Zelder O., Wittmann C. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl. Environ. Microbiol. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Reed J.L. RELATCH: relative optimality in metabolic networks explains robust metabolic and regulatory responses to perturbations. Genome Biol. 2012;13:R78. doi: 10.1186/gb-2012-13-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S., Zhang Z., Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanowski K., Volkmer B., Gerosa L., Haverkorn van Rijsewijk B.R., Schmidt A., Heinemann M., van Rijsewijk B.R., Schmidt A., Heinemann M. Functioning of a metabolic flux sensor in Escherichia coli. Proc. Natl. Acad. Sci. {Usa. } 2013;110:1130–1135. doi: 10.1073/pnas.1202582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S., Church G.M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte D.C., Koshland D.E. A protein with kinase and phosphatase activities involved in regulation of tricarboxylic acid cycle. Nature. 1982;300:458–460. doi: 10.1038/300458a0. [DOI] [PubMed] [Google Scholar]
- Lennen R.M., Herrgård M.J. Combinatorial strategies for improving multiple-stress resistance in industrially relevant Escherichia coli strains. Appl. Environ. Microbiol. 2014;80:6223–6242. doi: 10.1128/AEM.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-F., Yan J.-J., Lei H.-Y., Teng C.-H., Wang M.-C., Tseng C.-C., Wu J.-J. Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect. Immun. 2012;80:1815–1822. doi: 10.1128/IAI.06395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr R., Gätgens C., Gätgens J., Polen T., Kalinowski J., Frunzke J. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum. Metab. Eng. 2015;32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Martin V.J.J., Pitera D.J., Withers S.T., Newman J.D., Keasling J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Mazumdar S., Blankschien M.D., Clomburg J.M., Gonzalez R. Efficient synthesis of L-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb. Cell Fact. 2013;12:7. doi: 10.1186/1475-2859-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener J.K., Thodey K., Liang J.C., Smolke C.D. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab. Eng. 2012;14:212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J.M., Koza A., Campodonico M.A., Machado D., Seoane J.M., Palsson B.O., Herrgård M.J., Feist A.M. Multi-omics quantification of species variation of Escherichia coli links molecular features with strain phenotypes. Cell Syst. 2016;3:238–251. doi: 10.1016/j.cels.2016.08.013. (e12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi N., Grünberger A., Kohlheyer D., Bott M., Frunzke J. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab. Eng. 2012;14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Nègre D., Oudot C., Prost J.F., Murakami K., Ishihama A., Cozzone A.J., Cortay J.C. FruR-mediated transcriptional activation at the ppsA promoter of Escherichia coli. J. Mol. Biol. 1998;276:355–365. doi: 10.1006/jmbi.1997.1548. [DOI] [PubMed] [Google Scholar]
- Nikel P.I., Zhu J., San K.-Y., Méndez B.S., Bennett G.N. Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. J. Bacteriol. 2009;191:5538–5548. doi: 10.1128/JB.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm M.H.H. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 2010;10:21. doi: 10.1186/1472-6750-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H.H., Jensen J.K., Halkier B.A. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. 2014;111:201409523. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramseier T.M. Cra and the control of carbon flux via metabolic pathways. Res. Microbiol. 1996;147:489–493. doi: 10.1016/0923-2508(96)84003-4. [DOI] [PubMed] [Google Scholar]
- Ramseier T.W.M., Bledig S., Michotey V., Feghali R., Saier M.H.J., Saier M.H. The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol. Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Rau M.H., Calero P., Lennen R.M., Long K.S., Nielsen A.T. Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microb. Cell Fact. 2016;15:176. doi: 10.1186/s12934-016-0577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendzielorz G., Dippong M., Grünberger A., Kohlheyer D., Yoshida A., Binder S., Nishiyama C., Nishiyama M., Bott M., Eggeling L. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth. Biol. 2014;3:21–29. doi: 10.1021/sb400059y. [DOI] [PubMed] [Google Scholar]
- Semsey S., Krishna S., Sneppen K., Adhya S. Signal integration in the galactose network of Escherichia coli. Mol. Microbiol. 2007;65:465–476. doi: 10.1111/j.1365-2958.2007.05798.x. [DOI] [PubMed] [Google Scholar]
- Shimada T., Yamamoto K., Ishihama A. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 2011;193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedler S., Bringer S., Blank L.M., Bott M. Engineering yield and rate of reductive biotransformation in Escherichia coli by partial cyclization of the pentose phosphate pathway and PTS-independent glucose transport. Appl. Microbiol. Biotechnol. 2012;93:1459–1467. doi: 10.1007/s00253-011-3626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedler S., Schendzielorz G., Binder S., Eggeling L., Bringer S., Bott M. SoxR as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli. ACS Synth. Biol. 2014;3:41–47. doi: 10.1021/sb400110j. [DOI] [PubMed] [Google Scholar]
- Siedler S., Stahlhut S.G., Malla S., Maury J., Neves A.R. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab. Eng. 2014;21:2–8. doi: 10.1016/j.ymben.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de Las Heras A., Páez-Espino A.D., Durante-Rodríguez G., Kim J., Nikel P.I., Platero R., de Lorenzo V. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013;41:D666–D675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner A.J., Cooper R.A. The regulation of ribose-5-phosphate isomerisation in Escherichia coli K12. FEBS Lett. 1971;12:293–296. doi: 10.1016/0014-5793(71)80202-8. [DOI] [PubMed] [Google Scholar]
- Sørensen K.I., Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 1996;178:1003–1011. doi: 10.1128/jb.178.4.1003-1011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.-Y., Cirino P.C. Vol. 50. 2011. Design and application of a mevalonate-responsive regulatory protein; pp. 1084–1086. (Angew. Chemie Int. Ed). [DOI] [PubMed] [Google Scholar]
- Tang S.-Y., Qian S., Akinterinwa O., Frei C.S., Gredell J.A., Cirino P.C. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. J. Am. Chem. Soc. 2013;135:10099–10103. doi: 10.1021/ja402654z. [DOI] [PubMed] [Google Scholar]
- Taylor N.D., Garruss A.S., Moretti R., Chan S., Arbing M.A., Cascio D., Rogers J.K., Isaacs F.J., Kosuri S., Baker D., Fields S., Church G.M., Raman S. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods. 2015;13:177–183. doi: 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T., Miyazaki K., 2010. Substrate-induced gene expression screening: a method for high-throughput screening of metagenome libraries. In pp. 153–168. [DOI] [PubMed]
- Vermuri G.N., Altman E., Sangurdekar D.P., Khodursky A.B., Eiteman Overflow metabolism in Escherichia coli during steady-state grwoth: transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 2006;72(5):3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasblom J., Zuberi K., Rodriguez H., Arnold R., Gagarinova A., Deineko V., Kumar A., Leung E., Rizzolo K., Samanfar B., Chang L., Phanse S., Golshani A., Greenblatt J.F., Houry W.A., Emili A., Morris Q., Bader G., Babu M. Novel function discovery with GeneMANIA: a new integrated resource for gene function prediction in Escherichia coli. Bioinformatics. 2015;31:306–310. doi: 10.1093/bioinformatics/btu671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R., Church G.M. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert M.J., Adhya S. Isorepressor of the gal regulon in Escherichia coli. J. Mol. Biol. 1992;226:69–83. doi: 10.1016/0022-2836(92)90125-4. [DOI] [PubMed] [Google Scholar]
- Wetmore K.M., Price M.N., Waters R.J., Lamson J.S., He J., Hoover C.A., Blow M.J., Bristow J., Butland G., Arkin A.P., Deutschbauer A. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio. 2015;6:e00306–e00315. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Seo S.W., Jang S., Shin S.-I., Lim C.H., Roh T.-Y., Jung G.Y., Yadav V.G., Stephanopoulos G., Keasling J.D., Peralta-Yahya P.P., Nakagawa A., Pfleger B.F., Pitera D.J., Smolke C.D., Keasling J.D., Santos C.N., Stephanopoulos G., Isaacs F.J., Wang H.H. Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat. Commun. 2013;4:1413. doi: 10.1038/ncomms2404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material