Abstract
A high-resolution genetic linkage map is essential for a wide range of genetics and genomics studies such as comparative genomics analysis and QTL fine mapping. Crucian carp (Carassius auratus) is widely distributed in Eurasia, and is an important aquaculture fish worldwide. In this study, a high-density genetic linkage map was constructed for crucian carp using 2b-RAD technology. The consensus map contains 8487 SNP markers, assigning to 50 linkage groups (LGs) and spanning 3762.88 cM, with an average marker interval of 0.44 cM and genome coverage of 98.8%. The female map had 4410 SNPs, and spanned 3500.42 cM (0.79 cM/marker), while the male map had 4625 SNPs and spanned 3346.33 cM (0.72 cM/marker). The average recombination ratio of female to male was 2.13:1, and significant male-biased recombination suppressions were observed in LG47 and LG49. Comparative genomics analysis revealed a clear 2:1 syntenic relationship between crucian carp LGs and chromosomes of zebrafish and grass carp, and a 1:1 correspondence, but extensive chromosomal rearrangement, between crucian carp and common carp, providing evidence that crucian carp has experienced a fourth round of whole genome duplication (4R-WGD). Eight chromosome-wide QTL for body weight at 2 months after hatch were detected on five LGs, explaining 10.1–13.2% of the phenotypic variations. Potential candidate growth-related genes, such as an EGF-like domain and TGF-β, were identified within the QTL intervals. This high-density genetic map and QTL analysis supplies a basis for genome evolutionary studies in cyprinid fishes, genome assembly, and QTL fine mapping for complex traits in crucian carp.
Keywords: crucian carp, SNP, genetic linkage map, QTL, comparative genomics, candidate growth gene
Linkage mapping is an important area of genetic research, and high-density linkage maps provide a basic tool to gain insight into the genetic architecture of traits of evolutionary and economic interest (Naruse et al. 2000; Knapik et al. 1998). A high-density map can provide a lot of genetic information, such as chromosome structure, segregation distortion region, recombination rate, and recombination hotspots (Myers et al. 2005; Zhu et al. 2014; Guo et al. 2013; Shifman et al. 2006). Furthermore, it is an indispensable tool for fine-scale mapping of phenotypic traits of interest, candidate gene cloning, gene-centromere mapping, comparative genomic analysis, and genome assembly (Zhu et al. 2013, 2015; Peng et al. 2016; Xiao et al. 2015; Z. Wang et al. 2015; Tian et al. 2015; Shao et al. 2015; Li et al. 2015; Feng et al. 2015). SNPs are the most abundant genetic markers in the genomes of organisms, and have become the preferred markers for high-density map construction (Baird et al. 2008). The development of next-generation sequencing (NGS) technology provides the capacity for developing and genotyping large numbers of SNP markers rapidly and cost-effectively (Davey et al. 2011). Taking advantage of NGS, high-density linkage maps have been constructed using thousands of SNP markers in many aquaculture species, such as Atlantic salmon (Tsai et al. 2016), channel catfish (Li et al. 2015), Japanese flounder (Shao et al. 2015), Asian sea bass (Y. Wang et al. 2015), large yellow croaker (Xiao et al. 2015), Mexican tetra (Carlson et al. 2015), and oyster (Wang et al. 2016). Recently, a variety of genotyping-by-sequencing (GBS) methods have been developed for SNP discovery and genotyping, such as RAD (Baird et al. 2008), ddRAD (Peterson et al. 2012), GGRS (Chen et al. 2013), SLAF (Sun et al. 2013), and 2b-RAD (S. Wang et al. 2012). Among these methods, 2b-RAD is a simple and flexible method invented by S. Wang et al. (2012) that adopts type IIB restriction enzymes for genome-wide genotyping. Compared with other GBS methods, it allows screening of almost every restriction site in the genome, regulates genome coverage more flexibly, and has a simpler protocol for library preparation (S. Wang et al. 2012). 2b-RAD has been used successfully for constructing high-density maps and QTL fine mapping in several aquaculture species (Jiao et al. 2014; Shi et al. 2014; Cui et al. 2015; Tian et al. 2015; Dou et al. 2016; Fu et al. 2016; Palaiokostas et al. 2016).
Teleosts have undergone the third round (3R) of whole genome duplication (WGD) ∼350 million years ago (MYA), which is an additional round that mammals do not have (Jaillon et al. 2004; Meyer and Van de Peer 2005). The 3R-WGD may have facilitated the evolutionary innovations, and led to the diversity of teleosts (Taylor et al. 2003; Venkatesh 2003; Ravi and Venkatesh 2008). Recently, a variety of evidence (e.g., chromosome numbers, genome size, orthologous genes, and comparative genomes) indicates that some fish species have experienced a fourth round (4R) of WGD, such as some fishes of salmonids (Berthelot et al. 2014; Lien et al. 2016; Danzmann et al. 2008; Guyomard et al. 2012; Timusk et al. 2011; Brieuc et al. 2014; Allendorf and Thorgaard 1984), catastomids (Uyeno and Smith 1972; Zhou et al. 2002), Cobitidae (Ferris and Whitt 1977; Kusunoki et al. 1994; Kitagawa et al. 2003), and Cyprininae (J.T. Wang et al. 2012; David et al. 2003; Xu et al. 2014; Larhammar and Risinger 1994; Risinger and Larhammar 1993; Ohno et al. 1967; Zhang et al. 2013; Zhao et al. 2013; Henkel et al. 2012). The 4R-WGD provides an excellent opportunity for studying evolutionary patterns of vertebrate genomes after an autotetraploid WGD (Davidson et al. 2010; Lien et al. 2016). Comparative genome analysis based on whole-genome sequencing (WGS) have provided unambiguous evidence for WGD in some fishes (Berthelot et al. 2014; Lien et al. 2016; Xu et al. 2014; Glasauer and Neuhauss 2014). Comparative genomics using linkage maps is an alternative approach to investigating chromosome reduplication and chromosomal rearrangements after WGD events in nonmodel fish (Zhang et al. 2013; Zhao et al. 2013; Guyomard et al. 2012; Brieuc et al. 2014; Gharbi et al. 2006). For example, a clear 2:1 relationship of common carp linkage groups and zebrafish (Danio rerio) chromosomes revealed an additional genome reduplication (Zhang et al. 2013; Zhao et al. 2013), and comparative mapping studies between salmonids showed extensive chromosomal rearrangements and differentiation (Guyomard et al. 2012; Brieuc et al. 2014; Gharbi et al. 2006; Naish et al. 2013).
Crucian carp (Carassius auratus) is widely distributed in Eurasia and is one of the most important freshwater fish species for Chinese aquaculture. Previous karyotype studies demonstrated that crucian carp has a diploid chromosome number of 2n = 100, a number twice as much most of other cyprinid fishes (2n = 50 or 48) (Knytl et al. 2013; Ojima et al. 1979; Kobayasi et al. 1970). Recent studies have revealed diploid (2n = 100), triploid (3n = 150), and tetraploid (4n = 200) crucian carp in natural populations, and these three ploidy populations often coexist with each other in natural waters (Liu et al. 2001; Xiao et al. 2011; Gui and Zhou 2010). Thereby, it was believed that crucian carp may have experienced multiple successive rounds of chromosome doubling, which provided an excellent material for genome duplication and evolution studies (Zhang et al. 2015; Gui and Zhou 2010; Zhou et al. 2002). Moreover, as an important farmed fish, the global aquaculture production of crucian carp reached 2.91 million tons in 2015 (FAO, 2016). Growth rate is one of the most important traits for breeding programs in crucian carp (Zhou et al. 2000a; Hulata 1995). Several strains of triploid crucian carp have been developed in China via gynogenesis or interspecific hybridization between crucian carp and common carp, which has allowed great progress in breeding, and significantly increased the production of crucian carp (Gui and Zhou 2010; Zhou et al. 2000a,b, 2001; Liu et al. 2001, 2004; Hulata 1995). However, the genetic basis and architecture for growth modulation in crucian carp are still poorly understood because few genetic and genomic resources are available (Liao et al. 2013). It is well known that growth is a typical quantitative trait controlled by multiple genes known as quantitative trait loci (QTL), and may be influenced by environmental factors (Lynch and Walsh 1998; Mackay 2001; Mackay et al. 2009). Traditional selective breeding methods have encountered some difficulties such as uncertainty, extensive workload, being time-consuming, and being slow to take effect. Hence, molecular breeding methods, such as marker assisted selection (MAS) and genomic selection, are needed to accelerate breeding process in fish (Yue 2014; Liu and Cordes 2004; Tong and Sun 2015).
The objectives of this study include: (i) construction of a high-density SNP-based linkage map using 2b-RAD technology in diploid crucian carp; (ii) comparative genomics analysis between crucian carp and zebrafish (D. rerio), grass carp (Ctenopharyngodon idellus), and common carp (Cyprinus carpio). (iii) QTL fine mapping for body weight, and identification of candidate genes that may involve early growth of crucian carp. This study will provide a framework for further studies on genome evolution, comparative genomics, and fine-scale QTL mapping for economic traits in crucian carp.
Materials and Methods
Mapping family and DNA extraction
A total of 102 adult crucian carp was collected from a wild population in the Zhangdu Lake (Wuhan, China) and used as candidate broodfish in the production of mapping families. Genetic distances among these fish were evaluated using 20 previously reported microsatellite loci (Zheng et al. 2010). In April 2015, 17 candidate F1 full-sib families were established by crossing 14 sires and 17 dams via artificial fertilization. Larval fish of each family were raised in a plastic tank, and fed with brine shrimp (nauplii of the Artemia) twice a day. Body weights were measured and recorded for all families at the age of 2 months posthatch. One family with the highest genetic distance and largest within-family phenotypic variations in body weight was selected as the mapping panel for linkage map construction and QTL analysis for early growth in this study. A total of 160 progenies was randomly selected from this family, and a small piece of clipped fin was sampled from both parents and progenies and stored in 95% ethanol for DNA extraction. Genomic DNA was extracted from preserved fin tissues following a standard phenol-chloroform DNA extraction procedure (Sambrook and Russell 2001). The quality of DNA was checked by a NanoDrop 2000 spectrophotometer (Thermo Scientific), and 1% agarose gel electrophoresis. The concentrations of all DNA samples were adjusted to 50 ng/μl. All experimental animal programs involved in this study were approved by the Animal Care and Use Committee at the Institute of Hydrobiology, Chinese Academy of Sciences.
2b-RAD library construction and de novo genotyping
Before library preparation, the number of possible restriction sites was calculated based on crucian carp genome draft assembly (unpublished data). The 2b-RAD library was prepared following the protocol originally described by S. Wang et al. (2012) with minor modifications (Fu et al. 2016). Two parents and 160 offspring were used for the construction of 2b-RAD libraries. Briefly, 250 ng of genomic DNA was digested with 1 unit BcgI restriction enzyme (New England Biolabs) at 37° for 4 hr. The digested DNA was ligated at 16° for 8 hr with a 25 μl total volume reaction consisting of 1 unit T4 DNA ligase (New England Biolabs), 0.5 μM adapter 1 and adapter 2, 0.5 mM ATP (New England Biolabs). The ligation fragments were amplified in a 25 μl total volume consisting of 5 μl of ligated DNA, 0.5 μM P5 and P7 primer, 0.5 μM P4 and P6-BC primer, 0.75 mM dNTP (Shanghai Sangon, China), 5 μl 5× Phusion HF buffer, and 0.5 unit Phusion High-Fidelity DNA Polymerase (Thermo Scientific). The cycling conditions were: 98° for 30 sec; 98° for 20 sec, 63° for 50 sec, 72° for 30 sec for 15 cycles, 72° for 5 min. The amplification products were purified via retrieval from 8% polyacrylamide gels. All libraries were pooled with equal amount of products from each library to make a final library which was sequenced on a lane at the HiSeq2500 platform (Illumina). The raw read data were archived at the NCBI Sequence Read Archive (SRA) under accession number PRJNA327320.
Raw reads were first trimmed to remove adapter sequences, and the terminal 2-bp positions. Reads without restriction sites or containing long homopolymers (>10 bp), ambiguous bases (N), low-quality sequences (more than five positions with a quality <20) or mitochondrial origins were removed. The remaining trimmed reads with 32 bp in length were used for subsequent analysis. Filtered reads were analyzed with the software RADtyping program v1.0 (Fu et al. 2013) for de novo 2b-RAD genotyping. This software used stringent criteria in filtering candidate markers, and only those loci with at least four reads supporting were kept in the following analysis.
Linkage map construction
Only those markers that segregated in parents and could be genotyped in at least 80% of the offspring were used for further analysis. Markers that present significant segregation distortion in the χ2 goodness-of-fit tests (P < 0.05) were eliminated in the linkage analysis. Slightly distorted markers (P > 0.05) were also used for linkage analysis. A consensus linkage map was constructed by JoinMap 4.1 program (Van Ooijen 2006) with a threshold LOD score of 15.0. Male- and female-specific linkage map calculations were performed using the function of “Create Maternal and Paternal Population Nodes” in JoinMap 4.1, with a threshold LOD score of 10.0. The visualized linkage maps were drawn using MapChart v2.2 (Voorrips 2002). The linkage groups (LGs) of crucian carp were named according to their homologous groups of common carp and zebrafish based on the results of comparative genome analyses. The expected genetic map length was calculated in two ways: Ge1 (Fishman et al. 2001) and Ge2 (Chakravarti et al. 1991), and the average of these two indexes was used as the predicted total genetic map length (Ge). The recombination ratio of female to male was calculated by the ratios of mean distances of each LG in the female and male maps.
Comparative genome analysis
All mapped 2b-RAD sequences (32 bp) were first aligned against the crucian carp genome draft assembly (unpublished data). Those markers mapped at a single genome position were then extended by adding 100 nucleotide sequences from each side. A total of 5734 extended 2b-RAD sequences was searched against the genomes of zebrafish (Danio rerio, GRCz10), grass carp (Ctenopharyngodon idella), and common carp (C. carpio) using the basic local alignment search tool (BLASTN) with an e-value cutoff of 1e−10. If a single marker sequence aligned multiple targets at different positions, only the top hit (lowest e-value) alignment was retained. The genomic synteny was visualized using the software Circos v0.67 (Krzywinski et al. 2009).
QTL analysis for body weight
QTL mapping analysis was performed using the MapQTL 6.0 (Van Ooijen and Kyazma 2009) software program with a Multiple QTL Mapping (MQM) model. A mapping step size of 1 cM and five neighboring markers were used in QTL analysis. The genome-wide LOD threshold (significance level) or group-wide LOD threshold (suggestive level) were estimated using the permutation test (10,000 replicates) in MapQTL 6.0 with a confidence interval of 95%.
Identification of potential candidate genes
Potential candidate genes within each QTL region were hunted through comparative genomics. We performed sequence similarity searches (BLASTN) for all QTL-associated SNP markers against the whole genome sequences of crucian carp (unpublished data) and common carp (Xu et al. 2014). Only annotated genes closest to the peak of corresponding QTL region were regarded as candidate genes.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
2b-RAD genotyping
A total of 6.56, 3.98 and 177.44 million reads were produced by 2b-RAD sequencing for the female parents, and male parents and 160 progenies (1.11 million reads per progeny), respectively. After sequential quality filtering and sequence trimming, two parents’ reads were clustered into 124,367 representative reference tags, including 98,911 codominant tags (parent-shared), and 25,456 dominant tags (parent-specific). After removing the 4451 tags with insufficient sequencing depth, 96,328 codominant tags and 23,588 dominant tags were retained and used for constructing high-quality reference tags. Utilizing the constructed reference, a total of 14,732 polymorphic markers were detected, including 7310 codominant markers and 7422 dominant markers. After a Mendelian fit test (P > 0.05), and genotyping percentage (over 80% progenies) detection, 4073 codominant and 4693 dominant SNP markers were used to construct consensus map and sex-specific maps.
High-density linkage map
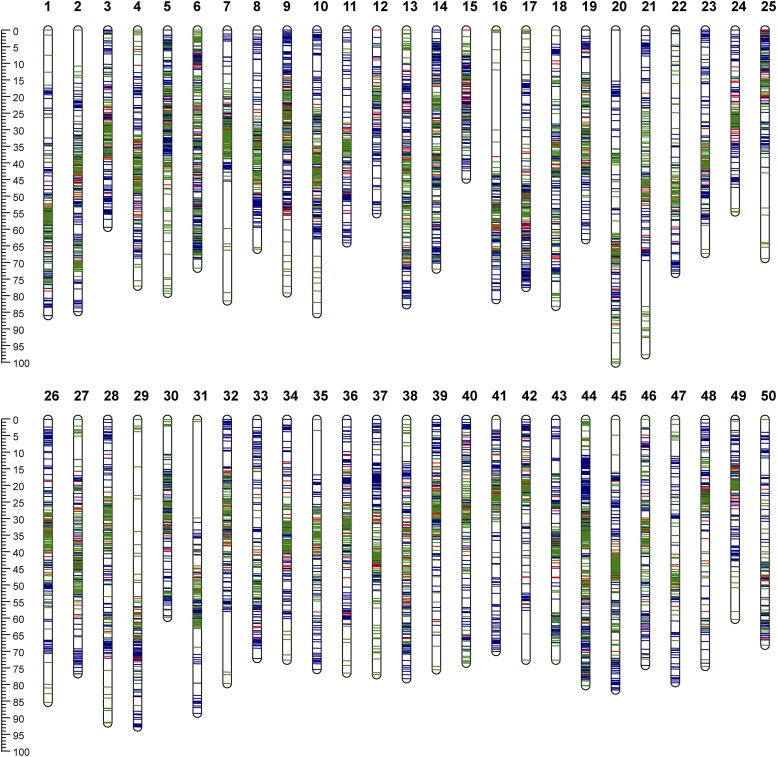
All polymorphic SNP markers were grouped into 50 linkage groups, which is consistent with the haploid chromosome number of crucian carp (2n = 100) (Knytl et al. 2013). The consensus map consisted of 8487 markers and spanned 3762.88 cM, with an average interval of 0.44 cM (Figure 1 and Table 1). The number of markers per group varied from 106 (LG 22 and LG 50) to 331 (LG 44), with an average of 169.74, and the length per group ranged from 44.95 cM (LG 15) to 100.34 cM (LG 20), with an average of 75.26 cM. A total of 589 shared markers (6.94%) were heterozygous in both parents, and the number of shared markers for each group ranged from six (LG 7 and LG 47) to 29 (LG 9), with an average of 11.78. A total of 607 slight segregation distortion markers (7.15%) were distributed across the linkage maps, and they mainly gathered in some LGs such as LG1, LG6, LG12, LG22, and LG50 (Table 1 and Supplemental Material, Table S1). The expected map length was calculated to be 3807.22 cM (Ge1) and 3809.784 cM (Ge2), and the average Ge of 3808.5 cM from these two estimation methods is taken as the expected genome length of crucian carp. The genome coverage of this consensus genetic map is then 98.8%.
Figure 1.
Genetic distances and marker distribution of 50 linkage groups in the consensus linkage map of crucian carp. Within each linkage group, blue, green, and red lines represent maternal heterozygous SNPs, paternal heterozygous SNPs, and SNPs heterozygous in both parents, respectively.
Table 1. Summary of statistics for the consensus linkage map, female-specific map, and male-specific map of crucian carp.
| Linkage Group | Consensus Map | Female-Specific Map | Male-Specific Map | F:M Ratio | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mapped Markers | Female-Specific Markers | Male-Specific Markers | Shared Markers | Distorted Markers | Genetic Length (cM) | Marker Interval (cM) | Mapped Markers | Distorted Markers | Genetic Length (cM) | Marker Interval (cM) | Mapped Markers | Distorted Markers | Genetic Length (cM) | Marker Interval (cM) | Map Length ratio | Recombination Rate Ratio | |
| 1 | 185 | 72 | 104 | 9 | 40 | 86.012 | 0.465 | 81 | 23 | 73.046 | 0.902 | 113 | 19 | 76.238 | 0.675 | 0.958 | 1.449 |
| 2 | 187 | 74 | 101 | 12 | 13 | 84.78 | 0.453 | 86 | 9 | 74.402 | 0.865 | 112 | 4 | 68.646 | 0.613 | 1.084 | 0.902 |
| 3 | 180 | 79 | 88 | 13 | 9 | 59.365 | 0.33 | 91 | 9 | 62.601 | 0.688 | 102 | 5 | 52.268 | 0.512 | 1.198 | 2.66 |
| 4 | 193 | 72 | 104 | 17 | 7 | 77.119 | 0.4 | 89 | 3 | 69.408 | 0.78 | 120 | 5 | 76.537 | 0.638 | 0.907 | 1.037 |
| 5 | 236 | 108 | 119 | 9 | 14 | 79.266 | 0.336 | 115 | 12 | 70.974 | 0.617 | 127 | 2 | 77.126 | 0.607 | 0.92 | 1.502 |
| 6 | 325 | 142 | 165 | 18 | 41 | 71.661 | 0.22 | 158 | 39 | 65.037 | 0.412 | 182 | 5 | 70.877 | 0.389 | 0.918 | 0.924 |
| 7 | 166 | 48 | 112 | 6 | 15 | 81.564 | 0.491 | 54 | 10 | 75.841 | 1.404 | 118 | 6 | 65.469 | 0.555 | 1.158 | 4.115 |
| 8 | 171 | 83 | 80 | 8 | 1 | 66.045 | 0.386 | 91 | 1 | 63.417 | 0.697 | 85 | 0 | 37.254 | 0.438 | 1.702 | 3.334 |
| 9 | 241 | 114 | 98 | 29 | 9 | 79.169 | 0.329 | 144 | 4 | 59.244 | 0.411 | 122 | 7 | 58.94 | 0.483 | 1.005 | 0.893 |
| 10 | 223 | 101 | 109 | 13 | 11 | 85.396 | 0.383 | 114 | 9 | 64.729 | 0.568 | 122 | 2 | 62.688 | 0.514 | 1.033 | 1.943 |
| 11 | 139 | 66 | 64 | 9 | 13 | 64.123 | 0.461 | 68 | 1 | 61.925 | 0.911 | 73 | 13 | 45.025 | 0.617 | 1.375 | 1.822 |
| 12 | 118 | 65 | 42 | 11 | 20 | 55.264 | 0.468 | 78 | 7 | 60.677 | 0.778 | 54 | 15 | 52.452 | 0.971 | 1.157 | 2.175 |
| 13 | 230 | 103 | 108 | 19 | 8 | 82.736 | 0.36 | 122 | 6 | 79.948 | 0.655 | 128 | 2 | 71.358 | 0.557 | 1.12 | 1.148 |
| 14 | 239 | 111 | 115 | 13 | 5 | 72.021 | 0.301 | 124 | 2 | 73.23 | 0.591 | 128 | 3 | 68.096 | 0.532 | 1.075 | 1.529 |
| 15 | 152 | 74 | 62 | 16 | 8 | 44.947 | 0.296 | 90 | 8 | 73.738 | 0.819 | 86 | 0 | 65.858 | 0.766 | 1.12 | 1.587 |
| 16 | 138 | 56 | 74 | 8 | 12 | 81.189 | 0.588 | 61 | 8 | 64.905 | 1.064 | 82 | 5 | 70.366 | 0.858 | 0.922 | 2 |
| 17 | 197 | 100 | 80 | 17 | 4 | 77.453 | 0.393 | 111 | 0 | 61.138 | 0.551 | 97 | 4 | 75.718 | 0.781 | 0.807 | 0.937 |
| 18 | 168 | 81 | 74 | 13 | 8 | 83.216 | 0.495 | 94 | 3 | 73.719 | 0.784 | 87 | 5 | 81.538 | 0.937 | 0.904 | 1.401 |
| 19 | 139 | 64 | 65 | 10 | 5 | 63.087 | 0.454 | 74 | 3 | 68.459 | 0.925 | 77 | 4 | 72.514 | 0.942 | 0.944 | 1.201 |
| 20 | 160 | 74 | 77 | 9 | 17 | 100.34 | 0.627 | 82 | 3 | 84.895 | 1.035 | 86 | 13 | 65.021 | 0.756 | 1.306 | 0.656 |
| 21 | 153 | 73 | 71 | 9 | 14 | 97.775 | 0.639 | 82 | 11 | 74.23 | 0.905 | 80 | 3 | 73.461 | 0.918 | 1.01 | 2.085 |
| 22 | 106 | 45 | 54 | 7 | 23 | 73.254 | 0.691 | 49 | 11 | 73.902 | 1.508 | 61 | 15 | 57.965 | 0.95 | 1.275 | 2.02 |
| 23 | 142 | 80 | 53 | 9 | 5 | 67.343 | 0.474 | 89 | 2 | 59.198 | 0.665 | 62 | 4 | 60.652 | 0.978 | 0.976 | 1.297 |
| 24 | 143 | 60 | 63 | 20 | 3 | 54.811 | 0.383 | 70 | 2 | 59.295 | 0.847 | 83 | 1 | 68.606 | 0.827 | 0.864 | 1.051 |
| 25 | 150 | 78 | 61 | 11 | 12 | 68.788 | 0.459 | 89 | 3 | 73.513 | 0.826 | 77 | 9 | 73.631 | 0.956 | 0.998 | 2.157 |
| 26 | 182 | 86 | 86 | 10 | 11 | 85.182 | 0.468 | 96 | 5 | 71.992 | 0.75 | 96 | 6 | 88.668 | 0.924 | 0.812 | 3.644 |
| 27 | 183 | 65 | 99 | 19 | 10 | 76.556 | 0.418 | 84 | 10 | 65.104 | 0.775 | 118 | 0 | 66.393 | 0.563 | 0.981 | 1.26 |
| 28 | 175 | 81 | 84 | 10 | 15 | 91.374 | 0.522 | 91 | 10 | 70.312 | 0.773 | 93 | 6 | 70.781 | 0.761 | 0.993 | 0.956 |
| 29 | 127 | 67 | 51 | 9 | 17 | 92.633 | 0.729 | 76 | 0 | 80.124 | 1.054 | 60 | 17 | 87.018 | 1.45 | 0.921 | 2.156 |
| 30 | 166 | 68 | 82 | 16 | 11 | 59.579 | 0.359 | 84 | 8 | 76.966 | 0.916 | 100 | 3 | 75.772 | 0.758 | 1.016 | 1.838 |
| 31 | 128 | 53 | 68 | 7 | 11 | 88.488 | 0.691 | 60 | 9 | 63.004 | 1.05 | 75 | 2 | 68.527 | 0.914 | 0.919 | 2.098 |
| 32 | 172 | 75 | 88 | 9 | 17 | 79.599 | 0.463 | 84 | 17 | 64.158 | 0.764 | 97 | 0 | 63.495 | 0.655 | 1.01 | 3.485 |
| 33 | 130 | 81 | 40 | 9 | 19 | 72.02 | 0.554 | 90 | 6 | 76.433 | 0.849 | 49 | 14 | 70.37 | 1.436 | 1.086 | 1.318 |
| 34 | 159 | 64 | 84 | 11 | 7 | 72.538 | 0.456 | 75 | 4 | 67.778 | 0.904 | 94 | 3 | 53.714 | 0.571 | 1.262 | 1.399 |
| 35 | 135 | 57 | 70 | 8 | 17 | 75.293 | 0.558 | 65 | 10 | 73.712 | 1.134 | 77 | 7 | 61.64 | 0.801 | 1.196 | 1.242 |
| 36 | 161 | 81 | 66 | 14 | 5 | 76.374 | 0.474 | 92 | 4 | 65.641 | 0.713 | 80 | 1 | 60.095 | 0.751 | 1.092 | 1.321 |
| 37 | 193 | 98 | 84 | 11 | 6 | 76.941 | 0.399 | 109 | 4 | 83.944 | 0.77 | 95 | 2 | 73.089 | 0.769 | 1.149 | 3.22 |
| 38 | 167 | 83 | 67 | 17 | 6 | 78.142 | 0.468 | 100 | 4 | 68.303 | 0.683 | 84 | 2 | 68.659 | 0.817 | 0.995 | 1.641 |
| 39 | 155 | 64 | 78 | 13 | 10 | 75.382 | 0.486 | 70 | 4 | 63.262 | 0.904 | 91 | 7 | 69.754 | 0.767 | 0.907 | 1.172 |
| 40 | 156 | 71 | 73 | 12 | 18 | 73.448 | 0.471 | 83 | 3 | 71.437 | 0.861 | 83 | 16 | 62.339 | 0.751 | 1.146 | 1.282 |
| 41 | 165 | 83 | 73 | 9 | 6 | 69.916 | 0.424 | 92 | 6 | 67.234 | 0.731 | 81 | 0 | 59.7 | 0.737 | 1.126 | 1.648 |
| 42 | 143 | 55 | 75 | 13 | 9 | 72.483 | 0.507 | 68 | 7 | 74.443 | 1.095 | 88 | 4 | 67.966 | 0.772 | 1.095 | 1.444 |
| 43 | 160 | 76 | 75 | 9 | 5 | 72.5 | 0.453 | 85 | 4 | 68.6 | 0.807 | 83 | 1 | 39.898 | 0.481 | 1.719 | 2.742 |
| 44 | 331 | 154 | 162 | 15 | 7 | 80.236 | 0.242 | 169 | 3 | 69.803 | 0.413 | 177 | 5 | 79.798 | 0.451 | 0.875 | 1.058 |
| 45 | 179 | 77 | 94 | 8 | 10 | 81.51 | 0.455 | 85 | 7 | 71.925 | 0.846 | 102 | 3 | 74.455 | 0.73 | 0.966 | 1.515 |
| 46 | 155 | 64 | 80 | 11 | 7 | 74.091 | 0.478 | 75 | 4 | 71.991 | 0.96 | 90 | 4 | 70.867 | 0.787 | 1.016 | 0.903 |
| 47 | 123 | 54 | 63 | 6 | 15 | 79.269 | 0.644 | 60 | 8 | 71.565 | 1.193 | 69 | 7 | 66.677 | 0.966 | 1.073 | 13.759 |
| 48 | 139 | 62 | 67 | 10 | 13 | 74.401 | 0.535 | 72 | 8 | 78.436 | 1.089 | 77 | 5 | 63.714 | 0.827 | 1.231 | 1.71 |
| 49 | 116 | 60 | 45 | 11 | 15 | 60.238 | 0.519 | 72 | 3 | 79.882 | 1.109 | 57 | 13 | 66.548 | 1.168 | 1.2 | 9.166 |
| 50 | 106 | 61 | 37 | 8 | 23 | 67.962 | 0.641 | 67 | 12 | 62.9 | 0.939 | 45 | 13 | 68.084 | 1.513 | 0.924 | 2.561 |
| Total | 8487 | 3863 | 4034 | 590 | 607 | 3762.879 | 0.443 | 4410 | 349 | 3500.42 | 0.794 | 4625 | 292 | 3346.325 | 0.724 | 1.046 | 2.127 |
The female map comprised 4410 SNPs and spanned 3500.42 cM, with an average marker interval of 0.79 cM (Table 1 and Figure S1). The number of markers per group varied from 49 (LG 22) to 169 (LG 44), with an average of 88.20, and the length ranged from 59.20 cM (LG 23) to 84.90 cM (LG 20), with an average of 70.01 cM. While the male map consisted of 4625 SNPs, and spanned 3346.33 cM with an average marker interval of 0.72 cM (Table 1 and Figure S2). The number of markers in each group varied from 45 (LG 50) to 182 (LG 44) with an average of 92.50, and the lengths of the LGs ranged from 37.25 cM (LG 8) to 83.75 cM (LG 26), with an average of 66.93 cM. The female and male maps have 349 (7.9%) and 292 (6.3%) distorted markers, respectively.
Marker orders are largely conserved between female map and male map, although some LGs showed minor intrachromosomal rearrangements (Figure S3). The average recombination ratio of female-to-male is 2.13:1 (Table 1). Of the 50 LGs, 43 showed higher recombination rate in males than females, and, in contrast, seven LGs showed lower recombination rate in males than females. Interestingly, significant differences in recombination ratios between the female and male maps were observed in LG47 (13.76:1) and LG49 (9.17:1) (Table 1 and Figure S3).
Comparative genome analysis
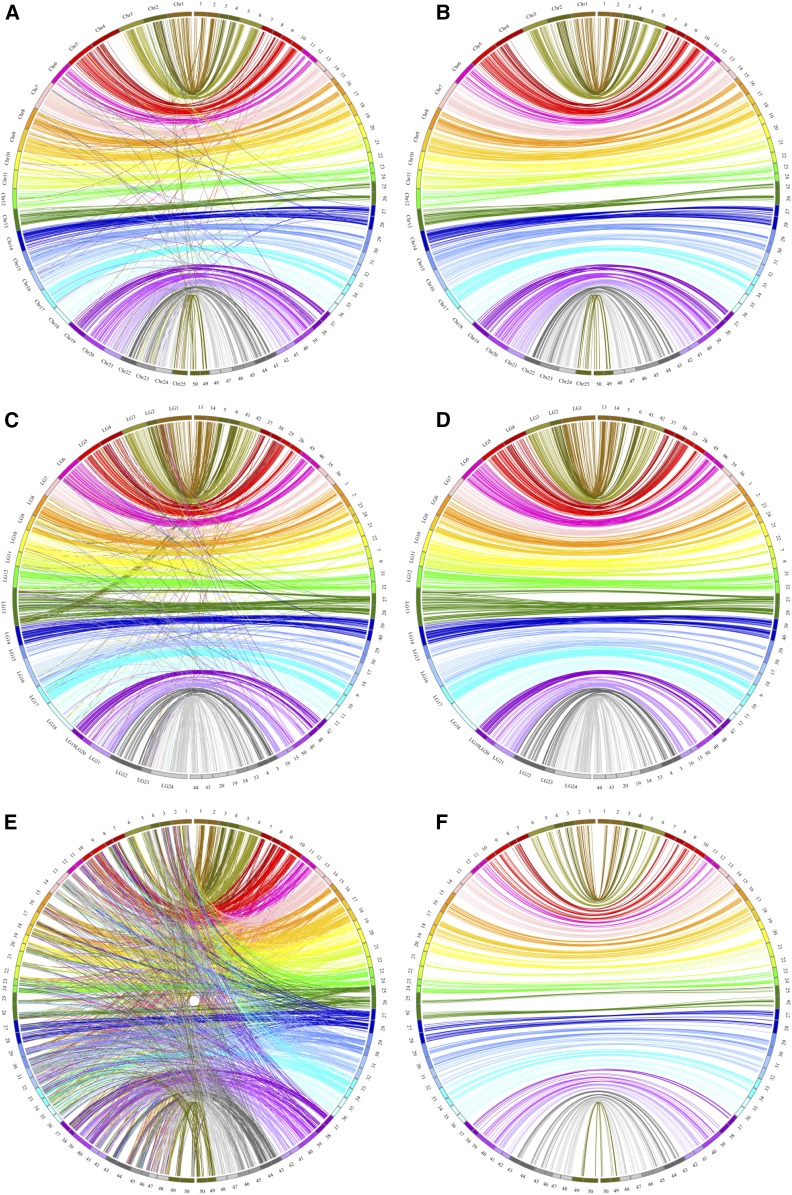
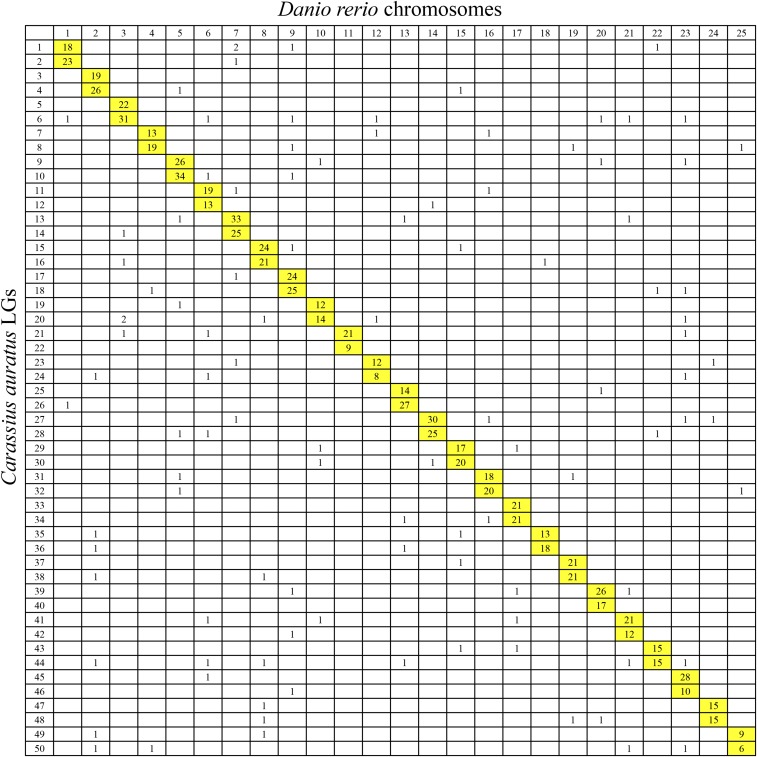
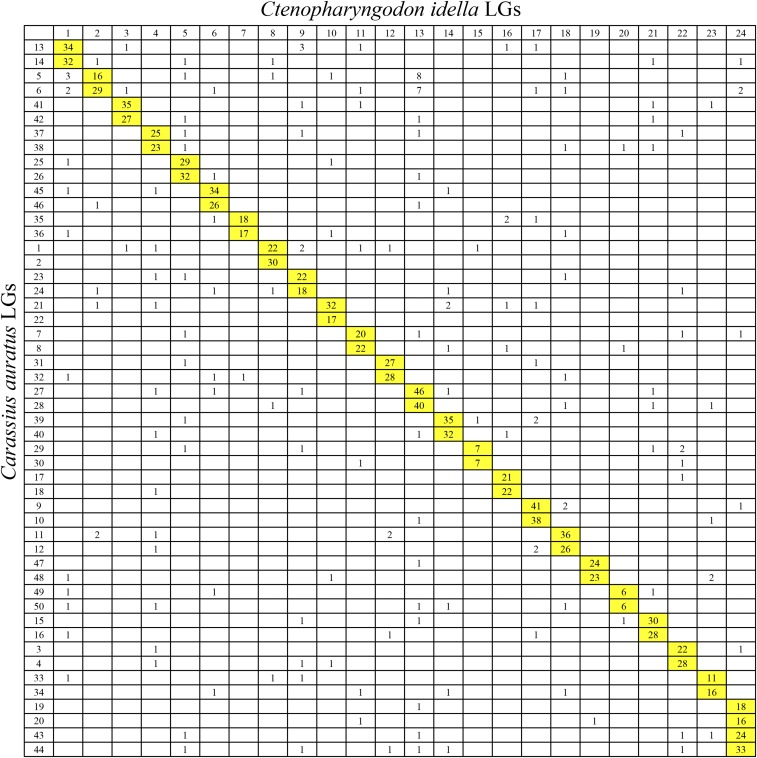
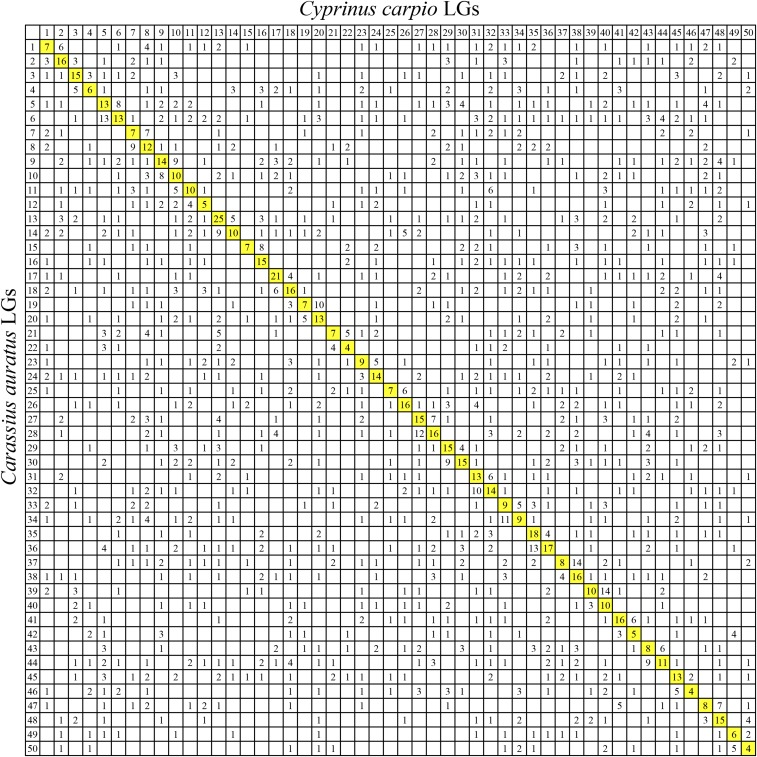
Comparative genomic analysis was performed between crucian carp LGs and zebrafish, grass carp, and common carp chromosomes. For 5734 extended sequences of SNP markers, a total of 1072 markers was uniquely aligned to the genome of zebrafish. The results showed that two LGs of crucian carp were homologous to one particular chromosome of zebrafish, suggesting a clear 2:1 relationship of crucian carp LGs and zebrafish chromosomes (Figure 2A and Figure 3). Of the 1072 orthologous pairs, 966 pairs (90.1%) were located into the conserved syntenic blocks, revealing highly conserved synteny between crucian carp and zebrafish genome (Figure 2B). Of note, 49% regions (37–75 Mb) on chromosome 4 of zebrafish had no homologous segments in crucian carp (Figure 2, A and B). A total of 1441 orthologous pairs between crucian carp and grass carp were identified, with 1267 (87.9%) mapped on those paired collinear blocks (Figure 2C). Comparison analysis showed a 2:1 synteny between crucian carp and grass carp LGs, except for LG 24 in grass carp, which was aligned to four LGs in crucian carp (LG19, LG20, LG43, and LG44) (Figure 2D and Figure 4). The comparative genomics results show that intensive chromosomal rearrangements were present between crucian carp and common carp (Figure 2E and Figure 5). Of the 2201 markers uniquely anchored to the chromosomes of common carp, only 574 (26.1%) 1:1 orthologous pairs were identified (Figure 2F).
Figure 2.
Circos diagram representing syntenic relationships between crucian carp and zebrafish (A and B), grass carp (C and D), and common carp (E and F). (B, D, and F) show only 1:2, 1:4 or 1:1 perfect orthologous pairs.
Figure 3.
Genomic synteny visualized using Oxford grids between crucian carp linkage groups and zebrafish chromosomes. The numbers in each cell represent number of homologous loci between crucian carp consensus linkage groups and zebrafish chromosomes. Homologous linkage groups and chromosomes are highlighted in yellow.
Figure 4.
Genomic synteny between crucian carp linkage groups and grass carp linkage groups visualized using Oxford grids. The numbers in each cell represent number of homologous loci between crucian carp consensus linkage groups and grass carp linkage groups. Homologous linkage groups are highlighted in yellow.
Figure 5.
Genomic synteny between crucian carp linkage groups and common carp chromosomes visualized using Oxford grids. The numbers in each cell represent number of homologous loci between crucian carp consensus linkage groups and common carp chromosomes. Homologous linkage groups and chromosomes are highlighted in yellow.
QTL for body weight
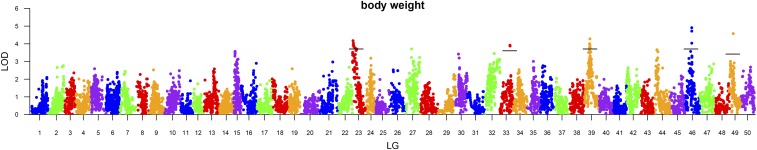
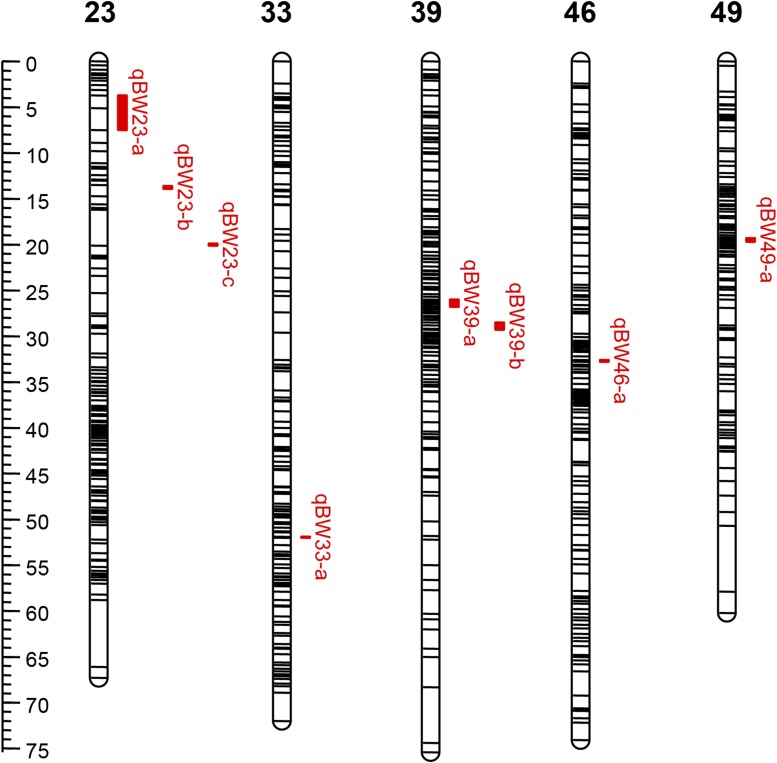
QTL fine mapping based on above high-density genetic linkage map showed that eight chromosome-wide QTL associated with body weight were identified. These QTL distribute on five LGs (LG23, LG33, LG39, LG46, and LG49), with LOD scores from 3.71 to 4.91, and the phenotypic variance explained (PVE) ranging from 10.1 to 13.2% (Figure 6, Figure 7, and Table 2). The highest PVE value was detected for qBW46-a on LG46 with the LG positions of 32.579–32.808 cM (Table 2). All QTL intervals in this study are <1 cM except qBW23-a on LG23 (3.765 cM), with an overall average 0.87 cM for eight QTL intervals. Each QTL interval harbors 1–3 SNP markers (Table 2).
Figure 6.
A genome scan of LOD profiles for body weight. The solid lines indicate the chromosome-wide significance thresholds.
Figure 7.
The distribution of eight QTL for body weight on five genetic linkage groups of crucian carp.
Table 2. Summary statistics of the QTL for body weight and uncovered candidate genes in crucian carp.
| QTL name | LG | Position (cM) | No. of SNPs | LOD | Chromosome -Wide Threshold | PVE (%) | Nearest Marker | Candidate Genes |
|---|---|---|---|---|---|---|---|---|
| qBW23-a | 23 | 3.694–7.459 | 3 | 4.17 | 3.6 | 11.3 | ref-33789_15 | Epidermal growth factor-like domain |
| qBW23-b | 23 | 13.039–13.467 | 1 | 3.77 | 3.6 | 10.3 | ref-102483_29 | Immunoglobulin-like |
| qBW23-c | 23 | 19.228–20.054 | 1 | 3.71 | 3.6 | 10.1 | ref-28586 | |
| qBW33-a | 33 | 51.928–52.005 | 2 | 3.92 | 3.6 | 10.7 | ref-68343_4 | Zinc finger, C2H2 |
| qBW39-a | 39 | 26.023–26.755 | 2 | 4 | 3.7 | 10.9 | ref-123717 | |
| qBW39-b | 39 | 28.471–29.264 | 3 | 4.28 | 3.7 | 11.6 | ref-96199 | Transforming growth factor β |
| qBW46-a | 46 | 32.579–32.808 | 3 | 4.91 | 3.7 | 13.2 | ref-38422_10 | Protein kinase, ATP binding site |
| qBW49-a | 49 | 19.534–19.65 | 1 | 4.58 | 3.4 | 12.3 | ref-103659 |
Potential candidate genes
BLASTN searches of QTL-associated SNP sequences against the genome sequences of crucian carp and common carp gave five potential candidate genes (Table 2). Markers in three QTL (qBW23-c, qBW39-a, and qBW49-a) failed to reveal any potential candidate genes because they were unable to align to any of the two genomes. Among those annotated genes with known biological functions (Table 2), epidermal growth factor-like domain (EGF-like domain) and transforming growth factor β (TGF-β) are two representatives.
Discussion
With the development of high-throughput sequencing technology and the advancement of GBS methods, constructing well-defined genetic linkage maps using thousands of SNP markers is now available in many nonmodel organisms, including aquaculture species. High-density SNP linkage maps have been constructed using 2b-RAD sequencing in several aquaculture animals, including Chlamys farreri (3806 SNPs) (Jiao et al. 2014), pearl oyster (3117 SNPs) (Shi et al. 2014), Chinese mitten crab (10,358 SNPs) (Cui et al. 2015), sea cucumber (7839 SNPs) (Tian et al. 2015), bighead carp (3121 SNPs) (Fu et al. 2016), and gilthead sea bream (Palaiokostas et al. 2016) (12,085 SNPs). The number of markers on a linkage map is usually determined by the choice of restriction enzymes, the number of restriction enzymes, the total number of enzyme cut sites, and the polymorphism rate across the genome (Andrews et al. 2016). In this work, taking advantage of 2b-RAD technology, we constructed a high-density linkage map containing 8487 SNP markers with a resolution of 0.44 cM for crucian carp. This robust genetic linkage map will contribute to a better understanding of the genome structure, function, and evolution of crucian carp. In addition, the linkage map will be highly valuable to fine mapping for more complex traits and chromosome assembly of WGS in crucian carp.
Sex differences in recombination rates have been reported in a number of teleosts (Zhu et al. 2014). In many fishes, females usually have a higher recombination frequency than males (Liu et al. 2013). Recombination ratio differences between female and male have been observed in zebrafish (2.74:1) (Singer et al. 2002), rainbow trout (1.68:1 and 3.25:1) (Rexroad et al. 2008; Sakamoto et al. 2000), Atlantic halibut (1.89–2.53:1) (Reid et al. 2007), turbot (1.6:1) (Bouza et al. 2007), barramundi (2.06:1) (Wang et al. 2007), and grass carp (2.03:1) (Xia et al. 2010). In this study, the average female to male recombination ratio of crucian carp was 2.13:1, which was similar to many other fish species. In the majority of female LGs, the recombination ratios were obviously higher than those of male LGs. However, significant male-biased recombination suppression was observed in LG47 and LG49. This is a very interesting phenomenon worthy of attention. Previous studies have suggested many factors, such as sex chromosomes, regions around centromeres and/or telomeres, large areas of repetitive DNA, and heterochromatin could influence recombination rate (Ninwichian et al. 2012; Jones et al. 2013). Recombination suppression between sex chromosomes is a common phenomenon in vertebrates, and is important in maintaining the stability of the sex-determining regions and leads to the degeneration of Y or W chromosomes (Charlesworth et al. 2005; Bergero and Charlesworth 2009). In three spine sticklebacks and medaka, recombination in the sex determination region is reduced in the male linkage map relative to the female linkage map (Peichel et al. 2004; Roesti et al. 2013; Matsuda et al. 1999; Naruse et al. 2000). It was reported that crucian carp has an XX/XY sex chromosome system (Yamamoto 1974), and has a pair of heteromorphic chromosomes that were taken as X and Y chromosomes (Ruiguang 1982). In our study, significant male-biased recombination suppression on LG47 and LG49 may suggest that these two LGs are potential sex chromosomes of the crucian carp genome. Further studies are required to confirm this hypothesis, and elucidate the genetic mechanism for sex determination in crucian carp and other cyprinid fishes.
A high-density genetic map constructed with sequence-based markers makes it possible for genome evolution studies in nonmodel species (Z. Wang et al. 2015; H. Liu et al. 2016). In this study, a detailed syntenic relationship was established between crucian carp LGs and zebrafish, grass carp, and common carp genomes via genetic maps and assembled genomes. In zebrafish, all chromosomal regions were covered by homologous loci of crucian carp except for half of Chromosome 4. The annotation information of zebrafish reference genome sequence indicated that this unique region harbored high gene duplication, high density of small nuclear RNAs (snRNAs), and may be related to sex chromosomes (Howe et al. 2013). This unique region presented obscure synteny with human, mouse, chicken, and common carp genomes (Howe et al. 2013; Xu et al. 2014), and our results were in accordance with those reports. The results of comparative mapping with zebrafish genome demonstrated a high level integrity of our linkage map. The conserved synteny with a clear 2:1 relationship between crucian carp LGs and zebrafish chromosomes was similar to that observed between common carp and goldfish (Peng et al. 2016; Xu et al. 2014; Zhang et al. 2013; Zhao et al. 2013; Kuang et al. 2016). In addition, a number of minor chromosome rearrangements were detected between crucian carp and zebrafish, which was similar to that reported in common carp (Xu et al. 2014). These findings suggested that crucian carp, similar to common carp, had undergone the 4R-WGD. Similarly, a clear 2:1 relationship was also observed between LGs of crucian carp and grass carp, except for LG24 of grass carp, which showed synteny to LG19, LG20, LG43, and LG44 of crucian carp. Previous studies indicated that LG24 of grass carp was orthologous to chromosome 10 and chromosome 22 of zebrafish (Z. Wang et al. 2015), so, after the 4R-WGD, grass carp LG24 is syntenic to four LGs of crucian carp.
Comparative analysis between crucian carp LGs, and common carp chromosomes demonstrated a 1:1 relationship, but with extensive chromosomal rearrangements. Crucian carp and common carp are two closely related species in the Cyprinidae, as they have the same number of chromosomes (2n = 100) and can produce hybrid offspring (Hulata 1995; Liu et al. 2004; S.J. Liu et al. 2001, 2016). Previous studies indicated that the common ancestor of crucian carp and common carp experienced the 4R-WGD ∼10.9–13.2 MYA, and the speciation occurred ranging from 8.1 to 12.9 MYA (Yuan et al. 2010; David et al. 2003; Chistiakov and Voronova 2009). A recent study confirmed that the 4R-WGD happened 8.2 MYA in common carp (Xu et al. 2014). Therefore, it was believed that crucian carp and common carp diverged from each other after the 4R-WGD event (Somamoto et al. 2005; Chistiakov and Voronova 2009). Then, the duplicated genes faced different destinations, such as nonfunctionalization, subfunctionalization, neofunctionalization, and gene dosage effects, which is very important for biological evolution, adaptation, speciation, and diversification (Glasauer and Neuhauss 2014; Lien et al. 2016). In this case, it can be speculated that chromosome structural difference between crucian carp and common carp could have occurred since they evolved in different directions. A similar genome structure has been seen in salmonids, in which common ancestor undergone the 4R-WGD 80 MYA, and then the genome experienced extensive chromosomal rearrangements (Berthelot et al. 2014; Lien et al. 2016; Brieuc et al. 2014; Guyomard et al. 2012; Gharbi et al. 2006; Naish et al. 2013; Timusk et al. 2011). Furthermore, the rediploidization process followed by the 4R-WGD would also resulted in significant genome rearrangements (Lien et al. 2016). Therefore, large genomic reorganizations between crucian carp and common carp may be due to the independent genome evolution and rediploidization process after genome duplication.
Growth is an important trait of interest in aquaculture species, and great efforts have been devoted to promoting the growth rate of crucian carp (Liu et al. 2001; Gui and Zhou 2010; Zhou et al. 2000a,b, 2001; Liu et al. 2004). However, for genetic improvement of quantitative traits, traditional breeding methods have encountered some bottlenecks and problems (Mackay et al. 2009). QTL fine mapping and positional cloning of candidate genes may have been an efficient approach for breeding programs in aquaculture animals, especially for quantitative traits (Peng et al. 2016; Yu et al. 2015; Xiao et al. 2015; Z. Wang et al. 2015; Tian et al. 2015; Shao et al. 2015; Li et al. 2015; Shi et al. 2014; H. Liu et al. 2016). In the present study, the high-density linkage map allowed us to perform QTL fine mapping for body weight, and discover potential candidate genes for early growth stage in crucian carp. As a result, eight chromosome-wide QTL associated with body weight were located in five different LGs, which reflected the complexity of this polygenic trait. However, it is worth noting that the average map length of eight QTL was 0.87 cM, and this relatively narrow genomic region would facilitate further validating and positional cloning of potential major genes for growth in crucian carp. Among five potential candidate genes identified by genomic synteny analysis, EGF-like domain and TGF-beta may be most promising because their biological functions are likely associated with early growth and development in vertebrates. For example, EGF regulates cell proliferation and growth in human, which play an important role in cellular organization and membrane repair when the tissue is destroyed (Engel 1989). TGF-β modulates cell proliferation, differentiation, apoptosis, and immune regulation (Sporn et al. 1986). Our QTL results and uncovered candidate growth genes would lay a foundation for genetic improvement for growth in crucian carp. Nevertheless, we must recognize that our QTL results could suffer from the limitation of the use of a single family, which may have higher power to detect family-specific or rare QTL, but could also trade off the detection power of common QTL shared across families. In the future, joint multiple population analysis will be needed to detect QTL shared among multiple families with a wider scope of inference. In addition, taking into account the Beavis effect, QTL effects tend to be overestimated as sample size is relatively small, and the genetic architecture of the character highly polygenic, as is the case with body weight.
Conclusion
In summary, a high-density linkage map of crucian carp was constructed by 2b-RAD method with 8487 SNPs (0.44 cM/marker). Comparative genomics among four cyprinid fishes (crucian carp, zebrafish, grass carp, and common carp) provides new insights into genome duplication and chromosomal rearrangements in crucian carp. Eight chromosome-wide QTL for body weight were detected at quite narrow regions of five LGs with the PVE values of 10.1–13.2%. A few potential candidate genes, e.g., EGF-like domain and TGF-β, whose biological functions are likely involved in genetic regulation of early growth, were identified from those QTL intervals. Our present study provides valuable genetic and genomic resources for further studies of comparative genomics, genome evolution, chromosome assembly, and QTL fine mapping in crucian carp.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.041376/-/DC1.
Acknowledgments
This research was supported by the Chinese Academy of Sciences (XDA08010405) and the State Key Laboratory of Freshwater Ecology and Biotechnology (2016FBZ05).
Author contributions: J.T. conceived and guided this research. H.L. carried out the experiments, performed data analysis, and wrote the manuscript. B.F. performed bioinformatics analysis. M.P., X.F. and X.Y. were involved in sample preparation and experiments. All authors read and approved the final manuscript.
Footnotes
Communicating editor: R. Houston
Literature Cited
- Allendorf F. W., Thorgaard G. H., 1984. Tetraploidy and the evolution of salmonid fishes, pp. 1–53 in Evolutionary Genetics of Fishes. Plenum Press, New York, NY. [Google Scholar]
- Andrews K. R., Good J. M., Miller M. R., Luikart G., Hohenlohe P. A., 2016. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 17: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102. [DOI] [PubMed] [Google Scholar]
- Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., et al. , 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5: 3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouza C., Hermida M., Pardo B. G., Fernandez C., Fortes G. G., et al. , 2007. A microsatellite genetic map of the turbot (Scophthalmus maximus). Genetics 177: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieuc M. S. O., Waters C. D., Seeb J. E., Naish K. A., 2014. A dense linkage map for chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 Bethesda 4: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B. M., Onusko S. W., Gross J. B., 2015. A high-density linkage map for Astyanax mexicanus using genotyping-by-sequencing technology. G3 Bethesda 5: 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., Lasher L. K., Reefer J. E., 1991. A maximum-likelihood method for estimating genome length using genetic-linkage data. Genetics 128: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Chen Q., Ma Y., Yang Y., Chen Z., Liao R., et al. , 2013. Genotyping by genome reducing and sequencing for outbred animals. PLoS One 8: e67500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov D. A., Voronova N. V., 2009. Genetic evolution and diversity of common carp Cyprinus carpio L. Cent. Eur. J. Biol. 4: 304–312. [Google Scholar]
- Cui Z., Hui M., Liu Y., Song C., Li X., et al. , 2015. High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann R. G., Davidson E. A., Ferguson M. M., Gharbi K., Koop B. F., et al. , 2008. Distribution of ancestral proto-actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon). BMC Genomics 9: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., et al. , 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12: 499–510. [DOI] [PubMed] [Google Scholar]
- David L., Blum S., Feldman M. W., Lavi U., Hillel J., 2003. Recent duplication of the, common carp (Cyprinus carpio L.) genome as revealed by analyses of microsatellite loci. Mol. Biol. Evol. 20: 1425–1434. [DOI] [PubMed] [Google Scholar]
- Davidson W. S., Koop B. F., Jones S. J., Iturra P., Vidal R., et al. , 2010. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 11: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J., Li X., Fu Q., Jiao W., Li Y., et al. , 2016. Evaluation of the 2b-RAD method for genomic selection in scallop breeding. Sci. Rep. 6: 19244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., 1989. EGF‐like domains in extracellular matrix proteins: localized signals for growth and differentiation? FEBS Lett. 251: 1–7. [DOI] [PubMed] [Google Scholar]
- FAO, 2016 The State of World Fisheries and Aquaculture 2016: Contributing to Food Security and Nutrition for All. Food and Agriculture Organization of the United Nations, Rome. [Google Scholar]
- Feng X., Wang X. H., Yu X. M., Zhang X. F., Lu C. Y., et al. , 2015. Microsatellite-centromere mapping in common carp through half-tetrad analysis in diploid meiogynogenetic families. Chromosoma 124: 67–79. [DOI] [PubMed] [Google Scholar]
- Ferris S. D., Whitt G. S., 1977. Duplicate gene-expression in diploid and tetraploid loaches (Cypriniformes, Cobitidae). Biochem Genet. 15: 1097–1112. [DOI] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B., Liu H., Yu X., Tong J., 2016. A high-density genetic map and growth related QTL mapping in bighead carp (Hypophthalmichthys nobilis). Sci. Rep. 6: 28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Dou J., Mao J., Su H., Jiao W., et al. , 2013. RADtyping: an integrated package for accurate de novo codominant and dominant RAD genotyping in mapping populations. PLoS One 8: e79960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi K., Gautier A., Danzmann R. G., Gharbi S., Sakamoto T., et al. , 2006. A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172: 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer S. M., Neuhauss S. C., 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289: 1045–1060. [DOI] [PubMed] [Google Scholar]
- Gui J., Zhou L., 2010. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci. China Life Sci. 53: 409–415. [DOI] [PubMed] [Google Scholar]
- Guo W. J., Tong J. G., Yu X. M., Zhu C. K., Feng X., et al. , 2013. A second generation genetic linkage map for silver carp (Hypophthalmichehys molitrix) using microsatellite markers. Aquaculture 412: 97–106. [Google Scholar]
- Guyomard R., Boussaha M., Krieg F., Hervet C., Quillet E., 2012. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 13: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel C. V., Dirks R. P., Jansen H. J., Forlenza M., Wiegertjes G. F., et al. , 2012. Comparison of the exomes of common carp (Cyprinus carpio) and zebrafish (Danio rerio). Zebrafish 9: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., et al. , 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulata G., 1995. A review of genetic improvement of the common carp (Cyprinus carpio L.) and other cyprinids by crossbreeding, hybridization and selection. Aquaculture 129: 143–155. [Google Scholar]
- Jaillon O., Aury J.-M., Brunet F., Petit J.-L., Stange-Thomann N., et al. , 2004. Genome duplication in the teleost fish tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. [DOI] [PubMed] [Google Scholar]
- Jiao W. Q., Fu X. T., Dou J. Z., Li H. D., Su H. L., et al. , 2014. High-resolution linkage and quantitative trait locus mapping aided by genome survey sequencing: building up an integrative genomic framework for a bivalve mollusc. DNA Res. 21: 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. B., Jerry D. R., Khatkar M. S., Raadsma H. W., Zenger K. R., 2013. A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14: 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa T., Watanabe M., Kitagawa E., Yoshioka M., Kashiwagi M., et al. , 2003. Phylogeography and the maternal origin of the tetraploid form of the Japanese spined loach, Cobitis biwae, revealed by mitochondrial DNA analysis. Ichthyol. Res. 50: 318–325. [Google Scholar]
- Knapik E. W., Goodman A., Ekker M., Chevrette M., Delgado J., et al. , 1998. A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. 18: 338–343. [DOI] [PubMed] [Google Scholar]
- Knytl M., Kalous L., Rab P., 2013. Karyotype and chromosome banding of endangered crucian carp, Carassius (Linnaeus, 1758) (Teleostei, Cyprinidae). Comp. Cytogenet. 7: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayasi H., Kawashima Y., Takeuchi N., 1970. Comparative chromosome studies in the genus Carassius, especially with a finding of polyploidy in the ginbuna (C. auratus langsdorfii). Jpn. J. Ichthyol. 17: 153–160. [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y. Y., Zheng X. H., Li C. Y., Li X. M., Cao D. C., et al. , 2016. The genetic map of goldfish (Carassius auratus) provided insights to the divergent genome evolutions in the Cyprinidae family. Sci. Rep. 6: 34849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunoki T., Arai K., Suzuki R., 1994. Production of viable gynogens without chromosome duplication in the spinous loach cobitis-biwae. Aquaculture 119: 11–23. [Google Scholar]
- Larhammar D., Risinger C., 1994. Molecular-genetic aspects of tetraploidy in the common carp Cyprinus carpio. Mol. Phylogenet. Evol. 3: 59–68. [DOI] [PubMed] [Google Scholar]
- Li Y., Liu S. K., Qin Z. K., Waldbieser G., Wang R. J., et al. , 2015. Construction of a high-density, high-resolution genetic map and its integration with BAC-based physical map in channel catfish. DNA Res. 22: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. L., Cheng L., Xu P., Lu G. Q., Wachholtz M., et al. , 2013. Transcriptome analysis of Crucian carp (Carassius auratus), an important aquaculture and hypoxia-tolerant species. PLoS One 8: e62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Koop B. F., Sandve S. R., Miller J. R., Kent M. P., et al. , 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Sun F., Li J., Xia J. H., Lin G., et al. , 2013. A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus x Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 14: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu B., Pang M., Feng X., Wang X., et al. , 2016. QTL fine mapping and identification of candidate genes for growth-related traits in bighead carp (Hypophthalmichehys nobilis). Aquaculture 465: 134–143. [Google Scholar]
- Liu S., Sun Y., Zhang C., Luo K., Liu Y., 2004. Production of gynogenetic progeny from allotetraploid hybrids red crucian carp × common carp. Aquaculture 236: 193–200. [Google Scholar]
- Liu S. J., Liu Y., Zhou G. J., Zhang X. J., Luo C., et al. , 2001. The formation of tetraploid stocks of red crucian carp × common carp hybrids as an effect of interspecific hybridization. Aquaculture 192: 171–186. [Google Scholar]
- Liu S. J., Luo J., Chai J., Ren L., Zhou Y., et al. , 2016. Genomic incompatibilities in the diploid and tetraploid offspring of the goldfish × common carp cross. Proc. Natl. Acad. Sci. USA 113: 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Cordes J., 2004. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238: 1–37. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA. [Google Scholar]
- Mackay T. F. C., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Sotoyama S., Hamaguchi S., Sakaizumi M., 1999. Male-specific restriction of recombination frequency in the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 73: 225–231. [Google Scholar]
- Meyer A., Van de Peer Y., 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27: 937–945. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Naish K. A., Phillips R. B., Brieuc M. S. O., Newton L. R., Elz A. E., et al. , 2013. Comparative genome mapping between chinook salmon (Oncorhynchus tshawytscha) and rainbow trout (O. mykiss) based on homologous microsatellite loci. G3 Bethesda 3: 2281–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K., Fukamachi S., Mitani H., Kondo M., Matsuoka T., et al. , 2000. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics 154: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninwichian P., Peatman E., Liu H., Kucuktas H., Somridhivej B., et al. , 2012. Second-generation genetic linkage map of catfish and its integration with the BAC-based physical map. G3 Bethesda 2: 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Muramoto J., Christian L., Atkin N. B., 1967. Diploid-tetraploid relationship among old-world members of the fish family Cyprinidae. Chromosoma 23: 1–9. [Google Scholar]
- Ojima Y., Ueda T., Narikawa T., 1979. A cytogenetic assessment on the origin of the gold-fish. Proc. Jpn. Acad., Ser. B 55(2): 58–63. [Google Scholar]
- Palaiokostas C., Ferraresso S., Franch R., Houston R. D., Bargelloni L., 2016. Genomic prediction of resistance to pasteurellosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. G3 Bethesda 6: 3693–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J., et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Peng W., Xu J., Zhang Y., Feng J., Dong C., et al. , 2016. An ultra-high density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci. Rep. 6: 26693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. K., Weber J. N., Kay E. H., Fisher H. S., Hoekstra H. E., 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V., Venkatesh B., 2008. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 18: 544–550. [DOI] [PubMed] [Google Scholar]
- Reid D. P., Smith C. A., Rommens M., Blanchard B., Martin-Robichaud D., et al. , 2007. A genetic linkage map of Atlantic halibut (Hippoglossus hippoglossus L.). Genetics 177: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad C. E., Palti Y., Gahr S. A., Vallejo R. L., 2008. A second generation genetic map for rainbow trout (Oncorhynchus mykiss). BMC Genet. 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risinger C., Larhammar D., 1993. Multiple loci for synapse protein snap-25 in the tetraploid goldfish. Proc. Natl. Acad. Sci. USA 90: 10598–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesti M., Moser D., Berner D., 2013. Recombination in the threespine stickleback genome—patterns and consequences. Mol. Ecol. 22: 3014–3027. [DOI] [PubMed] [Google Scholar]
- Ruiguang Z., 1982. Studies of sex chromosomes and C-banding karyotypes of two forms of Carassius auratus in Kunming Lake. Acta Genetica Sinica 9: 32–39. [Google Scholar]
- Sakamoto T., Danzmann R. G., Gharbi K., Howard P., Ozaki A., et al. , 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D., 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shao C., Niu Y., Rastas P., Liu Y., Xie Z., et al. , 2015. Genome-wide SNP identification for the construction of a high-resolution genetic map of Japanese flounder (Paralichthys olivaceus): applications to QTL mapping of Vibrio anguillarum disease resistance and comparative genomic analysis. DNA Res. 22: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. H., Wang S., Gu Z. F., Lv J., Zhan X., et al. , 2014. High-density single nucleotide polymorphisms linkage and quantitative trait locus mapping of the pearl oyster, Pinctada fucata martensii dunker. Aquaculture 434: 376–384. [Google Scholar]
- Shifman S., Bell J. T., Copley R. R., Taylor M. S., Williams R. W., et al. , 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4: 2227–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Perlman H., Yan Y. L., Walker C., Corley-Smith G., et al. , 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somamoto T., Yoshiura Y., Nakanishi T., Ototake M., 2005. Molecular cloning and characterization of two types of CD8 alpha from ginbuna crucian carp, Carassius auratus langsdorfii. Dev. Comp. Immunol. 29: 693–702. [DOI] [PubMed] [Google Scholar]
- Sporn M. B., Roberts A. B., Wakefield L. M., Assoian R. K., 1986. Transforming growth-factor-beta—biological function and chemical-structure. Science 233: 532–534. [DOI] [PubMed] [Google Scholar]
- Sun X. W., Liu D. Y., Zhang X. F., Li W. B., Liu H., et al. , 2013. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8: e58700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Braasch I., Frickey T., Meyer A., Van de Peer Y., 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 13: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M. L., Li Y. P., Jing J., Mu C., Du H. X., et al. , 2015. Construction of a high-density genetic map and quantitative trait locus mapping in the sea cucumber Apostichopus japonicus. Sci. Rep. 5: 14852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timusk E. R., Ferguson M. M., Moghadam H. K., Norman J. D., Wilson C. C., et al. , 2011. Genome evolution in the fish family salmonidae: generation of a brook charr genetic map and comparisons among charrs (Arctic charr and brook charr) with rainbow trout. BMC Genet. 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J. G., Sun X. W., 2015. Genetic and genomic analyses for economically important traits and their applications in molecular breeding of cultured fish. Sci. China Life Sci. 58: 178–186. [DOI] [PubMed] [Google Scholar]
- Tsai H. Y., Robledo D., Lowe N. R., Bekaert M., Taggart J. B., et al. , 2016. Construction and annotation of a high density SNP linkage map of the Atlantic salmon (Salmo salar) genome. G3 6: 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeno T., Smith G. R., 1972. Tetraploid origin of the karyotype of catostomid fishes. Science 175: 644–646. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J., 2006. JoinMap 4. Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Van Ooijen J., Kyazma B., 2009. MapQTL 6. Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species, Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Venkatesh B., 2003. Evolution and diversity of fish genomes. Curr. Opin. Genet. Dev. 13: 588–592. [DOI] [PubMed] [Google Scholar]
- Voorrips R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Wang C. M., Zhu Z. Y., Lo L. C., Feng F., Lin G., et al. , 2007. A microsatellite linkage map of barramundi, lates calcarifer. Genetics 175: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P., Li L., Zhang G. F., 2016. A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas × Crassostrea angulata) using genotyping-by-sequencing. G3 Bethesda 6: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. T., Li J. T., Zhang X. F., Sun X. W., 2012. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genomics 13: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wan Z. Y., Bai B., Huang S. Q., Chua E., et al. , 2015. Construction of a high-density linkage map and fine mapping of QTL for growth in Asian seabass. Sci. Rep. 5: 16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Meyer E., McKay J. K., Matz M. V., 2012. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9: 808. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lu Y., Zhang Y., Ning Z., Li Y., et al. , 2015. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 47: 625–631. [DOI] [PubMed] [Google Scholar]
- Xia J. H., Liu F., Zhu Z. Y., Fu J. J., Feng J. B., et al. , 2010. A consensus linkage map of the grass carp (Ctenopharyngodon idella) based on microsatellites and SNPs. BMC Genomics 11: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Zou T., Chen Y., Chen L., Liu S., et al. , 2011. Coexistence of diploid, triploid and tetraploid crucian carp (Carassius auratus) in natural waters. BMC Genet. 12: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Wang P., Zhang Y., Fang L., Liu Y., et al. , 2015. Gene map of large yellow croaker (Larimichthys crocea) provides insights into teleost genome evolution and conserved regions associated with growth. Sci. Rep. 5: 18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zhang X. F., Wang X. M., Li J. T., Liu G. M., et al. , 2014. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46: 1212–1219. [DOI] [PubMed] [Google Scholar]
- Yamamoto T.-O., 1974. A YY male goldfish from mating estrone-induced XY female and normal male. J. Hered. 66: 2–4. [DOI] [PubMed] [Google Scholar]
- Yu Y., Zhang X. J., Yuan J. B., Li F. H., Chen X. H., et al. , 2015. Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific White Shrimp Litopenaeus vannamei. Sci. Rep. 5: 15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. A., He Z. Z., Yuan X. N., Jiang X. Y., Sun X. W., et al. , 2010. Speciation of polyploid Cyprinidae fish of common carp, crucian carp, and silver crucian carp derived from duplicated Hox genes. J. Exp. Zoolog. B Mol. Dev. Evol. 314B(6): 445–456. [DOI] [PubMed] [Google Scholar]
- Yue G. H., 2014. Recent advances of genome mapping and marker‐assisted selection in aquaculture. Fish. 15: 376–396. [Google Scholar]
- Zhang J., Sun M., Zhou L., Li Z., Liu Z., et al. , 2015. Meiosis completion and various sperm responses lead to unisexual and sexual reproduction modes in one clone of polyploid Carassius gibelio. Sci. Rep. 5: 10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang Y., Zheng X., Kuang Y., Zhao Z., et al. , 2013. A consensus linkage map provides insights on genome character and evolution in common carp (Cyprinus carpio L.). Mar. Biotechnol. (NY) 15: 275–312. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang Y., Ji P. F., Zhang X. F., Zhao Z. X., et al. , 2013. A dense genetic linkage map for common carp and its integration with a BAC-based physical map. PLoS One 8: e63928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. H., Lu C. Y., Zhao Y. Y., Lee C., Cao D. C., et al. , 2010. A set of polymorphic trinucleotide and tetranucleotide microsatellite markers for silver crucian carp (Carassius auratus gibelio) and cross-amplification in crucian carp. Biochem. Genet. 48: 624–635. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Gui J., 2000a Analysis of genetic heterogeneity among five gynogenetic clones of silver crucian carp, Carassius auratus gibelio bloch, based on detection of RAPD molecular markers. Cytogenet. Genome Res. 88: 133–139. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Gui J. F., 2000b Genetic evidence for gonochoristic reproduction in gynogenetic silver crucian carp (Carassius auratus gibelio bloch) as revealed by RAPD assays. J. Mol. Evol. 51: 498–506. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang Y., Gui J.-F., 2001. Molecular analysis of silver crucian carp (Carassius auratus gibelio bloch) clones by SCAR markers. Aquaculture 201: 219–228. [Google Scholar]
- Zhou R., Cheng H., Tiersch T. R., 2002. Differential genome duplication and fish diversity. Rev. Fish Biol. Fish. 11: 331–337. [Google Scholar]
- Zhu C., Sun Y., Yu X., Tong J., 2013. Centromere localization for bighead carp (Aristichthys nobilis) through half-tetrad analysis in diploid gynogenetic families. PLoS One 8: e82950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Tong J., Yu X., Guo W., Wang X., et al. , 2014. A second-generation genetic linkage map for bighead carp (Aristichthys nobilis) based on microsatellite markers. Anim. Genet. 45: 699–708. [DOI] [PubMed] [Google Scholar]
- Zhu C. K., Tong J. O., Yu X. M., Guo W. J., 2015. Comparative mapping for bighead carp (Aristichthys nobilis) against model and non-model fishes provides insights into the genomic evolution of cyprinids. Mol. Genet. Genomics 290: 1313–1326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.