Abstract
In both Drosophila melanogaster and mammalian systems, epithelial structure and underlying cell polarity are essential for proper tissue morphogenesis and organ growth. Cell polarity interfaces with multiple cellular processes that are regulated by the phosphorylation status of large protein networks. To gain insight into the molecular mechanisms that coordinate cell polarity with tissue growth, we screened a boutique collection of RNAi stocks targeting the kinome for their capacity to modify Drosophila “cell polarity” eye and wing phenotypes. Initially, we identified kinase or phosphatase genes whose depletion modified adult eye phenotypes associated with the manipulation of cell polarity complexes (via overexpression of Crb or aPKC). We next conducted a secondary screen to test whether these cell polarity modifiers altered tissue overgrowth associated with depletion of Lgl in the wing. These screens identified Hippo, Jun kinase (JNK), and Notch signaling pathways, previously linked to cell polarity regulation of tissue growth. Furthermore, novel pathways not previously connected to cell polarity regulation of tissue growth were identified, including Wingless (Wg/Wnt), Ras, and lipid/Phospho-inositol-3-kinase (PI3K) signaling pathways. Additionally, we demonstrated that the “nutrient sensing” kinases Salt Inducible Kinase 2 and 3 (SIK2 and 3) are potent modifiers of cell polarity phenotypes and regulators of tissue growth. Overall, our screen has revealed novel cell polarity-interacting kinases and phosphatases that affect tissue growth, providing a platform for investigating molecular mechanisms coordinating cell polarity and tissue growth during development.
Keywords: cell polarity, Drosophila, phosphoprotein, kinase, phosphatase, Hippo, Wingless, Wnt, Ras, phosphoinositol, PI3K, nutrient sensing
Apical–basal polarization of the epithelium is essential to maintain tissue architecture and restrict organ growth (Elsum and Humbert 2013). Epithelial cell polarity arises due to the creation of distinct membrane domains (apical, basal, and basolateral) via the coordinated activity of three major polarity complexes, which are conserved from flies to humans. Specifically, epithelial cell polarity is coordinated by: (1) the Crumbs (CRB) complex, comprised of the transmembrane protein Crb and associated proteins Stardust and Patj, localized at the subapical region; (2) the Scribble module [Scribble (Scrib), Disc-large (Dlg), and Lethal (2) giant larvae (Lgl)], localized to septate junctions in Drosophila (basolaterally in mammals) to promote basolateral membrane identity; and (3) the Partitioning defective (PAR) complex [atypical protein kinase C (aPKC), Bazooka (PAR3), and PAR6)], which promotes the separation of basolateral and subapical membrane domains (Tepass 2012). Dynamic and reciprocal interactions between these polarity complexes determine cellular membrane identity and epithelial organization (McCaffrey and Macara 2011).
A critical determinant of cell polarity is the activity of the PAR complex and aPKC, which has dual roles in cell polarity. aPKC directly phosphorylates (1) Crb to allow binding of the Std/Patj complex (Sotillos et al. 2004) and (2) Lgl to result in exclusion from the apical membrane (Betschinger et al. 2005; Plant et al. 2003). Additionally, these apical–basal cell polarity regulators also control tissue growth. Drosophila lgl, scrib, and dlg are termed “junctional scaffold” neoplastic tumor suppressor genes, mutations of which are associated with loss of cell polarity and characterized by imaginal disc epithelial and neural tissue overgrowth, impaired differentiation, and the formation of transplantable tumors (Hariharan and Bilder 2006). Despite detailed knowledge of the molecular interactions between the Crb, PAR, and Lgl complexes in the establishment and maintenance of cell polarity, how these mutually exclusive polarity modules interact to coordinate epithelial organization with tissue growth is less well understood. Thus, we have been investigating how these polarity regulators control tissue growth, and have discovered that Lgl’s, but not Scrib’s or Dlg’s, role in tissue growth control occurs via regulation of signaling pathways and that this function is independent of Lgl’s role in cell polarity (Doggett et al. 2011, 2015; Grzeschik et al. 2007, 2010a; Parsons et al. 2014a; Portela et al. 2015; Richardson and Portela 2017). Of particular relevance here, our previous studies have shown that loss of lgl and the concomitant increase in aPKC activity, or increased levels of Crb, impair the Hippo tissue growth control pathway and are associated with ectopic cell proliferation, decreased apoptosis, and subsequent tissue overgrowth in the Drosophila eye (Grzeschik et al. 2010a,b; Parsons et al. 2010, 2014a,b; Portela et al. 2015; Richardson and Portela 2017).
To gain insights into the relationship between epithelial structure and organ growth, we utilized cell polarity phenotypes in the adult Drosophila eye and undertook a boutique genetic screen using RNA interference (RNAi). Due to the critical role phosphorylation plays in regulating the activity of numerous cellular signaling processes and growth pathways, we screened a collection of kinase and phosphatase RNAi lines. By screening for modification of the adult eye phenotypes due to overexpression/activation of Crb or aPKC (using GMR > crbintra or GMR > aPKCCA), we identified 185/365 genes that were capable of modifying these phenotypes. To further explore the ability of these cell polarity modifier genes to regulate tissue growth, we extended our analysis to screen for modification of a tissue overgrowth phenotype associated with knockdown of lgl in the adult Drosophila wing [using en > lgl-RNAi (lgli)]. From this secondary screen of the 185 genes from the primary screen, we identified 18 genes that also modified the en > lgli adult Drosophila wing size, compared with en > alone. Of the 18 genes that modified cell polarity phenotypes in the adult Drosophila eye and wing, several modulated signaling pathways involved in tissue growth control, such as the Hippo, Wingless/Wnt, and inositol phosphate signaling pathways. We also identified stress responsive genes, such as members of the nutrient sensing or AMP-activated protein kinases genes SIK2 and SIK3 (Shackelford and Shaw 2009). In summary, our genetic screens identified genes and biological processes that provide entry points to investigate the molecular mechanisms that coordinate epithelial structure and tissue growth control during development.
Materials and Methods
Drosophila stocks
We used the GAL4/UAS system for tissue-specific expression of transgenes (Brand and Perrimon 1993): glass multimer repeat [GMR - GAL4 (II)] and engrailed [en - GAL4 or en - GAL4, UAS - GFP (II)] express GAL4 predominantly in larval eye or wing discs, respectively [obtained from the Bloomington Drosophila Stock Center (BDSC)]. We constructed three cell polarity stocks: GMR - GAL4 was recombined with UAS - aPKCCAAXWT (Sotillos et al. 2004) or UAS - crbintra-38.1.2b (Klebes and Knust 2000), and the en > lgli stock was generated by standard genetic techniques. UAS - lgl - RNAi51249 was obtained from the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al. 2007), and we have shown that it efficiently depletes Lgl and can be rescued by the human ortholog Hugl1 (Grzeschik et al. 2010a).
Two independent stocks of each genotype were used to screen for modifiers.
UAS - SIK3K70M and null allele SIK372 were obtained from M. Montminy, Salk Institute for Biological Studies, California.
UAS - 40D (UAS40D) was obtained from the VDRC (60101).
naked-lacZ (P(ry[+7.2]=PZ)nkd[04869a]) and independent SIK3 RNAi stock TRiP.JF03002 were obtained from the BDSC (#25111 and #28366 respectively).
Genetic screens
Genetic screens are described in Figure 1 and Figure 3. Two virgin females from GMR > aPKCCA or GMR > crbintra stocks were crossed to two males carrying UAS - RNAi transgenes. Two virgin females from en > lgli stocks were crossed to two males from UAS - RNAi lines that showed a modification of the GMR > aPKCCA and/or GMR > crbintra phenotype. RNAi fly stocks were obtained from the National Institutes of Genetics (NIG-Fly, Japan) or the VDRC (Supplemental Material, Table S1 in File S1). At least 30 adult F1 flies were scored for each cross, representative images are shown. All flies were raised on standard cornmeal agar food at 25° unless stated otherwise. RNAi lines that modified GMR > aPKCCA adult eyes or en > lgli adult wings were rescreened at 18° or room temperature, respectively, as both phenotypes were weaker at lower temperatures.
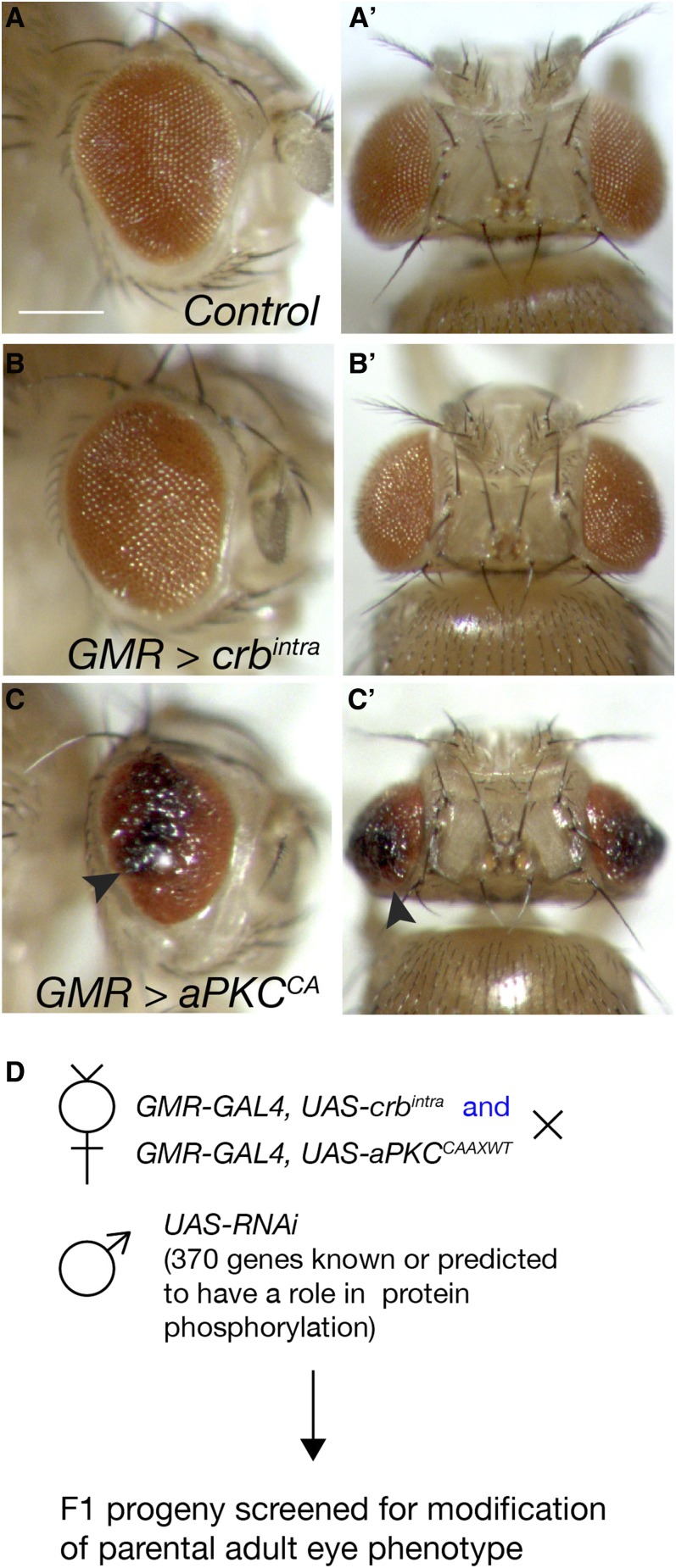
Figure 1.
Overexpression of cell polarity components in the developing Drosophila eye alters adult eye structure. (A–C’) Adult female eye images, side and dorsal views, respectively. Posterior to the left in (A–C). White bar, 250 μM. (A and A’) Control (GMR-GAL4) adult eyes displaying highly organized, regular, lattice-like organization of ommatidia to form the adult retina. (B and B’) GMR > crbintra adult eyes are slightly larger and rougher than control eyes. (C and C’) GMR > aPKCCA adult eyes are smaller, areas of the retina show disruptions in retinal architecture and necrosis (arrowheads). (D) Genetic scheme of F1 cell polarity modifier screen. Virgin females expressing components of cell polarity complexes in the developing eye (GMR > crbintra or GMR > aPKCCA) were crossed to males carrying UAS-RNAi transgenes to deplete proteins with roles in protein phosphorylation. F1 progeny were scored for modification of parental adult eye phenotypes. RNAi, RNA interference; UAS, upstream activating sequence.
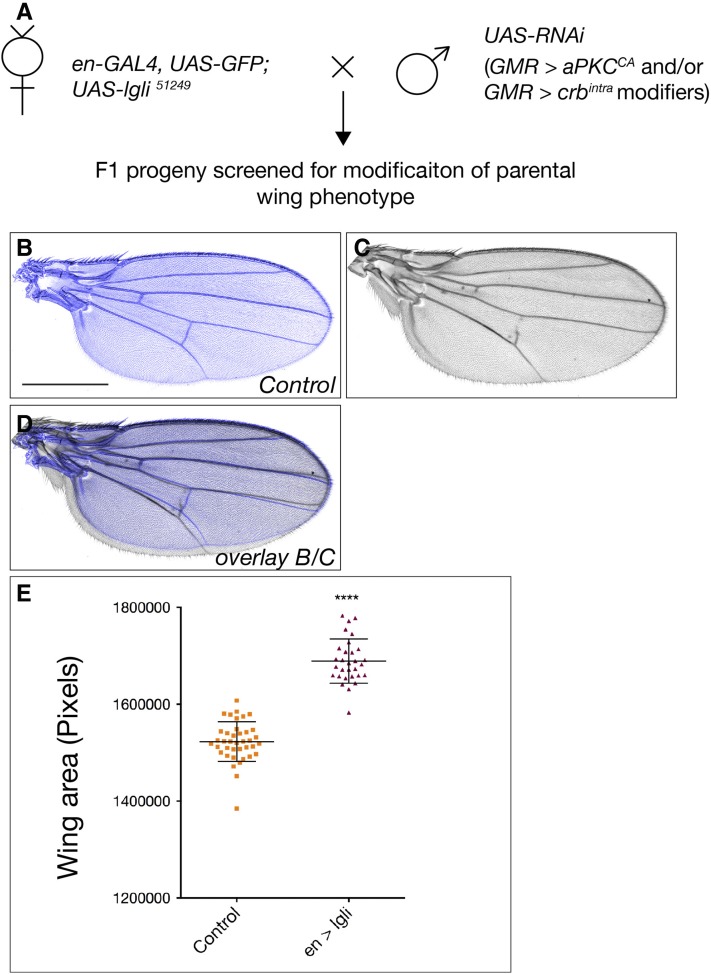
Figure 3.
Depletion of lgl in the developing Drosophila wing increases adult wing size. (A) Genetic scheme of F1 cell polarity modifier screen. Virgin females expressing RNAi depletion of Lgl (en > lgli) were crossed to males carrying UAS-RNAi transgenes corresponding to the 185 genes identified in the primary screen. F1 progeny were scored for modification of parental adult wing size. Black bar, 500 μM. (B, C, and D) Adult female wings, anterior up, proximal to the left. (B) Control adult wing. (C) Adult wing en > UAS-GFP; UAS-lgl-RNAi (VDRC 51249) displayed increased growth. (D) Overlay of (B) and (C) highlighting en > lgli overgrowth (gray). (E) Quantification of total wing area in control (en > GFP, RFP) flies compared to en > lgli. Results represent individual wings SD. **** P < 0.0001. RNAi, RNA interference; UAS, upstream activating sequence.
Analysis of adult eye phenotypes
F1 progeny from crosses of GMR > aPKCCA or GMR > crbintra to UAS - RNAi were scored for modification of parental eye phenotypes. Adult eyes showing modification were imaged with a Scitec Infinity1 camera. Images were processed through Adobe Photoshop CS2 and Adobe Illustrator CS2.
Analysis of wing size
F1 progeny from crosses of en > lgli to UAS - RNAi were scored for modification of parental wing phenotype. Adult wings were mounted in Canada Balsam/Xylene (Sigma) and imaged with an Olympus Stereomicroscope connected to a Scitec Infinity1 camera. Total wing area was measured with Adobe Photoshop CS2. Wing images were processed using Adobe Photoshop and Illustrator CS6.
Statistical analysis
All statistical tests were performed separately for each data set of wing sizes. Probability values were calculated using an unpaired t-test with Welch’s correction to reject the null hypothesis (variation of wing size through random, independent actions of UAS - RNAi transgenes) in Graph Pad Prism. P < 0.05 was considered statistically significant.
Gene ontology (GO) term (biological function) analysis
To functionally annotate gene lists and identify enriched GO term classes from genetic modifiers, the Princeton University web based tool, generic GO term finder, was used.
Signaling pathway analysis
The online pathway annotation tools DAVID (Database for Annotation, Visualization and Integrated Discovery, https://david.ncifcrf.gov), PANTHER (Protein ANalysis THrough Evolutionary Relationships, http://www.pantherdb.org), and KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg) were used to place modifier genes into functional groups.
Immunohistochemistry
For analysis of third-instar larval wing discs, discs were dissected in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, washed in PBS + 0.1% Triton X-100 (PBT), and blocked in PBT + 2% normal goat serum. Antibodies used were mouse anti-β−galactosidase (1:500; Rockland), rabbit anti-Lgl (1:500; Dennis Strand, Johannes Gutenberg University, Germany), and Alexa Fluor-conjugated 561, (1:500; Abcam). Confocal images were taken with an Olympus FV 1000, processed through Fiji and Adobe Photoshop CS6, and assembled in Adobe Illustrator CS6.
Data availability
Drosophila stocks and antibodies are available upon request or from stock centers as listed in the Materials and Methods. File S1 contains four supplemental figures and seven supplemental tables.
Results and Discussion
Primary adult eye screen
The primary RNAi screen for modifiers of cell polarity phenotypes was conducted using the adult Drosophila eye, which has a regular, lattice-like structure due to the repetitive organization of groups of epithelial retinal cells (Figure 1A). The organized structure of the Drosophila retina makes it sensitive to defects in epithelial structure and tissue growth, and therefore an ideal system for genetic modifier screens. When compared with control (GMR - GAL4/+, Figure 1, A and A’), overexpression of the transmembrane–intracellular domain of crb (UAS - crbintra) in the posterior region of the developing larval and pupal eye (using GMR - GAL4) results in weak overgrowth and lens defects in the adult eye (Figure 1, B and B’) (Grzeschik and Knust 2005; Grzeschik et al. 2010a; Johnson et al. 2002; Robinson et al. 2010). Expression of membrane-tethered constitutively-active aPKC (aPKCCAAXWT, hereafter referred to as aPKCCA) via GMR - GAL4 resulted in a small and rough adult eye with necrosis (arrowhead Figure 1, C and C’). We and others have previously demonstrated the utility of GMR > crbintra (crbintra) and GMR > aPKCCA (aPKCCA) to detect genes capable of modifying cell polarity phenotypes (Grzeschik et al. 2010a; Ogawa et al. 2009; Parsons et al. 2010, 2014a,b; Robinson et al. 2010). Thus, we conducted an F1 modifier screen to detect genes capable of altering the morphology and/or growth of the crbintra and/or aPKCCA adult eye phenotypes to identify novel factors connecting cell polarity to tissue architecture and growth.
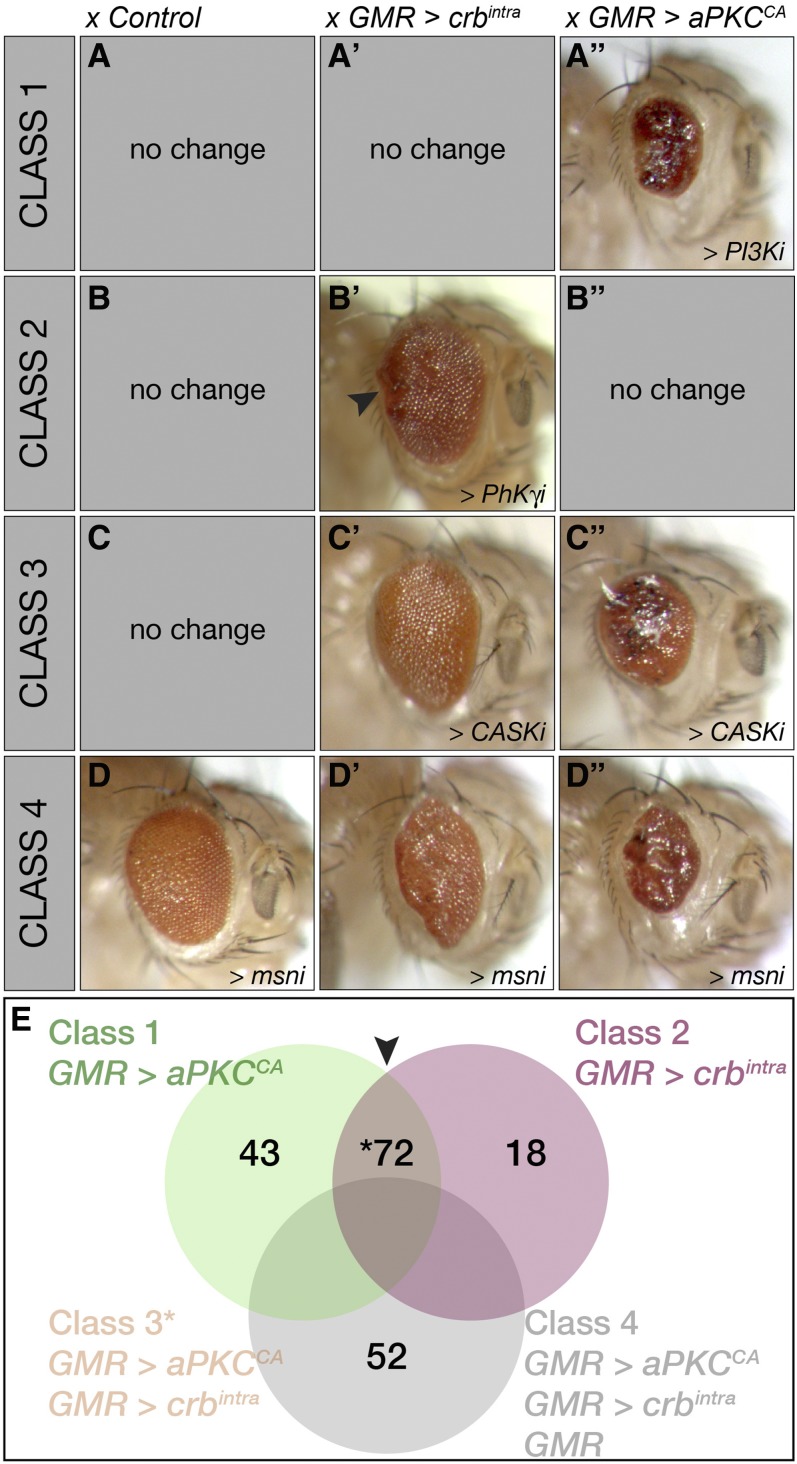
Since epithelial structure and cell polarity are integrated with cellular networks controlled by phosphorylation, we focused on genes predicted to encode kinases, phosphatases, and associated factors (Tables S1 and S2 in File S1, respectively). To identify those factors capable of genetically interacting with crb and/or aPKC, we conducted an F1 screen for modifiers of the crbintra and/or aPKCCA adult eye phenotypes using transgenic UAS - RNAi hairpin lines targeting 365 kinases or phosphatases (Figure 1D). From this screen, we identified 185 genes that modified the adult Drosophila cell polarity eye phenotypes. The 185 genes identified were grouped into four classes based on their interaction with crbintra and/or aPKCCA and/or GMR – GAL4 alone. Class 1 genes only modified aPKCCA [e.g., Dp110 (Pi3K92E); Figure 2, A–A”]. Class 2 genes only modified crbintra [e.g., calcium calmodulin regulated kinase PhKγ; Figure 2, B–B”). Class 3 genes modified both aPKCCA and crbintra but did not generate a phenotype with GMR - GAL4 alone (e.g., membrane-associated guanylate kinase CASK; Figure 2, C–C”). Class 4 hits not only interacted with with crbintra, aPKCCA, but their knockdown alone with GMR - GAL4 resulted in a visible phenotype [e.g., JNK pathway member misshapen (msn); Figure 2, D–D”]. Because these RNAi lines crossed to GMR - GAL4 generate an eye phenotype, it is possible that some of these genes may be false positive Crb and aPKC modifiers; however, so as to not miss any Crb and aPKC interactors, we proceeded with the analysis of this class of interactors. Of the 185 genes identified, 43 belonged to Class 1, 18 belonged to Class 2, 72 belonged to Class 3, and 52 were in Class 4 (Figure 2E). For a full list of genes identified in the screen and a brief description of their phenotype alone and/or phenotypic modification of crbintra and/or aPKCCA see Table S3 in File S1. The high proportion of genetic interactions (51% of genes screened) suggests that phosphoprotein networks play in important role in epithelial organization and/or tissue growth control together with aPKC and/or Crb in Drosophila eye development. Interestingly, the CMGC family of kinases (CDK, MAPK, GS3K, and CLK), comprising protein kinases involved in the MAPK cascade and mitotic cell cycle (Table 1), is the most highly enriched, suggesting that these kinases might play important roles in aPKC and/or Crb function.
Figure 2.
Classification of adult Drosophila eye cell polarity modifiers. (A–D”) Side view, adult female eyes, posterior to the left. Classification of genetic modifiers was based on genetic interaction with GMR > crbintra and/or GMR > aPKCCA and if RNAi expression alone via GMR-GAL4 also resulted in an aberrant eye phenotype. Classes of interactors are indicated. (A–A”) Class 1: modifier genes only interacted with GMR > aPKCCA. (A) Adult eyes expressing UAS-PI3Ki (VDRC 38985) showed no interaction with GMR-GAL4 or (A’) GMR > crbintra but modified (A”) GMR > aPKCCA to generate a small, glassy eye with decreased necrotic areas. (B–B”) Class 2: modifier genes only interacted with GMR > crbintra. (B) Adult eyes expressing UAS-PhKγi (VDRC 33054) showed no interaction with GMR-GAL4 or (B”) GMR > aPKCCA but modified (B’) GMR > crbintra to generate a larger eye with slight ruffling at the posterior edge (arrowhead). (C–C”) Class 3: modifier genes interacted with both GMR > crbintra and GMR > aPKCCA. (C) Adult eyes expressing UAS-CASKi (VDRC 104793 and 34184) showed a normal phenotype with GMR – GAL4 alone but modified (C’) GMR > crbintra to generate slightly larger eyes and (C”) GMR > aPKCCA to reduce necrosis. (D–D”) Class 4: genes modified all three parental phenotypes. (D) Adult eyes expressing UAS-msni (NIG 1697R-1) interact with GMR-GAL4 to generate glassy eyes. (D’) UAS-msni also modified GMR > crbintra to produce elongated rough adult eyes and (D”) modified GMR > aPKCCA to generate small, glassy eyes with reduced areas of necrosis. (E) Venn diagram depicting the number of genes in each Class. Green shading denotes Class 1, GMR > aPKCCA. Pink shading represents Class 2, GMR > crbintra. Brown central region (arrowhead) overlapping Class 1 and 2 represents Class 3 genes that modified both GMR > crbintra and GMR > aPKCCA. Gray shading denotes Class 4, genes that interacted with GMR > crbintra and GMR > aPKCCA and also produced an aberrant eye phenotype when expressed alone via GMR-GAL4. RNAi, RNA interference; UAS, upstream activating sequence; VDRC, Vienna Drosophila RNAi Center.
Table 1. Frequency analysis of GO terms Class 1–4 modifiers of GMR > crbintra and GMR > aPKCCA.
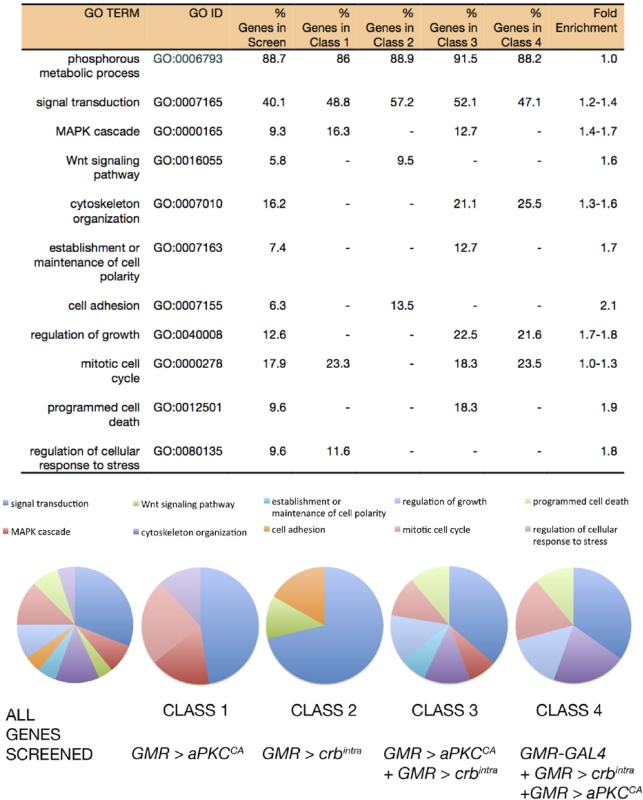 |
The frequency of GO terms in Classes 1–4 is shown as a percentage of genes within the individual class that associate with the GO term. Fold enrichment was calculated by dividing the percentage of genes associated in individual classes by the percentage of genes in the total number of genes screened. Fold enrichment range reflects lowest to highest enrichment from all four Classes. Similarities and differences in GO term frequency between Classes are depicted graphically in pie charts (see Legend for color coding of GO terms analyzed). Individual pie charts for Classes 1–4 lack the range of GO terms observed in the total pool of genes screened, indicating that the cell polarity phenotypes selectively interacted with modifiers. Moreover, each Class has different GO term association patterns suggesting that the cell polarity phenotypes are not equivalent and therefore may impact different signaling networks.
GO analysis of adult Drosophila eye cell polarity modifier genes
As the starting population of genes for this boutique screen was highly enriched for GO terms, such as phosphorous metabolic process genes (323/365, 88.7%), it was not possible to detect GO term enrichment for highly represented terms from Class 1–4 gene sets. Nevertheless, examination of GO terms with lower levels of enrichment between Classes 1–4, compared with the starting pool of genes screened, revealed modest enrichment of GO terms between the starting pool and modifier genes (Table 1). Classes 1–4 all showed different distributions for the 10 GO terms analyzed (Table 1). Genes associated with the GO term “establishment or maintenance of cell polarity” were only found in Class 3 (Table 1), highlighting the sensitivity of both the aPKCCA and crbintra adult eye phenotypes to changes in cell polarity network activity. Although all classes showed association with the GO term “signal transduction,” only classes with aPKCCA modifiers showed enrichment for MAPK signaling pathway genes. Furthermore, depletion of genes associated with “regulation of cellular response to stress” was only observed in Class 1 (aPKCCA). Taken together, these genetic screens demonstrated that cell polarity network activity was sensitive to several cellular processes, including proliferation, stress, and signaling pathways; however, distinct cell polarity modules may have different sensitivities and responses to these inputs.
Secondary adult wing screen
Examination of gene lists corresponding to RNAi lines that modified aPKCCA and/or crbintra (Classes 1–4) revealed numerous genetic interactors that might be new genes involved in cell polarity regulation of tissue growth (Table S3 in File S1). To confirm these genetic interactions, as well as to reveal genes involved in linking cell polarity regulation to tissue growth, we conducted a secondary genetic screen where we upregulated aPKC and Crb in the developing wing by knocking down Lgl. Due to the antagonistic interaction between Lgl and aPKC, knockdown of Lgl results in increased aPKC activity (Betschinger et al. 2005), which in turn phosphorylates and activates the Crb complex (Fletcher et al. 2012; Sotillos et al. 2004). We used the wing epithelium, rather than the eye, since it is easier to quantify effects on tissue growth in the adult wing than in the eye, as well as to reveal genes that interact with deregulated Lgl/aPKC and Crb in another epithelial tissue. To knockdown Lgl, we used the engrailed (en)-GAL4 driven expression of a UAS-lgl-RNAi line, which is expressed in the posterior compartment of the developing wing from embryogenesis (Figure 3A). In this wing model, we quantified altered tissue growth by measuring adult wing size. Depletion of Lgl in the posterior half of the developing wing disc [en - GAL4 driven UAS - lgl-RNAi (en > lgli)] (Figure S1 in File S1) resulted in a 10% increase in total adult wing area (Figure 3, B and C, overlayed in Figure 3D, and quantified in Figure 3E). Thus, we conducted a secondary screen of the 185 cell polarity modifier hits for those able to modify the wing overgrowth due to Lgl depletion.
Of the 185 positive hits identified in the adult eye screen, 93 also interacted with en > lgli or en-GAL4 alone (Figure 4A). en > lgli interactors were broadly classified into three groups (Table S4 in File S1). Class 1 comprised 41 genes that resulted in pupal lethality with en > lgli and/or en – GAL4 alone, which precluded the specific effect of the gene knockdown on the wing growth being analyzed. Class 2 included 10 genes that produced adults following coknockdown of lgl, but the crumpled wing phenotype precluded measurement. The most informative class, Class 3, comprised 42 genes, where either codepletion with lgl and/or depletion alone altered adult wing size. Essentially all Class 3 genes, except Tao-1 and SIK3, suppressed adult wing growth associated with lgl depletion.
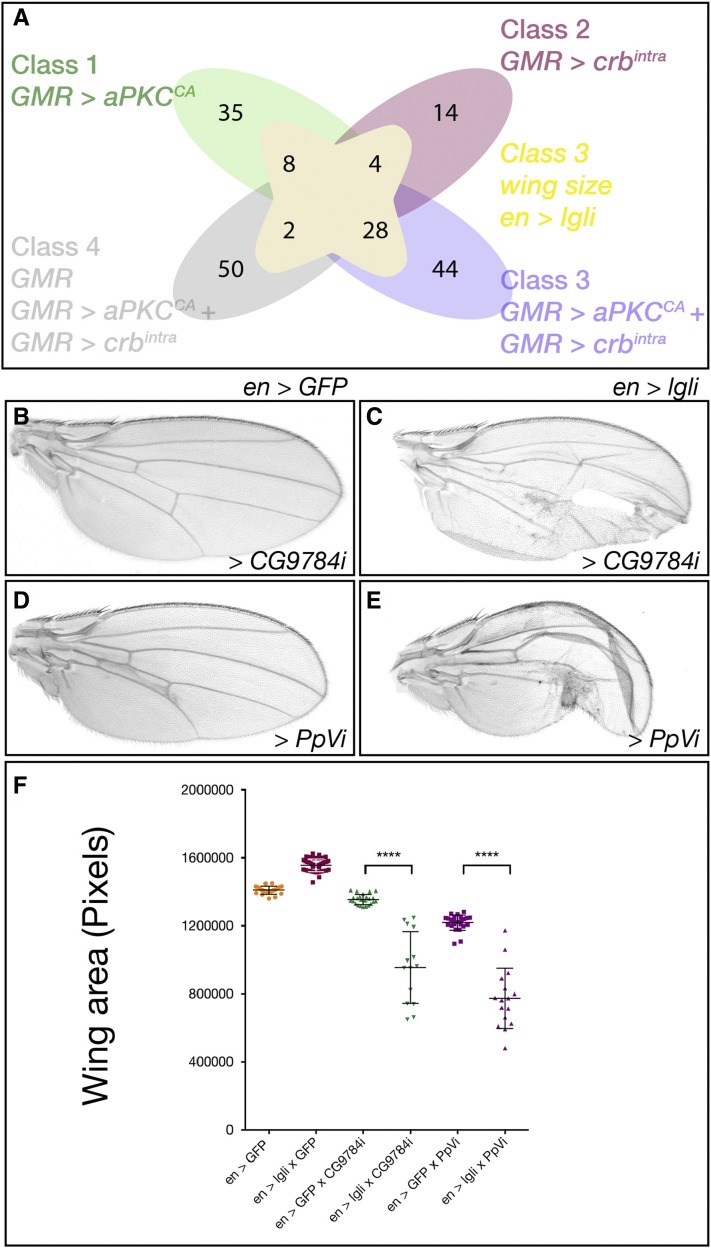
Figure 4.
Modifier genes of en >lgli adult wing size. (A) Venn diagram depicting the number of modifier genes in Classes 1–4 from the adult eye screen (total 185) and overlap between modifier genes identified in en > lgli wing size Class 3 (total 42). Central yellow shading denotes en > lgli wing size Class 3 modifiers. Green shading denotes eye Class 1, GMR > aPKCCA. Pink shading denotes eye Class 2, GMR > crbintra. Purple denotes eye Class 3 genes that modified: GMR > crbintra and GMR > aPKCCA. Gray shading denotes eye Class 4, genes that interacted with GMR > crbintra and GMR > aPKCCA but also show a phenotype when expressed alone via GMR-GAL4. The overlap with en >lgli (yellow) indicates the number of genes from each eye Class that also modified en >lgli wing size Class 3. (B–E) Adult female wings, anterior up, proximal to the left. (B) en > lgli specific modifier gene: en > GFP; CG9784i (VDRC 30098) wings show no change in wing size. (C) en > lgli/CG9784i have decreased wing growth and holes. (D) en > lgli modifier gene: en > GFP; PpVi (VDRC 101997) wings are slightly smaller than the control (<5%) yet significantly impacted (E) en >lgli adult wing growth. (F) Quantification of total wing area (A–D) in control animals compared to en > GFP and en > lgli. Results represent individual wings SD. **** P < 0.0001. VDRC, Vienna Drosophila RNAi Center.
After we had completed our screens, we were alerted to a recent report suggesting that ∼25% of the VDRC KK RNAi collection can generate false positive enhancement of impaired Hippo pathway signaling, due to ectopic expression of the tiptop (tio) transcription factor gene from the 40D insertion site (Vissers et al. 2016). To determine if aberrant tio expression might be influencing our screen results, we tested the polarity phenotypes with the tio tester stock (40DUAS). We observed modification of the GMR > crbintra, en - GAL4, and en > lgli, but not GMR - GAL4 or GMR > aPKCCA, with the tio tester stock (40DUAS) (Figure S2 in File S1). We note that in some instances where two or more RNAi lines for a given gene were tested for modification of crbintra and aPKCCA, the genetic interaction produced opposite results [e.g., wunen (wun), Table S3 in File S1] this may be due to a false positive interaction of the KK line with crbintra, off-target effects, or the ability of different RNAis to efficiently suppress target genes. As KK lines interacting with both crbintra and lgli may represent false positives, we intersected crbintra and lgli modifiers, which revealed four interactors: CG1830 (PhKγ), CG10417, CG8866, and CG32484 (Sk2) (Figure 4A and Table S5 in File S1). Three of these interactors, CG10417, CG8866, and CG32484 (Sk2), were identified with KK lines and may represent false positives that need further verification by testing with independent RNAi lines.
Systematic analysis of wing sizes of en > lgli compared with en – GAL4 interactors in Class 3 revealed four subclasses: Subclass 3.1 interactors only modified en > lgli; Subclass 3.2 modifiers affected en – GAL4 and en > lgli wing size equivalently; Subclass 3.3 en > lgli wings were smaller than en – GAL4 modified wings; and Subclass 3.4 en > lgli wing sizes were larger than en – GAL4 wings. Since genes in Subclass 3.2 modified en – GAL4 and en > lgli wing size equivalently, these genes were ruled out as being specific en > lgli interactors, leaving 18 genes in the remaining classes as Lgl modifiers. Additionally, as both en - GAL4 and en > lgli crossed to 40DUAS wings displayed an ∼10% decrease in wing size (Figure S2 in File S1), 15 Subclass 3.2 wing modifiers (where equivalent reduction in en > lgli and en wing growth was observed) may also be false positives (indicated with *, Table S6 in File S1). In summary, the en > lgli screen identified 18 kinases and phosphatases where knockdown of the modifier gene only showed modification of wing growth with lgli but not with en, or modified en > lgli wing size more than en alone (Table S6 in File S1). These genes include CG9784 (IPP, lipid phosphatase) and PpV (predicted Wnt pathway regulator; Swarup et al. 2015), which dramatically modified Lgl-depleted wings, but had little effect or ∼10% reduced growth following knockdown alone (Figure 4, B–E, respectively, quantified in Figure 4F). Thus, although screening the VDRC KK RNAi collection can generate false positive genetic interactions and some modifier genes require further validation (see Table S6 in File S1), we have identified 18 kinase/phosphatase genes that might coordinate cell polarity cues and tissue growth signals during organ development.
Overlap between modifier genes that specifically affected adult eye cell polarity phenotypes (Classes 1–3) and the en > lgli wing size (Classes 3.1, 3.3, and 3.4) (Figure S3 and Tables S6 and S7 in File S1) revealed that 15 of the Lgl wing size modifier genes interacted with both aPKC and Crb, suggesting that these genes were general cell polarity tissue growth regulators. Three genes, Btk29A, Ror, and PpY-55A, did not interact with Crb, suggesting that these genes might be specific for the Lgl–aPKC axis of the tissue growth regulatory pathway. For simplicity, we will henceforth refer to these 18 genes as cell polarity–tissue growth interactors.
Cell polarity–tissue growth-interacting genes are associated with many signaling pathways
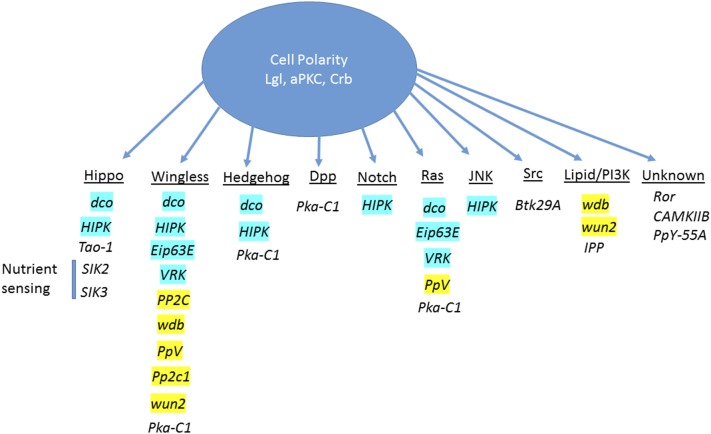
Analysis of the 18 cell polarity–tissue growth interactors (Classes 3.1, 3.3, and 3.4) for their links to signaling pathways (Figure 5 and Table S7 in File S1), revealed that many were associated with signaling pathways that Lgl, aPKC, or Crb have previously been shown to regulate in tissue growth control: the Hippo, Notch, and JNK pathways (Grzeschik et al. 2010a; Parsons et al. 2014a,b; Portela et al. 2015; Sun and Irvine 2011; Zhu et al. 2010). Notably, other Lgl-interacting genes were associated with signaling pathways not previously linked to the negative regulation of tissue growth by Lgl: Wg, Decapentaplegic (Dpp), Hedgehog (Hh), Src, Ras, and lipid signaling/PI3K (Figure 5 and Table S7 in File S1). However, three Lgl-interacting genes, the protein kinase genes Ror and CAMKIIB and the phosphatase gene PpY-55A, have been poorly studied and have not yet been associated with known signaling pathways.
Figure 5.
Summary of cell polarity tissue growth interactors and the signaling pathways they are associated with. Genes from the wing size Class 3 that modified en > lgli, greater than en-GAL4 alone (Classes 3.1, 3.3, and 3.4), are listed under the signaling pathways that they are associated with. Genes highlighted in blue or yellow were identified as kinases or phosphatase regulators of Wnt signaling respectively (Swarup et al. 2015). JNK, Jun kinase; PI3K, phospho-inositol-3-kinase.
Several of the cell polarity–tissue growth interactors affected the Hippo signaling pathway; depletion of Tao-1 (Class 3.4) (Boggiano et al. 2011; Poon et al. 2011), Hipk (Class 3.3) (Chen and Verheyen 2012), dco (Class 3.1) (Milton et al. 2010; Sopko et al. 2009), SIK2 (Class 3.3), and SIK3 (Class 3.4) (Wehr et al. 2013) modified the Lgl, aPKC, and Crb cell polarity phenotypes. Interestingly, the founding members of the Hippo pathway, hpo and wts (Class 3.1) (Harvey and Tapon 2007), also interacted with en > lgli, resulting in pupal lethality, although at lower temperatures hpo-RNAi produced adults with wing size defects (Table S4 in File S1). Since Lgl, aPKC, and Crb are known to regulate the Hippo pathway (Chen et al. 2010; Grzeschik et al. 2010a; Ling et al. 2010; Parsons et al. 2010; Robinson et al. 2010), the identification of Hippo pathway regulators in our en > lgli screen demonstrates that this phenotype is sensitive to modifier genes that regulate tissue growth.
We have previously demonstrated that Notch signaling is impaired in lgl mutant eye epithelial tissue and contributes to the tissue growth effects (Parsons et al. 2014a; Portela et al. 2015), and Crb has also been shown to regulate Notch signaling in the eye (Richardson and Pichaud 2010). HIPK, identified as an lgl, aPKC, and crb interactor, in addition to its regulation of Hippo pathway signaling, also acts a positive regulator of Notch signaling in eye development (Lee et al. 2009). HIPK also regulates JNK signaling in the Drosophila wing epithelium (Huang et al. 2011), and in this tissue Lgl depletion-mediated cell polarity and tissue growth effects are JNK-dependent (Sun and Irvine 2011; Zhu et al. 2010). Thus, HIPK’s role in regulating JNK signaling in the wing tissue and in Notch signaling in the eye tissue might also underlie its genetic interactions with lgl, aPKC, and crb. However, although core members of the JNK pathway, Bsk, Tak1, Takl2, and Misshapen (Msn), were identified as modifiers of the GMR > aPKC or GMR > aPKC and GMR > crb eye phenotypes (Table S3 in File S1), knockdown of Tak1 or Msn were lethal with en > lgli and en>, precluding analysis of their specific interaction with lgl, while Bsk or Takl2 knockdown did not noticeably modify the en > lgli phenotype (Table S4 in File S1).
Our analysis of the Lgl-interacting genes revealed several novel signaling pathways (Dpp, Wg, Hh, Src, Ras, and lipid-PI3K) not previously implicated in the negative regulation of tissue growth by Lgl, aPKC, or Crb cell polarity regulators, which we will detail below.
Lgl regulates Dpp [Bone Morphogenetic Protein (BMP)] signaling in the wing epithelium by promoting the secretion of the Dpp morphogen (Arquier et al. 2001). Pka-C1, which genetically interacts with lgl, aPKC, and crb phenotypes, negatively regulates Dpp signaling (Li et al. 1995), so may genetically interact with Lgl, by affecting Dpp signaling. However, since Dpp positively regulates wing tissue growth (Brumby and Richardson 2005) and Lgl depletion would be expected to decrease Dpp signaling, this is unlikely to directly account for the wing size increase. Furthermore, knockdown of Pka-C1 should lead to increased Dpp signaling, as well as Hh and Wg signaling in the wing epithelium (Li et al. 1995), and therefore it is difficult to understand how Pka-C1 leads to a reduction in en > lgli wing growth. Interestingly, Pka-C1 promotes Ras-induced stem cell proliferation in the malpighian tubules (Zeng et al. 2010), and therefore reduced Pka-C1 might also reduce Ras signaling in the wing leading to the reduced tissue growth of en > lgli wings; however, further investigation is required to investigate this possibility. Interestingly, there is cross talk between the Dpp and Hippo pathways that might also impact on this interaction (Oh and Irvine 2011; Rogulja et al. 2008).
The Wg (Wnt) signaling pathway was associated with many of the Lgl interactors. The Wg/Wnt pathway regulates many biological processes, including proliferation and differentiation, to coordinate organ growth and planar cell polarity (Clevers and Nusse 2012). Interestingly, as measured by increased expression of the naked (nkd)-lacZ Wg signaling reporter (Tyler and Baker 2007; Zeng et al. 2000), we found that Wg signaling was upregulated in lgl27S3 mutant clones relative to the surrounding wild-type cells in larval eye discs (Figure S4 in File S1). Consistent with Lgl-modulating Wg signaling, two cell polarity tissue growth interactor genes are implicated in Wg signaling [discs overgrown (dco) (Class 3.1) (Klein et al. 2006), and cAMP-dependent protein kinase 1 (Pka-C1) (Class 3.3) (Li et al. 1995)]. Furthermore, comparison between the cell polarity tissue growth interactors, and kinase and phosphatase genes recently predicted to regulate Wg signaling in Drosophila (Swarup et al. 2015), revealed dco and three other kinase genes [VRK (Class 3.3), Hipk (Class 3.3), and Eip63E (Class 3.1)], as well as five phosphatases genes [wun2, PP2c1, PpV, wdb (all Class 3.3), and PP2C (Class 3.1)] (Figure 5 and Tables S6 and S7 in File S1). Therefore, these genes might regulate Wg signaling to modify the Lgl, aPKC, or Crb phenotypes. Consistent with the association of these interactors with the Wg pathway, previous studies have revealed genetic interactions between the Wg signaling pathway and Lgl/aPKC in Drosophila embryo epithelial morphogenesis (Dollar et al. 2005; Kaplan et al. 2009, 2011; Kaplan and Tolwinski 2010) and in Xenopus (Choi and Sokol 2009). Thus, cell polarity regulation of Wg signaling might be important in coordinating epithelial structure and organ growth. However, it should also be noted that the Wg signaling pathway can cross talk to the Hippo pathway in tissue growth control during wing development (Zecca and Struhl 2010), and therefore the effect of these cell polarity tissue growth interactors on the Wg pathway might indirectly affect the Hippo pathway to modulate the Lgl, aPKC, or Crb phenotypes.
Other signaling pathways associated with the cell polarity–tissue growth interactor genes, were Hh (dco, HIPK, and Pka-C1), Src (Btk29A), Ras (dco, PpV, Eip63E, VRK, and Pka-C1), lipid-PI3K [wdb and CG9784 (IPP)], and wun2 (Figure 5 and Tables S6 and S7 in File S1). With the cell polarity tissue growth interactors that are associated with Hh signaling, dco (Class 3.1; Shi et al. 2014), HIPK (Class 3.3; Swarup and Verheyen 2011), and Pka-C1 (Class 3.3; Kiger and O’Shea 2001), two of these genes are also regulators of the Hippo pathway, and indeed Hh signaling has been linked to Hippo pathway regulation (Kagey et al. 2012). Thus, the link between these cell polarity tissue growth interactors and the Hh pathway may ultimately affect the Hippo pathway in the modulation of Lgl, aPKC, or Crb phenotypes. Likewise, the link between the cell polarity tissue growth interactors and the Src and Ras pathways might be also related to Hippo signaling (Enomoto and Igaki 2013; Reddy and Irvine 2013), as detailed below.
Btk29A (Class 3.1) is regulated by Src signaling in tissue growth (Read et al. 2004), and interestingly Src signaling has previously been shown to interact with Lgl mutant cell extrusion and invasion phenotypes in the wing epithelium (Ma et al. 2013). Recent studies have revealed that overexpression of Src regulates tissue growth via JNK-dependent repression of the Hippo pathway (Enomoto and Igaki 2013), and thus the suppression of the en > lgli wing overgrowth by Btk29A might be due to restored Hippo pathway signaling.
Cell polarity tissue growth interactors were also associated with the Ras pathway: dco (Class 3.1), PpV (Class 3.3), Eip63E (Class 3.1 (Friedman et al. 2011), VRK (Class 3.3) (Ashton-Beaucage et al. 2014), and Pka-C1 (Class 3.3) (Zeng et al. 2010). Since elevated Ras signaling is a driver of tissue growth through promoting cell proliferation and inhibiting apoptosis (Brumby and Richardson 2005), but has been also linked to Hippo pathway impairment (Reddy and Irvine 2013), these interacting genes might indirectly affect Hippo signaling to modulate the Lgl, aPKC, and Crb phenotypes by regulating Ras signaling.
Cell polarity tissue growth interactors were also associated with lipid/PI3K signaling (Tables S6 and S7 in File S1): CG9784 (IPP, encoding a Inositol phosphate phosphatase, which is involved in membrane trafficking and lipid signaling, Class 3.1) (De Craene et al. 2017; Erneux et al. 2016; Liu and Bankaitis 2010; Morrison et al. 2000), wdb (encoding PP2A-B’ subunit, which modulates PI3K-Akt signaling, Class 3.3) (Vereshchagina et al. 2008), and wun2 (encoding a lipid phosphatase involved in glycerolipid metabolism, which is required for septate junction formation in the larval tracheal system, Class 3.3) (Ile et al. 2012). Of relevance, deregulation of phospholipid metabolism is linked to cell polarity in both mammalian cells and Drosophila (Claret et al. 2014; Shewan et al. 2011). Furthermore, Lgl has been recently shown to bind to PIP2 and PI4P phospholipids, which targets Lgl in Drosophila and mammalian cells to the plasma membrane (Dong et al. 2015). Thus, our data supports the growing body of research that links plasma membrane lipid domains and cell polarity. However, PI3K phosphorylates the phospho-inositol PIP2 to generate PIP3, which signals via Akt to regulate mTor (mechanistic target of rapomycin) activity in tissue growth control (Yu and Cui 2016), and mTor signaling has been recently shown to modulate target gene accessibility of the Hippo pathway effector Yki (Parker and Struhl 2015). Thus, these lipid/PI3K signaling regulators might also indirectly affect the Hippo pathway to mediate their interactions with the Lgl, aPKC, or Crb phenotypes.
Altogether, our analyses reveal several known and novel signaling pathways linking cell polarity modules to multiple regulatory networks controlling tissue growth. Many of these signaling pathways are also linked to the regulation of the Hippo pathway, therefore their interaction with Lgl, aPKC, and/or Crb may reflect their modulation of Hippo signaling. However, the precise mode by which these cell polarity–tissue growth-interacting genes modulate these signaling pathways, and in turn how they might be regulated by the polarity regulators, requires further investigation.
Cell polarity complexes and Salt Inducible Kinase 3 interact to restrict tissue growth
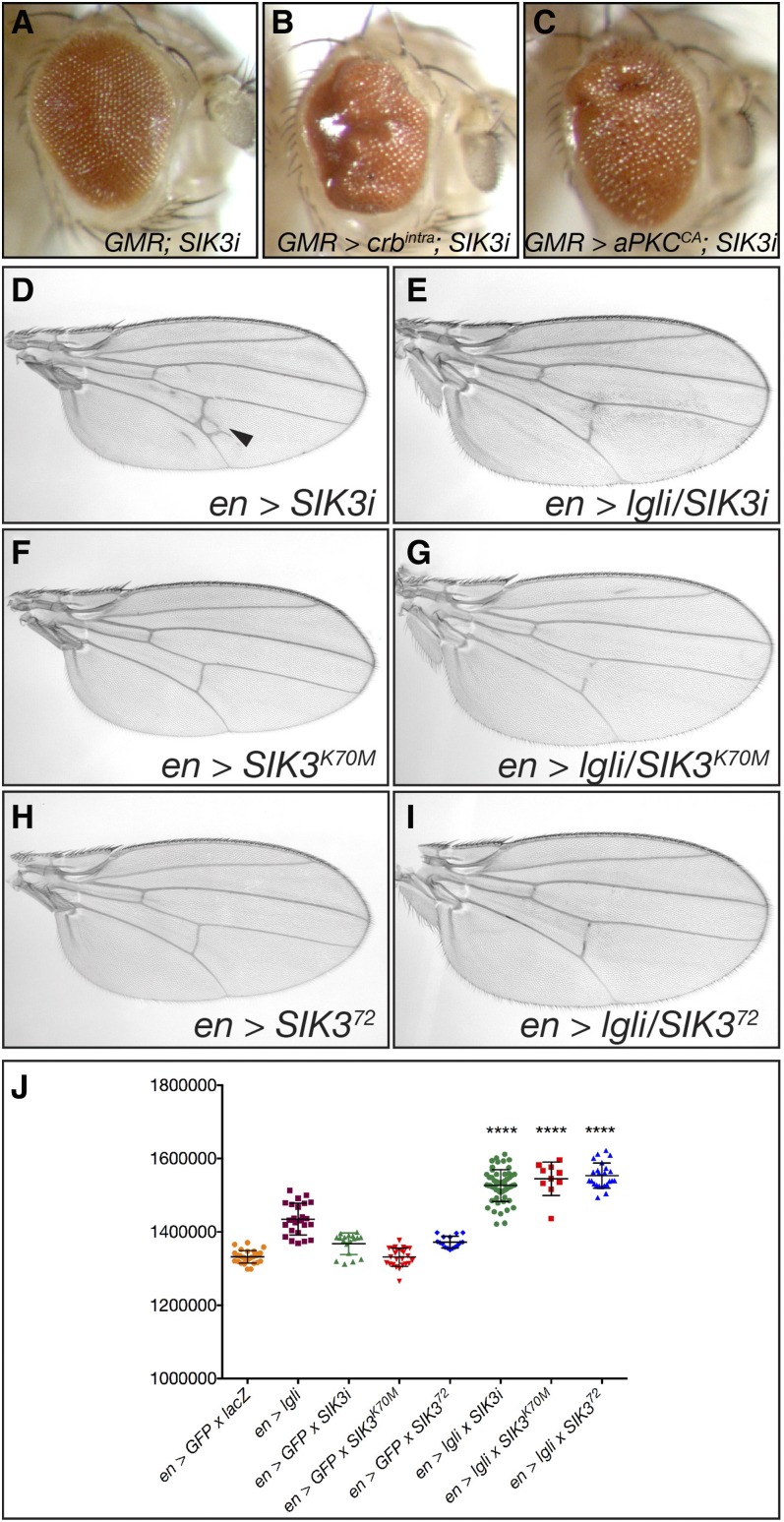
Of particular interest, our cell polarity gene screens identified two members of the AMP-related kinase family, Salt Inducible Kinase 3 (SIK3) (Class 3.4) and SIK2 (Class 3.3), which have important roles in nutrient-dependent signaling (Choi et al. 2011, 2015; Wang et al. 2011), but had not previously been connected to cell polarity regulation. SIK3 was identified in our screen as a negative regulator of tissue growth affecting aPKC, Crb, and Lgl phenotypes (Tables S4, S6, and S7 in File S1). Although depletion of SIK3 alone did not noticeably affect adult eye morphology or size (Figure 6A), SIK3 knockdown caused glassiness and bulging of retinal tissue in adult eyes in the crbintra or aPKCCA background (Figure 6, B and C respectively, compared with Figure 1, C and D). Wing size was unaltered following depletion of SIK3 alone in the posterior wing compartment via RNAi (en – GAL4 driven SIK3i), expression of a kinase dead transgene (SIK3K70M), or reduced SIK3 gene dosage using SIK3 heterozygotes (null allele SIK372/+). However, reduced SIK3 activity, in combination with lgl depletion, resulted in a significant increase in wing size (compare Figure 6, D and E, Figure 6, F and G, and Figure 6, H and I; quantified in Figure J). Thus, SIK3 modulates the activity of the cell polarity complexes and growth pathways to restrict tissue growth.
Figure 6.
SIK3 genetically interacts with cell polarity complexes to negatively regulate organ size. (A–C) Adult female eyes, posterior is to the left. (D–I) Adult female wings, anterior up, proximal to the left. (A) Expression of UAS-SIK3-RNAi (independent RNAi TRiP.JF03002) with GMR-GAL4 in the developing eye has no effect on adult eye morphology. (B) Expression of UAS-SIK3 RNAi in conjunction with GMR > crbintra or (C) GMR > aPKCCA at 18° increases adult eye size. (D) Reduction of SIK3 activity by RNAi depletion en > GFP; UAS-SIK3-RNAi, (F) overexpression of kinase dead transgene en > GFP; UAS-SIK3K70M, or (H) null allele en > GFP/ SIK372 has no effect on adult wing size (< 1%). (E) In conjunction with reduced lgl activity, en > lgli decreased SIK3 activity by RNAi depletion with UAS-SIK3-RNAi. (G) Overexpression of kinase dead transgene (UAS-SIK3K70M) or (I) null allele SIK372 significantly increased wing size. (J) Quantification of total wing area (D–I). In D, the arrowhead indicates ectopic cross veins. Results represent individual wings SD. **** P < 0.0001. RNAi, RNA interference
Intriguingly, our data support a requirement for SIK2 activity in promoting normal and lgli-dependent wing growth (Tables S4, S6, and S7 in File S1), while reduced SIK3 activity increased wing growth in the Lgl loss-of-function background. In Drosophila, Salt Inducible kinases have been implicated in tissue growth via Hippo pathway signaling (Wehr et al. 2013). SIK2 and SIK3 phosphorylate and inactivate Salvador (SAV), a core component of the Hippo kinase complex, leading to activation of the Yki transcriptional program and increased tissue growth (Wehr et al. 2013). Furthermore, codepletion of SIK2 and SIK3 reduced tumor growth in a Drosophila tumor model (activated Src + RasV12) (Hirabayashi and Cagan 2015). Given the observation that SIK2 and SIK3 differentially modify Lgl-dependent tissue growth, but are both required for Hippo pathway inactivation and Src + RasV12-driven tumor growth, future studies are required to determine how the SIKs interact with Lgl to control wing growth.
Conclusions
Genetic screens in Drosophila remain a powerful tool for identifying and unraveling gene function in specific signaling pathways and cellular processes. We undertook a boutique genetic screen, specifically using RNAi lines targeting kinases and phosphatases, to identify novel signaling pathways involved in the regulation of epithelial structure with tissue growth. Analysis of the hits that genetically interacted with lgl, aPKC, and/or crb cell polarity genes revealed that they were associated with Hippo, Notch, JNK, Dpp, Hh, Wg, Ras, lipid/PI3K, and unknown signaling pathways. Future studies determining the molecular relationships between cell polarity proteins and the modifiers identified will be required to determine whether these interactions are direct. Recent advances in proteomics through the generation of the Drosophila and human Protein Interaction Map, and studies coupling genetic manipulation to the analysis of kinase–phosphatase networks, will considerably advance our capacity to define the in vivo function of these genes (Guruharsha et al. 2011; Huttlin et al. 2015; Sopko et al. 2014). Of relevance to the novel pathways revealed in this study, further exploration of the roles of SIK2 and SIK3 in Drosophila development, and mammalian cancer models, is required to unravel the intricacies between cell polarity protein complexes and nutrient sensing kinases in normal development and cancer.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043513/-/DC1.
Acknowledgments
We thank all members of our laboratories for helpful discussions during the course of this study. We thank FlyBase for their wealth of information. This work was funded by a Contributing to Australian Scholarship and Science Foundation Science and Medicine Grant (SM-14-5398) awarded to L.M.P. and National Health and Medical Research Council (NHMRC) Project grant 628401 awarded to H.E.R. and N.G. H.E.R. was supported by a NHMRC fellowship (1020056), and funds from La Trobe University and the La Trobe Institute of Molecular Science.
Footnotes
Communicating editor: C. Gonzalez
Literature Cited
- Arquier N., Perrin L., Manfruelli P., Semeriva M., 2001. The Drosophila tumor suppressor gene lethal(2)giant larvae is required for the emission of the Decapentaplegic signal. Development 128: 2209–2220. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D., Udell C. M., Gendron P., Sahmi M., Lefrancois M., et al. , 2014. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 12: e1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Eisenhaber F., Knoblich J. A., 2005. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr. Biol. 15: 276–282. [DOI] [PubMed] [Google Scholar]
- Boggiano J. C., Vanderzalm P. J., Fehon R. G., 2011. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brumby A. M., Richardson H. E., 2005. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer 5: 626–639. [DOI] [PubMed] [Google Scholar]
- Chen C. L., Gajewski K. M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., et al. , 2010. The apical-basal cell polarity determinant crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA 107: 15810–15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Verheyen E. M., 2012. Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr. Biol. 22: 1582–1586. [DOI] [PubMed] [Google Scholar]
- Choi S., Kim W., Chung J., 2011. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J. Biol. Chem. 286: 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Lim D. S., Chung J., 2015. Feeding and fasting signals converge on the LKB1–SIK3 pathway to regulate lipid metabolism in Drosophila. PLoS Genet. 11: e1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.-C., Sokol S. Y., 2009. The involvement of lethal giant larvae and Wnt signaling in bottle cell formation in Xenopus embryos. Dev. Biol. 336: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret S., Jouette J., Benoit B., Legent K., Guichet A., 2014. PI(4,5)P2 produced by the PI4P5K SKTL controls apical size by tethering PAR-3 in Drosophila epithelial cells. Curr. Biol. 24: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Clevers H., Nusse R., 2012. Wnt/β-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- De Craene J. O., Bertazzi D. L., Bar S., Friant S., 2017. Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int. J. Mol. Sci. 18: E634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Doggett K., Grusche F. A., Richardson H. E., Brumby A. M., 2011. Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Dev. Biol. 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett K., Turkel N., Willoughby L. F., Ellul J., Murray M. J., et al. , 2015. BTB-zinc finger oncogenes are required for Ras and Notch-driven tumorigenesis in Drosophila. PLoS One 10: e0132987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollar G. L., Weber U., Mlodzik M., Sokol S. Y., 2005. Regulation of Lethal giant larvae by Dishevelled. Nature 437: 1376–1380. [DOI] [PubMed] [Google Scholar]
- Dong W., Zhang X., Liu W., Chen Y. J., Huang J., et al. , 2015. A conserved polybasic domain mediates plasma membrane targeting of Lgl and its regulation by hypoxia. J. Cell Biol. 211: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsum I. A., Humbert P. O., 2013. Localization, not important in all tumor-suppressing properties: a lesson learnt from Scribble. Cells Tissues Organs 198: 1–11. [DOI] [PubMed] [Google Scholar]
- Enomoto M., Igaki T., 2013. Src controls tumorigenesis via JNK-dependent regulation of the Hippo pathway in Drosophila. EMBO Rep. 14: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erneux C., Ghosh S., Ramos A. R., Edimo W. E., 2016. New functions of the inositol polyphosphate 5-phosphatases in cancer. Curr. Pharm. Des. 22: 2309–2314. [DOI] [PubMed] [Google Scholar]
- Fletcher G. C., Lucas E. P., Brain R., Tournier A., Thompson B. J., 2012. Positive feedback and mutual antagonism combine to polarize crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 22: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Friedman A. A., Tucker G., Singh R., Yan D., Vinayagam A., et al. , 2011. Proteomic and functional genomic landscape of receptor tyrosine kinase and Ras to extracellular signal-regulated kinase signaling. Sci. Signal. 4: rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N. A., Knust E., 2005. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development 132: 2035–2045. [DOI] [PubMed] [Google Scholar]
- Grzeschik N. A., Amin N., Secombe J., Brumby A. M., Richardson H. E., 2007. Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev. Biol. 311: 106–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F., Richardson H. E., 2010a Lgl, aPKC, and crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20: 573–581. [DOI] [PubMed] [Google Scholar]
- Grzeschik N. A., Parsons L. M., Richardson H. E., 2010b Lgl, the SWH pathway and tumorigenesis: it’s a matter of context & competition! Cell Cycle 9: 3202–3212. [DOI] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan I. K., Bilder D., 2006. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu. Rev. Genet. 40: 335–361. [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N., 2007. The Salvador–Warts–Hippo pathway—an emerging tumour-suppressor network. Nat. Rev. Cancer 7: 182–191. [DOI] [PubMed] [Google Scholar]
- Hirabayashi S., Cagan R. L., 2015. Salt-inducible kinases mediate nutrient-sensing to link dietary sugar and tumorigenesis in Drosophila. eLife 4: e08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Du G., Chen H., Liang X., Li C., et al. , 2011. Drosophila Smt3 negatively regulates JNK signaling through sequestering Hipk in the nucleus. Development 138: 2477–2485. [DOI] [PubMed] [Google Scholar]
- Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., et al. , 2015. The BioPlex network: a systematic exploration of the human interactome. Cell 162: 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ile K. E., Tripathy R., Goldfinger V., Renault A. D., 2012. Wunen, a Drosophila lipid phosphate phosphatase, is required for septate junction-mediated barrier function. Development 139: 2535–2546. [DOI] [PubMed] [Google Scholar]
- Johnson K., Grawe F., Grzeschik N., Knust E., 2002. Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr. Biol. 12: 1675–1680. [DOI] [PubMed] [Google Scholar]
- Kagey J. D., Brown J. A., Moberg K. H., 2012. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mech. Dev. 129: 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. A., Tolwinski N. S., 2010. Spatially defined Dsh-Lgl interaction contributes to directional tissue morphogenesis. J. Cell Sci. 123: 3157–3165 [DOI] [PubMed] [Google Scholar]
- Kaplan N. A., Liu X., Tolwinski N. S., 2009. Epithelial polarity: interactions between junctions and apical-basal machinery. Genetics 183: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. A., Colosimo P. F., Liu X., Tolwinski N. S., 2011. Complex interactions between GSK3 and aPKC in Drosophila embryonic epithelial morphogenesis. PLoS One 6: e18616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger J. A., Jr., O’Shea C., 2001. Genetic evidence for a protein kinase A/cubitus interruptus complex that facilitates processing of cubitus interruptus in Drosophila. Genetics 158: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebes A., Knust E., 2000. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila. Curr. Biol. 10: 76–85. [DOI] [PubMed] [Google Scholar]
- Klein T. J., Jenny A., Djiane A., Mlodzik M., 2006. CKIɛ/discs overgrown promotes both Wnt-Fz/β-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 16: 1337–1343. [DOI] [PubMed] [Google Scholar]
- Lee W., Andrews B. C., Faust M., Walldorf U., Verheyen E. M., 2009. Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev. Biol. 325: 263–272. [DOI] [PubMed] [Google Scholar]
- Li W., Ohlmeyer J. T., Lane M. E., Kalderon D., 1995. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell 80: 553–562. [DOI] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., Yu J., Huang J., et al. , 2010. The apical transmembrane protein crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to expanded. Proc. Natl. Acad. Sci. USA 107: 10532–10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bankaitis V. A., 2010. Phosphoinositide phosphatases in cell biology and disease. Prog. Lipid Res. 49: 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Shao Y., Zheng H., Li M., Li W., et al. , 2013. Src42A modulates tumor invasion and cell death via Ben/dUev1a-mediated JNK activation in Drosophila. Cell Death Dis. 4: e864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey L. M., Macara I. G., 2011. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 21: 727–735. [DOI] [PubMed] [Google Scholar]
- Milton C. C., Zhang X., Albanese N. O., Harvey K. F., 2010. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development 137: 735–743. [DOI] [PubMed] [Google Scholar]
- Morrison D. K., Murakami M. S., Cleghon V., 2000. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 150: F57–F62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Ohta N., Moon W., Matsuzaki F., 2009. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J. Cell Sci. 122: 3242–3249. [DOI] [PubMed] [Google Scholar]
- Oh H., Irvine K. D., 2011. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell 20: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J., Struhl G., 2015. Scaling the Drosophila wing: TOR-dependent target gene access by the Hippo pathway transducer Yorkie. PLoS Biol. 13: e1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. M., Grzeschik N. A., Allott M. L., Richardson H. E., 2010. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 4: 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. M., Portela M., Grzeschik N., Richardson H. E., 2014a Lgl regulates Notch signaling via endocytosis, independently of the apical aPKC-Par6-Baz polarity complex. Curr. Biol. 24: 2073–2084. [DOI] [PubMed] [Google Scholar]
- Parsons L. M., Grzeschik N. A., Richardson H. E., 2014b lgl regulates the Hippo pathway independently of Fat/Dachs, Kibra/Expanded/Merlin and dRASSF/dSTRIPAK. Cancers (Basel) 6: 879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant P. J., Fawcett J. P., Lin D. C., Holdorf A. D., Binns K., et al. , 2003. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5: 301–308. [DOI] [PubMed] [Google Scholar]
- Poon C. L., Lin J. I., Zhang X., Harvey K. F., 2011. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21: 896–906. [DOI] [PubMed] [Google Scholar]
- Portela M., Parsons L. M., Grzeschik N. A., Richardson H. E., 2015. Regulation of Notch signaling and endocytosis by the Lgl neoplastic tumor suppressor. Cell Cycle 14: 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. D., Bach E. A., Cagan R. L., 2004. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase, and STAT pathways. Mol. Cell. Biol. 24: 6676–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. V., Irvine K. D., 2013. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell 24: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson E. C., Pichaud F., 2010. Crumbs is required to achieve proper organ size control during Drosophila head development. Development 137: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H. E., Portela M., 2017. Tissue growth and tumorigenesis in Drosophila: cell polarity and the Hippo pathway. Curr. Opin. Cell Biol. 48: 1–9. [DOI] [PubMed] [Google Scholar]
- Robinson B. S., Huang J., Hong Y., Moberg K. H., 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr. Biol. 20: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D., Rauskolb C., Irvine K. D., 2008. Morphogen control of wing growth through the fat signaling pathway. Dev. Cell 15: 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford D. B., Shaw R. J., 2009. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9: 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewan A., Eastburn D. J., Mostov K., 2011. Phosphoinositides in cell architecture. Cold Spring Harb. Perspect. Biol. 3: a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Li S., Li S., Jiang A., Chen Y., et al. , 2014. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc. Natl. Acad. Sci. USA 111: E5651–E5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Silva E., Clayton L., Gardano L., Barrios-Rodiles M., et al. , 2009. Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr. Biol. 19: 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Foos M., Vinayagam A., Zhai B., Binari R., et al. , 2014. Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in Drosophila embryos. Dev. Cell 31: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S., Diaz-Meco M. T., Caminero E., Moscat J., Campuzano S., 2004. DaPKC-dependent phosphorylation of crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Irvine K. D., 2011. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev. Biol. 350: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Verheyen E. M., 2011. Drosophila homeodomain-interacting protein kinase inhibits the Skp1-Cul1-F-box E3 ligase complex to dually promote Wingless and Hedgehog signaling. Proc. Natl. Acad. Sci. USA 108: 9887–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Pradhan-Sundd T., Verheyen E. M., 2015. Genome-wide identification of phospho-regulators of Wnt signaling in Drosophila. Development 142: 1502–1515. [DOI] [PubMed] [Google Scholar]
- Tepass U., 2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28: 655–685. [DOI] [PubMed] [Google Scholar]
- Tyler D. M., Baker N. E., 2007. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 305: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereshchagina N., Ramel M. C., Bitoun E., Wilson C., 2008. The protein phosphatase PP2A-B’ subunit Widerborst is a negative regulator of cytoplasmic activated Akt and lipid metabolism in Drosophila. J. Cell Sci. 121: 3383–3392. [DOI] [PubMed] [Google Scholar]
- Vissers J. H., Manning S. A., Kulkarni A., Harvey K. F., 2016. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat. Commun. 7: 10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Moya N., Niessen S., Hoover H., Mihaylova M. M., et al. , 2011. A hormone-dependent module regulating energy balance. Cell 145: 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M. C., Holder M. V., Gailite I., Saunders R. E., Maile T. M., et al. , 2013. Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat. Cell Biol. 15: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. S., Cui W., 2016. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 143: 3050–3060. [DOI] [PubMed] [Google Scholar]
- Zecca M., Struhl G., 2010. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 8: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Wharton K. A., Jr., Mack J. A., Wang K., Gadbaw M., et al. , 2000. Naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403: 789–795. [DOI] [PubMed] [Google Scholar]
- Zeng X., Singh S. R., Hou D., Hou S. X., 2010. Tumor suppressors Sav/Scrib and oncogene Ras regulate stem-cell transformation in adult Drosophila malpighian tubules. J. Cell. Physiol. 224: 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Xin T., Weng S., Gao Y., Zhang Y., et al. , 2010. Activation of JNK signaling links lgl mutations to disruption of the cell polarity and epithelial organization in Drosophila imaginal discs. Cell Res. 20: 242–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Drosophila stocks and antibodies are available upon request or from stock centers as listed in the Materials and Methods. File S1 contains four supplemental figures and seven supplemental tables.