Abstract
Epilepsy has many causes and comorbidities affecting as many as 4% of people in their lifetime. Both idiopathic and symptomatic epilepsies are highly heritable, but genetic factors are difficult to characterize among humans due to complex disease etiologies. Rodent genetic studies have been critical to the discovery of seizure susceptibility loci, including Kcnj10 mutations identified in both mouse and human cohorts. However, genetic analyses of epilepsy phenotypes in mice to date have been carried out as acute studies in seizure-naive animals or in Mendelian models of epilepsy, while humans with epilepsy have a history of recurrent seizures that also modify brain physiology. We have applied a repeated seizure model to a genetic reference population, following seizure susceptibility over a 36-d period. Initial differences in generalized seizure threshold among the Hybrid Mouse Diversity Panel (HMDP) were associated with a well-characterized seizure susceptibility locus found in mice: Seizure susceptibility 1. Remarkably, Szs1 influence diminished as subsequent induced seizures had diminishing latencies in certain HMDP strains. Administration of eight seizures, followed by an incubation period and an induced retest seizure, revealed novel associations within the calmodulin-binding transcription activator 1, Camta1. Using systems genetics, we have identified four candidate genes that are differentially expressed between seizure-sensitive and -resistant strains close to our novel Epileptogenesis susceptibility factor 1 (Esf1) locus that may act individually or as a coordinated response to the neuronal stress of seizures.
Keywords: genetics, epilepsy, preclinical model, neuronal plasticity, complex traits
Epilepsies in humans are highly heritable syndromes that are modified by complex interactions between genes and the environment. In the US, ∼2.2 million people will develop some form of epilepsy during their lifetime (England et al. 2012). Despite diverse treatment options, largely focused on seizure suppression, individuals with epilepsy suffer multiple quality of life issues, refractory seizures, comorbid complications, and a higher incidence of sudden death (Gupta et al. 2016). Understanding interactions between genetic seizure susceptibility loci and environmental stressors induced by repeated seizures will uncover novel mechanisms of epilepsy pathogenesis. This information will improve therapeutic options by focusing treatment on epileptogenesis (Ferraro 2016).
Identification of gene by environment interactions for individuals with epilepsy is limited by complex disease etiologies and the inability to identify at risk populations prior to disease onset. Consequently, the majority of epilepsy genome-wide association studies (GWAS) carried out to date compare affected populations to seizure-naive controls rather than following epilepsy progression in a population from a seizure-naive to an epileptic state (International League Against Epilepsy Consortium on Complex Epilepsies 2014). Experimental seizure paradigms can overcome such limitations to discover the genetic basis of epileptogenesis.
Preclinical epilepsy models have implicated multiple gene mutations as contributors to seizure disorders. This success is due in part to strong genetic influences that segregate among rodent models of seizure susceptibility (Engstrom and Woodbury 1988; Kosobud and Crabbe 1990). Unfortunately, most of these models utilized either acute seizure measurements or status epilepticus as quantitative measures of seizure susceptibility (Grone and Baraban 2015). Alternatively, animal models that measure epileptogenesis and other models of brain plasticity are typically designed to minimize genetic background effects (Grone and Baraban 2015). Notably, while numerous hyper-excitability factors are now known, genetic factors influencing epileptogenesis remain largely uncharacterized.
We have utilized a repeated-flurothyl-induced seizure model delivered to the Hybrid Mouse Diversity Panel (HMDP) to identify genetic susceptibility factors that modify epileptogenesis (Papandrea et al. 2009). Genetic differences in initial flurothyl-induced seizures were consistent with previous studies showing the strongest quantitative trait loci (QTL) mapping to the Szs1 locus (Ferraro et al. 1997, 2004; Chaix et al. 2007). Surprisingly, as additional seizures were administered to this population, the effects of Szs1 diminished, and genetic associations on distal chromosome 4 garnered influence over seizure susceptibility. We attribute the shift in susceptibility to seizure-related changes in brain plasticity that are modified by gene(s) that may mitigate the effects of Szs1. This hypothesis is supported by interactions between Szs1 and our novel epileptogenesis susceptibility locus on chromosome 4; Epileptogenesis susceptibility factor 1 (Esf1). Systems genetics analysis of the Esf1 region has uncovered a coregulated set of expression QTL (eQTL) that have been implicated in neurological stress responses that are now also implicated in epileptogenesis.
Materials and Methods
Animals
The rationale for using the HMDP and its advantages with respect to statistical power and genetic resolution have been previously described (Bennett et al. 2010). The inbred strains of mice that were used for this study include the following: Balb/cJ (Balb; n = 8); BTBR T+Ltpr3tf/J (BTBR; n = 6); BXD#/TyJ (30 strains, average n/strain ≤6); BXD##/RwwJ (18 strains, average n/strain ≤4); C57BL/6J (B6; n = 10); C57BL/10SNJ (10SNJ; n = 12); C57BL/10J (10J; n = 8); C57BL/6NJ (6NJ; n = 16); C57BLKS/J (KS/J; n = 15); CAST/EiJ (CAST; n = 6); CBA/J (CBA; n = 17); CE/J (CE; n = 11); DBA/2J (D2; n = 14); FVB/NJ (FVB; n = 12); LG/J (LG; n = 19); MRL/MpJ (MRL; n = 6); NOD/ShiLtJ (NOD; n = 13); NZW/LacJ (NZW; n = 10); PWD/PhJ (PWD; n = 9), and Sm/J (Sm; n = 13). Male and female mice were obtained from the Jackson Laboratories (Bar Harbor, ME) and (1) were acclimated to the animal facility for 1 wk before seizure testing commenced, or (2) were further bred at the Wadsworth Center and exposed to the repeated-flurothyl seizure model. Mice were housed on a 12 hr light–dark cycle with ad libitum access to food and water.
A [B6×D2] F2 cross was generated by intercrossing [B6×D2] F1 parents. Both male and female F2 mice were exposed to the repeated-flurothyl seizure model as described below (Papandrea et al. 2009; Kadiyala et al. 2016). Mice (n = 150) from this cohort were genotyped with the Mouse Universal Genotyping Array (http://genomics.neogen.com/en/mouse-universal-genotyping-array) and mapping (regression analysis) was performed using R/qtl (http://www.rqtl.org; Broman et al. 2003).
All testing was performed under approval of the Institutional Animal Care and Use Committees of the Wadsworth Center, Albany Medical College, and Rensselaer Polytechnic Institute in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Repeated-flurothyl seizure model
Six- to seven-week-old male and female mice were placed in a closed chamber and exposed to an inhaled 10% flurothyl solution [bis(2,2,2-trifluoroethyl) ether diluted in 95% ethanol] infused via a syringe pump onto a gauze pad suspended at the top of the chamber at a rate of 100 μl/min (Papandrea et al. 2009; Kadiyala et al. 2015; Ferland, 2017). Once a generalized seizure was observed, indicated by a loss of postural control, the top of the chamber was removed to stop flurothyl exposure and allowing the animal to recover. The latency to the loss of postural control (measured in seconds) was recorded as the generalized seizure threshold (GST). Mice received a single flurothyl-induced seizure once a day for 8 consecutive days during the induction phase. The induction phase was followed by a 28-d incubation phase where no induced seizures were administered. After the incubation phase, mice were given one additional flurothyl-induced retest seizure. The reduction in daily GST in certain HMDP strains over the induction phase (kindling) was quantified by plotting the eight induction phase GSTs over the seizure number and calculating the slope of these data.
Efficient Mixed-Model Association (EMMA)
EMMA is a statistical test for association mapping that can correct for genetic relatedness due to population structure (Bennett et al. 2010). The calculations for genome-wide false discovery rate of 5% in the HMDP is P < 4.4 × 10−6 (Bennett et al. 2010). The implementation of EMMA used here is available online at http://mouse.cs.ucla.edu/emma. All association studies were carried out with male seizure data. A limited set of female data for select HMDP strains were collected, but did not show sufficient differences from their male counterparts to warrant further study in a genome-wide analysis. These data are available upon request.
RNA extraction
Brains (n = 5 per group) were isolated from 11- to 12-wk-old B6 and D2 seizure-naive or seizure-exposed mice 90 min after completion of the repeated-flurothyl model as in Kadiyala et al. (2015). Brains were incubated in RNAlater at 4° overnight and subsequently stored at −20° until dissection. Cerebellum and hippocampus were dissected from whole brains, weighed, and processed with the RNeasy Lipid Tissue Mini Kit (Qiagen) using on-column DNAse digestions during mRNA purification. Total RNA concentrations were measured via Nanodrop and samples were aliquoted and stored at −80°.
cDNA synthesis
One microgram of total RNA was primed with random hexamers using the NEB Protoscript first strand cDNA synthesis kit per the manufacturer’s protocol. Ten-fold dilutions were prepared for quantitative PCR (qPCR).
Quantitative PCR
Relative RNA expression levels were quantified using the following Primetime premade 5′ nuclease assays from Integrated DNA Technologies: Actb (Loading control, NM_007393, exons 4–5, Mm.PT.58.33257376); Per3 (NM_011067, exons 4–6, Mm.PT.58.12973804); Park7 (NM_020569, exons 2–3, Mm.PT.58.7064397); Camta1 (NM_001081557, Exons 1–3, Mm.PT.58.15110314); and Vamp3 (NM_009498, exons 1–3, Mm.PT.58.32746859). Primer sequences are available upon request or from the manufacturer (www.idtdna.com/site/order/qpcr/predesignedassay). Real-time 5× HOT FirePol qPCR master mix purchased from Mango Biotechnology was used to perform PCR reactions according to the manufacturer’s recommendations (Solis BioDyne) using a 7500 Fast Real-Time PCR system (Applied Biosystems). All qPCR reactions were quantified using the relative standard curve approach based on the manufacturer’s recommendations. Candidate genes Camta1, Park7, Per3, and Vamp3 were normalized to β actin prior to determining B6:D2 expression ratios that exceeded a P value <0.05 assuming unequal variance between the populations.
Statistics
Statistical analyses evaluating genetic interactions between GSTs by trial were carried out by either one-way ANOVA or Student’s t-tests using JMP version 10.0 (SAS Institute). Groups were defined for interaction studies between Szs1 and Esf1 based on their genotype at single nucleotide polymorphisms (SNPs) rs8259388 for Szs1 and rs13478053 for Esf1. Correlational and eQTL mapping tools using BXD strains are freely available using GeneNetwork (http://www.genenetwork.org).
Candidate gene selection for Esf1
Cis-eQTLs adjacent to Esf1 were selected using the tools available on GeneNetwork. We initially used expression data from the UTHSC BXD Aged Hippocampus data generated from Affymetrix mouse gene 1.0 ST exon level arrays (dataset GN392, kindly made available by R. Williams) for correlational analyses with retest GST values. Additional datasets used to confirm these analyses included GN72 provided by Genome Explorations Inc. for the NIAAA as part of an SBIR grant to Dr. David Patel; GN46 that was generated by the UTHSC-SJCRH cerebellum transcriptome profiling consortium; and published datasets GN206 and GN281 (Overall et al. 2009; Mozhui et al. 2012). Candidate genes were chosen for correlational study based on their presence within a 2-Mb interval encompassing rs32163108 based on it having the highest significance for retest GST. Individual exons were interrogated for each gene in the surrounding 2-Mb interval using their relative expression among 68 of the 97 available BXD lines in this dataset. Expression values for individual exons from candidate loci were used in a mapping analysis with the PyLMM algorithm as implemented within GeneNetwork (Lippert et al. 2011). All exons having an SNP with a Likelihood Ratio Score (LRS) >10 within 20 Mb of rs32163108 were deemed to have cis effects for the expression change being interrogated. All genomic positions are listed relative to build GRCm38/mm10, except where noted. All exons showing differential expression were evaluated for SNPs that could impact hybridizations by interrogating the variant table for the respective gene using tools available at ensemble (http://www.ensembl.org) or the UTHSC Genome Browser (http://ucscbrowserbeta.genenetwork.org).
Data availability
All seizure threshold data are available from the GeneNetwork website under accession numbers 18963–18981 (http://www.genenetwork.org/).
Results
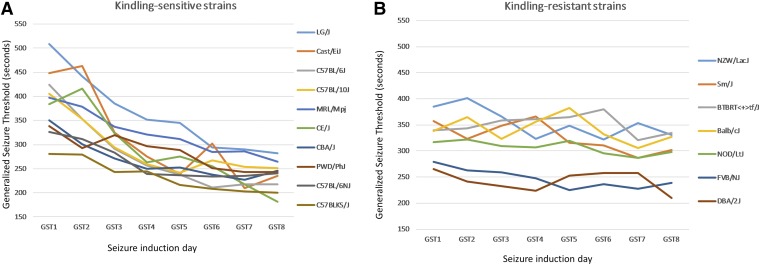
We previously reported that certain phenotypes quantified in the repeated-flurothyl seizure model are dissociable between B6, D2, and [B6×D2] F1 hybrids, suggesting that independent genetic factors mediated different aspects of seizure response (Papandrea et al. 2009a,b). Applying this repeated seizure model to a larger population of 17 inbred strains revealed three subgroups by plotting their daily GST response times over the eight induction trials (Figure 1). Most of the strains performed similar to B6 mice showing significant reductions in GST latency over the eight induction trials (i.e., kindling-sensitive strains, Figure 1A) (Papandrea et al. 2009). Others behaved like D2 mice with statistically indistinguishable GST latencies over induction trials (Figure 1B). These kindling-resistant strains further clustered into two distinct groups (P < 0.0001), based on their average daily GST scores of 275 sec for the most sensitive groups (D2 and FVB mice) compared to 350 sec for the intermediate groups (BTBR, Balb, NZW, and Sm mice, Figure 1B).
Figure 1.
Inbred strain survey of GST over eight flurothyl-induced seizures. Latency to the loss of postural control [generalized seizure threshold (GST)] among several inbred strains of mice of diverse ancestry (n ≥ 6 per strain, see Materials and Methods). (A) Kindling-sensitive inbred strains were identified by calculating the slope of their daily GST (seconds) plotted against the induction phase seizure and selecting strains with a score <−10. (B) Kindling-resistant strains were classified by having a slope with the same score >−10. Note the clustering of kindling-resistant strains into two distinct groups (P < 0.001).
We considered the possibility that the locus responsible for the separation in average GST between the kindling-resistant subgroups was due to Szs1; the strongest seizure susceptibility locus observed to date between B6 and D2 strains (Ferraro et al. 1997, 2004). However, haplotypes of kindling-resistant strains varied across the Szs1 interval [as defined by Ferraro et al. (2004)] with no obvious region of correlation when these strains are aligned using the mouse phylogeny viewer [data not shown (Yang et al. 2011)]. Notably, the missense mutation reported to be responsible for Szs1 effects in Kcnj10 (Buono et al. 2004) was present in all kindling-resistant strains with the exception of BTBR. The clustering of kindling-resistant strains into two groups further supports a multigene model for genetic susceptibility during repeated-flurothyl-induced epileptogenesis and indicates additional uncharacterized modifiers other than Szs1 are present in these strains.
Repeated seizures shift genetic susceptibility over induction trials
We evaluated an additional 58 HMDP strains (Ghazalpour et al. 2012) with repeated seizures. Adding BXD recombinant inbred lines increased statistical power, while also providing access to publicly available expression data on GeneNetwork (Williams and Mulligan 2012). Our intent was to determine if previously mapped acute seizure susceptibility QTL, identified among B6, D2, or BXD mouse strains (Neumann and Collins 1991; Ferraro et al. 1997), were shared with flurothyl-induced seizures and to determine if repeatedly induced seizures influenced these genetic associations.
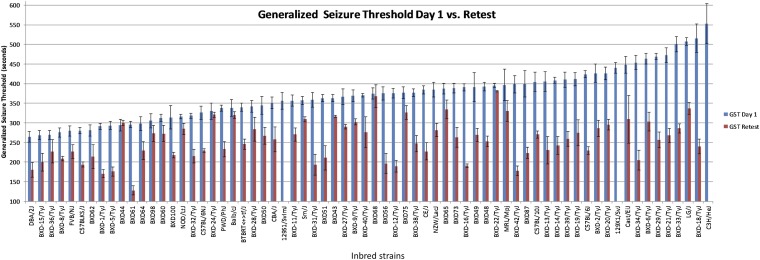
There were diverse phenotypic responses across HMDP strains for initial and subsequent GSTs over seizure inductions (Figure 2 and Table 1). Mice exposed to the repeated-flurothyl seizure model had reduced interstrain variance upon retest compared to initial GST, but showed strong heritability at all seizure time points (e.g., GST1 average: 375 sec; SD: 83.63 sec and retest GST average: 256 sec; SD 71 sec). We calculated the slope of induction phase GSTs (seizure trials 1–8) as a quantitative surrogate for the kindling effect seen previously (Samoriski and Applegate 1997). Kindling also had strong heritability and differences were highly significant (P < 0.0001) between strains (Table 1).
Figure 2.
HMDP responses to initial and retest seizures spanning the repeated-flurothyl seizure model. Latency to loss of postural control (GST) for 68 HMDP strains surveyed for our association analyses. Blue bars indicate the initial GST (day 1) average for each strain with error bars showing the SEM. Red bars indicate retest GST averages for the strain with error bars showing the SEM. GST values are given in seconds.
Table 1. GST values for HMDP strains over repeated-flurothyl seizures.
| Strain | GST1 Mean | GST1 SE | GST2 Mean | GST2 SE | GST3 Mean | GST3 SE | GST4 Mean | GST4 SE | GST5 Mean | GST5 SE | GST6 Mean | GST6 SE | GST7 Mean | GST7 SE | GST8 Mean | GST8 SE | GST Retest Mean | GST Retest SE | Kindling Mean | Kindling SE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 129S1/SvImJ | 356 | 21 | 410 | 10 | 425 | 28 | 379 | 18 | 368 | 23 | 335 | 29 | 312 | 11 | 364 | 5 | −9 | 1 | ||
| 129X1/SvJ | 440 | 14 | 406 | 20 | 394 | 21 | 405 | 50 | 325 | 43 | 298 | 39 | 368 | 12 | 394 | 48 | −9 | 6 | ||
| A/J | 397 | 57 | 519 | 7 | 402 | 98 | 351 | 91 | 421 | 99 | 425 | 153 | 422 | 110 | 586 | 68 | 621 | 21 | 4 | 31 |
| Balb/cJ | 338 | 22 | 365 | 23 | 323 | 35 | 356 | 24 | 382 | 15 | 332 | 21 | 306 | 22 | 327 | 25 | 320 | 8 | −5 | 3 |
| BTBRT<+>tf/J | 340 | 10 | 343 | 17 | 358 | 13 | 360 | 8 | 365 | 19 | 380 | 9 | 321 | 16 | 334 | 19 | 246 | 13 | −1 | 3 |
| BXD1/TyJ | 291 | 8 | 283 | 11 | 243 | 10 | 210 | 7 | 204 | 9 | 195 | 8 | 192 | 14 | 183 | 12 | 170 | 11 | −16 | 2 |
| BXD100/RwwJ | 314 | 29 | 300 | 11 | 282 | 7 | 274 | 11 | 241 | 15 | 240 | 11 | 214 | 18 | 205 | 17 | 218 | 7 | −16 | 4 |
| BXD11/TyJ | 357 | 14 | 321 | 10 | 325 | 9 | 292 | 12 | 270 | 7 | 260 | 16 | 245 | 11 | 248 | 5 | 270 | 17 | −17 | 2 |
| BXD12/TyJ | 375 | 12 | 289 | 8 | 281 | 9 | 257 | 8 | 228 | 9 | 187 | 5 | 238 | 8 | 220 | 10 | 189 | 15 | −20 | 2 |
| BXD13/TyJ | 405 | 25 | 367 | 8 | 312 | 14 | 298 | 15 | 232 | 16 | 217 | 16 | 175 | 12 | 234 | 16 | 231 | 35 | −30 | 2 |
| BXD14/TyJ | 409 | 7 | 384 | 18 | 275 | 10 | 311 | 18 | 247 | 7 | 277 | 15 | 255 | 12 | 234 | 22 | 243 | 22 | −22 | 2 |
| BXD15/TyJ | 269 | 12 | 264 | 12 | 247 | 17 | 203 | 16 | 202 | 15 | 209 | 12 | 207 | 13 | 218 | 22 | 200 | 23 | −9 | 2 |
| BXD16/TyJ | 391 | 9 | 307 | 9 | 298 | 14 | 258 | 13 | 253 | 13 | 238 | 9 | 230 | 16 | 224 | 12 | 191 | 4 | −21 | 3 |
| BXD18/TyJ | 516 | 36 | 399 | 28 | 379 | 37 | 324 | 9 | 291 | 14 | 262 | 13 | 243 | 4 | 254 | 14 | 240 | 19 | −36 | 3 |
| BXD19/TyJ | 411 | 17 | 423 | 50 | 327 | 30 | 306 | 10 | 320 | 36 | 299 | 22 | 265 | 30 | 314 | 38 | 275 | 33 | −18 | 6 |
| BXD2/TyJ | 426 | 23 | 350 | 10 | 304 | 13 | 330 | 18 | 317 | 22 | 314 | 21 | 317 | 20 | 305 | 27 | 286 | 21 | −9 | 3 |
| BXD20/TyJ | 426 | 16 | 330 | 18 | 292 | 18 | 289 | 20 | 281 | 20 | 273 | 16 | 262 | 13 | 267 | 19 | 296 | 13 | −18 | 2 |
| BXD21/TyJ | 473 | 18 | 315 | 14 | 258 | 5 | 235 | 10 | 259 | 10 | 238 | 15 | 263 | 15 | 265 | 10 | 268 | 18 | −21 | 2 |
| BXD22/TyJ | 395 | 5 | 370 | 7 | 331 | 5 | 346 | 12 | 254 | 36 | 339 | 19 | 326 | 7 | 291 | 11 | 381 | 2 | −12 | 1 |
| BXD24/TyJ | 330 | 21 | 304 | 12 | 266 | 22 | 299 | 13 | 303 | 9 | 303 | 12 | 286 | 8 | 265 | 12 | 321 | 7 | −5 | 3 |
| BXD27/TyJ | 366 | 20 | 361 | 15 | 326 | 8 | 320 | 12 | 304 | 12 | 310 | 15 | 284 | 9 | 285 | 9 | 290 | 6 | −12 | 2 |
| BXD28/TyJ | 342 | 15 | 363 | 17 | 296 | 11 | 280 | 18 | 268 | 18 | 253 | 9 | 232 | 17 | 247 | 8 | 284 | 30 | −17 | 2 |
| BXD29/TyJ | 469 | 8 | 394 | 19 | 352 | 19 | 331 | 9 | 325 | 13 | 290 | 28 | 260 | 24 | 270 | 22 | 257 | 20 | −27 | 2 |
| BXD31/TyJ | 359 | 19 | 295 | 12 | 247 | 9 | 209 | 17 | 198 | 17 | 197 | 15 | 178 | 17 | 168 | 21 | 193 | 27 | −25 | 4 |
| BXD32/TyJ | 318 | 7 | 265 | 11 | 268 | 9 | 288 | 15 | 265 | 17 | 259 | 6 | 257 | 9 | 256 | 20 | 216 | 17 | −6 | 2 |
| BXD33/TyJ | 501 | 19 | 398 | 19 | 309 | 16 | 262 | 15 | 233 | 7 | 216 | 10 | 217 | 9 | 221 | 8 | 286 | 12 | −38 | 3 |
| BXD34/TyJ | 454 | 18 | 285 | 16 | 258 | 3 | 229 | 12 | 220 | 11 | 219 | 12 | 234 | 13 | 233 | 13 | 205 | 25 | −23 | 2 |
| BXD36/TyJ | 269 | 12 | 227 | 10 | 190 | 10 | 196 | 5 | 202 | 8 | 215 | 10 | 222 | 6 | 218 | 13 | 227 | 30 | −4 | 2 |
| BXD38/TyJ | 377 | 10 | 338 | 10 | 268 | 10 | 234 | 8 | 217 | 6 | 219 | 15 | 193 | 18 | 172 | 11 | 247 | 21 | −28 | 2 |
| BXD39/TyJ | 410 | 19 | 377 | 13 | 363 | 14 | 305 | 13 | 320 | 10 | 335 | 16 | 332 | 14 | 340 | 13 | 259 | 19 | −9 | 2 |
| BXD40/TyJ | 371 | 5 | 324 | 10 | 262 | 9 | 236 | 17 | 179 | 14 | 193 | 18 | 212 | 20 | 199 | 16 | 276 | 40 | −24 | 2 |
| BXD42/TyJ | 398 | 22 | 314 | 14 | 284 | 16 | 264 | 12 | 249 | 17 | 225 | 10 | 248 | 11 | 228 | 11 | 178 | 13 | −20 | 3 |
| BXD43/RwwJ | 363 | 9 | 325 | 41 | 352 | 13 | 349 | 31 | 283 | 10 | 289 | 16 | 264 | 32 | 292 | 12 | 317 | 3 | −13 | 3 |
| BXD44/RwwJ | 295 | 15 | 279 | 14 | 274 | 14 | 259 | 17 | 264 | 25 | 245 | 19 | 210 | 10 | 207 | 10 | 300 | 7 | −12 | 2 |
| BXD48/RwwJ | 393 | 11 | 354 | 25 | 345 | 17 | 345 | 21 | 343 | 20 | 331 | 25 | 235 | 9 | 251 | 16 | 253 | 13 | −19 | 3 |
| BXD49/RwwJ | 391 | 37 | 289 | 17 | 288 | 18 | 286 | 27 | 288 | 10 | 261 | 18 | 283 | 17 | 259 | 20 | 269 | 17 | −12 | 3 |
| BXD5/TyJ | 293 | 10 | 263 | 14 | 243 | 11 | 222 | 5 | 242 | 11 | 229 | 8 | 229 | 11 | 226 | 15 | 176 | 12 | −10 | 2 |
| BXD50/RwwJ | 345 | 23 | 308 | 20 | 214 | 18 | 243 | 9 | 252 | 10 | 256 | 18 | 248 | 23 | 236 | 12 | 268 | 21 | −11 | 3 |
| BXD51/RwwJ | 363 | 9 | 309 | 16 | 265 | 39 | 220 | 24 | 207 | 25 | 200 | 1 | 228 | 15 | 254 | 23 | 211 | 31 | −16 | 2 |
| BXD55/RwwJ | 422 | 99 | 388 | 23 | 208 | 16 | 246 | 40 | 208 | 19 | 218 | 7 | 188 | 12 | 191 | 5 | 334 | 111 | −31 | 6 |
| BXD56/RwwJ | 375 | 17 | 345 | 28 | 312 | 25 | 297 | 19 | 198 | 30 | 301 | 25 | 170 | 40 | 243 | 21 | 196 | 26 | −23 | 3 |
| BXD6/TyJ | 463 | 13 | 442 | 44 | 436 | 48 | 377 | 38 | 367 | 43 | 287 | 22 | 263 | 8 | 241 | 10 | 303 | 24 | −16 | 5 |
| BXD60/RwwJ | 313 | 9 | 306 | 19 | 229 | 15 | 271 | 18 | 281 | 10 | 309 | 12 | 247 | 22 | 211 | 13 | 273 | 21 | −9 | 2 |
| BXD61/RwwJ | 296 | 8 | 268 | 10 | 253 | 16 | 261 | 23 | 251 | 22 | 204 | 4 | 197 | 34 | 223 | 28 | 128 | 12 | −12 | 2 |
| BXD62/RwwJ | 281 | 14 | 309 | 18 | 316 | 16 | 261 | 12 | 256 | 1 | 269 | 5 | 240 | 24 | 241 | 23 | 214 | 30 | −9 | 4 |
| BXD64/RwwJ | 298 | 17 | 267 | 32 | 285 | 17 | 283 | 12 | 279 | 21 | 222 | 23 | 158 | 27 | 215 | 3 | 229 | 23 | −16 | 4 |
| BXD65/RwwJ | 387 | 14 | 356 | 9 | 407 | 23 | 356 | 12 | 366 | 23 | 318 | 22 | 295 | 14 | 319 | 16 | 335 | 24 | −12 | 1 |
| BXD68/RwwJ | 375 | 14 | 337 | 16 | 334 | 12 | 359 | 10 | 310 | 23 | 316 | 15 | 356 | 12 | 328 | 34 | 368 | 30 | −4 | 4 |
| BXD73/RwwJ | 389 | 12 | 322 | 22 | 277 | 24 | 273 | 8 | 233 | 15 | 262 | 14 | 238 | 11 | 219 | 9 | 263 | 25 | −20 | 3 |
| BXD75/RwwJ | 377 | 15 | 310 | 29 | 322 | 23 | 315 | 15 | 326 | 20 | 268 | 31 | 303 | 10 | 348 | 17 | 326 | 18 | −5 | 4 |
| BXD8/TyJ | 276 | 12 | 250 | 14 | 207 | 14 | 229 | 22 | 194 | 12 | 188 | 13 | 188 | 14 | 178 | 7 | 209 | 5 | −13 | 2 |
| BXD87/RwwJ | 400 | 34 | 287 | 12 | 288 | 33 | 220 | 15 | 229 | 14 | 220 | 19 | 212 | 10 | 200 | 11 | 224 | 14 | −23 | 4 |
| BXD9/TyJ | 370 | 15 | 346 | 12 | 323 | 12 | 304 | 13 | 283 | 11 | 234 | 13 | 238 | 14 | 220 | 12 | 303 | 8 | −21 | 2 |
| BXD98/RwwJ | 306 | 18 | 310 | 29 | 234 | 17 | 266 | 17 | 250 | 10 | 234 | 9 | 251 | 19 | 219 | 10 | 273 | 21 | −11 | 2 |
| C3H/HeJ | 553 | 51 | 473 | 87 | 575 | 330 | 241 | 213 | 297 | 392 | −35 | |||||||||
| C57BL/10J | 405 | 24 | 352 | 20 | 294 | 14 | 259 | 12 | 242 | 11 | 267 | 9 | 254 | 11 | 252 | 9 | 271 | 9 | −19 | 3 |
| C57BL/6J | 424 | 9 | 353 | 9 | 291 | 6 | 256 | 6 | 238 | 10 | 211 | 6 | 217 | 9 | 218 | 7 | 229 | 10 | −28 | 1 |
| C57BL/6NJ | 326 | 17 | 311 | 14 | 284 | 11 | 239 | 12 | 237 | 10 | 234 | 9 | 235 | 7 | 241 | 15 | 230 | 5 | −13 | 3 |
| C57BLKS/J | 281 | 7 | 280 | 17 | 243 | 7 | 244 | 16 | 216 | 6 | 209 | 6 | 203 | 6 | 201 | 9 | 194 | 6 | −13 | 1 |
| Cast/EiJ | 448 | 21 | 463 | 45 | 325 | 24 | 275 | 28 | 240 | 32 | 303 | 50 | 210 | 20 | 235 | 23 | 309 | 61 | −34 | 4 |
| CBA/J | 351 | 15 | 301 | 15 | 272 | 19 | 250 | 16 | 252 | 16 | 238 | 15 | 227 | 17 | 245 | 16 | 258 | 32 | −15 | 2 |
| CE/J | 384 | 10 | 416 | 22 | 323 | 17 | 263 | 14 | 275 | 13 | 255 | 33 | 217 | 17 | 182 | 19 | 228 | 22 | −31 | 4 |
| DBA/2J | 265 | 13 | 241 | 11 | 233 | 9 | 224 | 12 | 252 | 12 | 258 | 17 | 257 | 14 | 210 | 19 | 180 | 19 | −4 | 3 |
| FVB/NJ | 280 | 13 | 263 | 14 | 259 | 10 | 248 | 10 | 225 | 11 | 236 | 9 | 227 | 11 | 239 | 9 | 227 | 17 | −7 | 3 |
| LG/J | 508 | 10 | 441 | 19 | 385 | 18 | 352 | 12 | 345 | 20 | 294 | 7 | 290 | 9 | 282 | 8 | 337 | 15 | −32 | 2 |
| MRL/Mpj | 397 | 40 | 379 | 21 | 337 | 15 | 321 | 23 | 311 | 27 | 285 | 12 | 287 | 24 | 265 | 6 | 331 | 26 | −18 | 3 |
| NOD/LtJ | 317 | 6 | 322 | 9 | 309 | 11 | 307 | 15 | 320 | 12 | 295 | 16 | 287 | 15 | 298 | 17 | 285 | 14 | −6 | 3 |
| NZW/LacJ | 385 | 19 | 401 | 20 | 366 | 25 | 323 | 19 | 348 | 17 | 322 | 11 | 354 | 11 | 331 | 9 | 282 | 16 | −9 | 2 |
| PWD/PhJ | 338 | 8 | 293 | 30 | 320 | 10 | 297 | 18 | 289 | 9 | 251 | 13 | 243 | 14 | 243 | 18 | 233 | 19 | −16 | 4 |
| Sm/J | 357 | 10 | 324 | 20 | 348 | 8 | 367 | 15 | 316 | 11 | 310 | 10 | 286 | 9 | 302 | 11 | 309 | 6 | −8 | 3 |
| Average | 373 | 18 | 337 | 19 | 304 | 17 | 286 | 17 | 271 | 18 | 263 | 17 | 252 | 16 | 257 | 16 | 262 | 20 | −16 | 3 |
| h2 | 0.55 | 0.71 | 0.54 | 0.75 | 0.70 | 0.79 | 0.62 | 0.36 | 0.50 | 0.94 |
Approximately 4% of HMDP strains responded to flurothyl-induced seizures with high incidences of severe myoclonic seizures leading to brainstem seizures (Papandrea et al. 2009). These mice have pronounced jerking seizure incidents but do not lose postural control, which was our phenotypic criterion for GST onset. Such responses typically lead to increased flurothyl dosing while waiting for the animal to have a clonic seizure often leading directly to brainstem seizures (Papandrea et al. 2009). Any strains with >25% of animals exhibiting brainstem seizures on their 1st or 2nd trial were excluded from GWAS studies.
Initial flurothyl-induced GSTs are consistent with previous models of seizure susceptibility
We used association analysis, correcting for genetic relatedness between strains with EMMA (Bennett et al. 2010). Daily GST response differences (in seconds) among HMDP mice were tested for association with 105 SNPs having minor allele frequencies >0.05. Previous studies have established genome-wide significance for HMDP studies at 4.1 × 10−6 (Bennett et al. 2010).
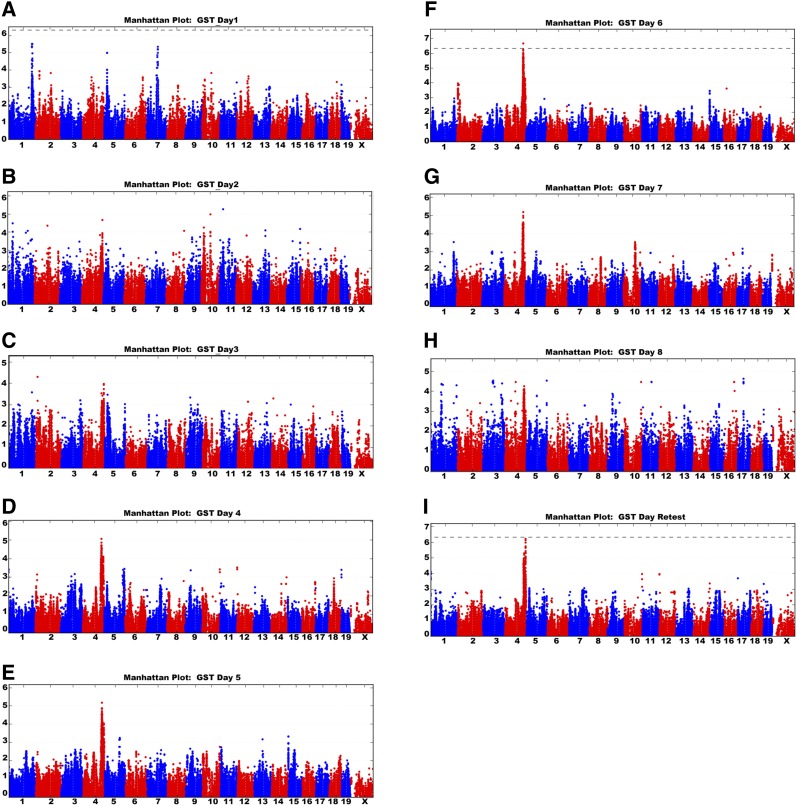
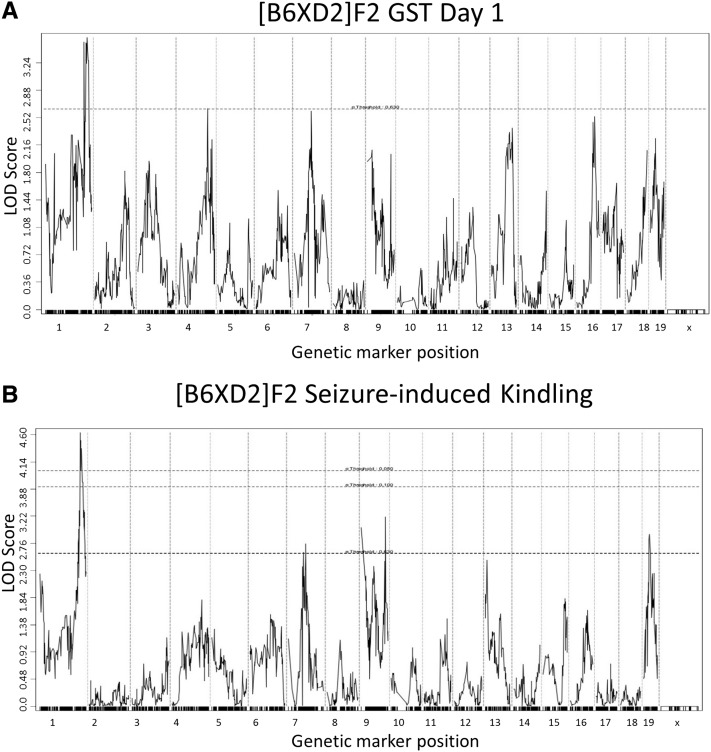
Day 1 GST values among HMDP strains were associated with SNPs on mouse chromosome 1 that overlapped with the previously designated QTL Szs1 (Ferraro et al. 2004; Chaix et al. 2007). However, this QTL, in addition to another on chromosome 5 near Szs6 and a novel locus on mouse chromosome 7, did not reach the genome-wide significance cutoff (Figure 3 and Table 2). A [B6×D2] F2 intercross was also exposed to the repeated-flurothyl seizure model that was genotyped with the Mouse Universal Genotyping Array (Morgan et al. 2015). Multiple markers closely linked to the Szs1 locus reached a suggestive genome-wide significance in this independent cross achieving a maximal LOD of 3.47 (Figure 4) confirming the similarities between our approach and alternative seizure studies.
Figure 3.
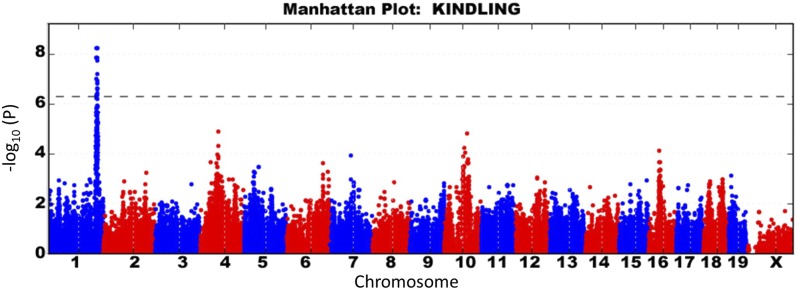
Genome-wide association results in the HMDP demonstrate a shift in seizure susceptibility over repeated-flurothyl-induced seizures. Manhattan plots for the seizure susceptibility associations over repeated seizures. (A) Acute seizure susceptibility associated with multiple suggestive associations >10−4 including chromosomes 1, 5, and 7. (B–E) Manhattan plots of day 2 to day 5 GST scores variance show a progression of associations to Distal Chromosome 4. (F) By the 6th induction seizure (day 6), associations exceeding genome-wide significance are detected on distal chromosome 4 near 139 Mb (GRCm38). (G and H) Manhattan plots of day 7 and day 8 GST variance. (I) Distal chromosome 4 associations persist until a retest seizure (given 4 wk after the induction period) with the five most significant SNP associations occurring in Camta1 (151 Mb). Dotted line represents genome-wide significance (P < 4.4 × 10−6).
Table 2. Top five SNP associations for kindling, GST1, GST6, and retest GST.
| Phenotype | Position (bp)a | −logP | SNP ID | M.A.F.b | Flanking Genes Within 500 kb of SNPc |
|---|---|---|---|---|---|
| Kindling | 1:174420753 | 5.86E−09 | rs3707910 | 0.1433 | Igfs8, Atp1a2, Kcnj9, Kcnj10, Pigm, Igsf9, Slamf9, Tafln2, Cfap45, Slamf8, Fcrl6, Dusp23, Crp |
| 1:175151112 | 5.86E−09 | rs8242481 | 0.182 | Olfr1408, Olfr1406, Olfr218, Mptx2, Olfr1404, Olfr418, Ackr1, Cadm3, Aim2, Gm4995 | |
| 1: 174797513 | 6.06E−09 | rs30463665 | 0.25 | Dusp23, Crp, Apcs, SNORA17, Olfr16 | |
| 1: 176533310 | 1.40E−08 | rs2020486 | 0.22 | Olfr417, Olfr248, Olfr414, olfr220, Fmn2 | |
| 1: 174239958 | 1.41E−08 | rs30472229 | 0.25 | Copa, Pex19, Dcaf8, Pea15a, Casaq1, Igfs8, Atp1a2, Kcnj9, Kcnj10, Pigm | |
| GST day 1 | 1: 174420753 | 3.15E−06 | rs3707910 | 0.143 | Igfs8, Atp1a2, Kcnj9, Kcnj10, Pigm, Igsf9, Slamf9, Tagln2, Cfap45, Slamf8, Fcrl6, Dusp23 |
| 1: 175151112 | 3.15E−06 | rs8242481 | 0.182 | Olfr16, Olfr1408, Olfr1406, Olfr218, Mptx2, Olfr1404, Olfr418, Ackr1, Cadm3, Aim2 | |
| 1: 174455423 | 4.22E−06 | rs31561177 | 0.4 | Igfs8, Atp1a2, Kcnj9, Kcnj10, Pigm, Igsf9, Slamf9, Tafln2, Cfap45, Slamf8, Fcrl6, Dusp23, Crp | |
| 7: 87409991 | 4.61E−06 | rs4226715 | 0.2 | ###Rik, Zfp710, idh2, Cib1, Ttll13, Ngrn, Rccd1, Prc1, Unc45a, Vps33b, Man2a2, Fes, Furin, Blm | |
| 1: 174239958 | 4.98E−06 | rs30472229 | 0.25 | Ncstn, Copa, Pex19, Dcaf8, Pea15a, Casaq1, Igfs8, Atp1a2, Kcnj9, Kcnj10, Pigm, Igsf9, Slamf9 | |
| GST day 6 | 4: 138646868 | 2.29E−07 | rs32914632 | 0.2 | Otud3, Pla2g2a, Pla2g2e, Rnf186, Tmco4, Htr6, Nbl1, Minos, Capzb, Pqlc2 |
| 4: 138129729 | 5.93E−07 | rs32007081 | 0.25 | Cda, Fam43b, Mul1, ABO41806, Camk2n1, Vwa5b1, Ubxn10, Pla2g2c, Pla2g25 | |
| 4: 137042287 | 6.80E−07 | rs27577055 | 0.25 | Wnt4, Cdc42, Cela3a, Cela 3b, Hspg2, Ldlrad2, ####Rik, Usp48, Rap1gap | |
| 4: 137353761 | 9.71E−07 | rs8256572 | 0.33 | Ldlrad2, ####Rik, Usp48, Rap1gap, Rap1gapcs, Alpl, Ece1, Eif4g3 | |
| 4: 138291047 | 1.13E−06 | rs27575102 | 0.4 | ABO41806, Camk2n1, Vwa5b1, Ubxn10, Pla2g2c, pla2g25, Otud3, Pla2g2a | |
| GST retest | 4: 150729074 | 6.53E−07 | rs32163108 | 0.4 | Camta1 |
| 4: 150886138 | 7.71E−07 | rs32757832 | 0.25 | Camta1 | |
| 4: 150659779 | 1.11E−06 | rs13478053 | 0.5 | Vamp3, Camta1 | |
| 4: 150681054 | 1.97E−06 | rs32590839 | 0.667 | Uts2, Per3, Vamp3, Camta1 | |
| 4: 150745317 | 2.26E−06 | rs32180816 | 0.5 | Camta1 |
Position relative to NCBI Build 37 genome assembly.
Minor allele frequency.
Genes in bold are considered the best candidates for influence over trait.
Figure 4.
Regression analysis of [B6×D2] F2 intercross. (A) GST1 susceptibility was used as a quantitative trait to confirm the presence of a distal suggestive QTL corresponding to Szs1. (B) The slope of induction phase GSTs was used as a quantitative measure of kindling that also identified the Szs1 locus.
GWAS analysis performed with day 2 GST data greatly diminished significance associations across the genome causing drops in associations observed on the previous day (Figure 3). By day 3, the genome-wide level of significant associations continued to decline compared to day 1 GST associations, as novel SNP associations increased on distal chromosome 4. The nonsignificant chromosome 4 associations persisted in subsequent induction phase GST Manhattan plots (Figure 3) achieving genome-wide significance on day 6 of the induction phase (Figure 3).
We hypothesized that QTL effects over the eight induction trials were dynamic because of homeostatic plasticity responses to induced seizures (Turrigiano and Nelson 2004). Repeated seizures exacerbated environmental variance limiting our ability to detect significant QTL in early induction trials. As the CNS adapted to the heightened state of excitability that was elicited by the induction phase seizures, a window of significant associations was revealed in the latter half of this period.
Seizure-induced kindling effects are also associated with Szs1
To better understand the genetic basis for the seizure-induced kindling phenomenon, we calculated the slope of induction phase GSTs over 8 d to evaluate kindling effects as a quantitative trait (Table 1). In this manner, we hoped to minimize day to day environmental influence on seizure susceptibility while enhancing the genetic effects in seizure-induced brain plasticity. Association analysis of the kindling effect revealed SNPs that once again overlapped with the Szs1 locus (Figure 5 and Table 2), but this time exceeding genome-wide significance (maximum −log p 5.86 e−09). The effects of Szs1 on kindling were also confirmed in our [B6×D2] F2 cross, achieving a maximum genome-wide significance of P < 0.05 at a LOD of 4.60 (Figure 4).
Figure 5.
Genome-wide association results in the HMDP for flurothyl kindling. Using the slope of induction phase GSTs as a measure of kindling, distal chromosome 1 associations, overlapping with Szs1, were identified as the strongest genetic effect. Dotted line represents genome-wide significance (P < 4.4 × 10−6).
Completion of the repeated-flurothyl seizure model reveals novel associations on chromosome 4
Like flurothyl, repeated doses of kainic acid induce both short-term and long-term effects on epileptogenesis in mice (Tse et al. 2014). We have demonstrated that eight flurothyl-induced generalized seizures led to a transient spontaneous seizure state similar to kainic acid epileptogenesis (Kadiyala et al. 2016), but unlike kainic acid, flurothyl induces no obvious neuronal death and is a generalized seizure model (Kadiyala et al. 2016; Kadiyala and Ferland 2017). To understand how such generalized spontaneous seizures could further influence genetic susceptibility, we left flurothyl-exposed mice undisturbed for 28 d, retesting their seizure thresholds with an additional flurothyl-induced seizure on the final day (i.e., retest seizure). As shown in Figure 3, retest GST was associated with significant SNP associations in the distal portion of chromosome 4, with the top five SNPs residing within the Calmodulin Binding Transcription Activator 1 (Camta1) gene (Figure 3 and Table 2).
Chromosome 4 GWAS associations were detected at two distinct time points and locations during the repeated-flurothyl seizure model. We first detected a locus with maximal effect (−log p 2.29 e−07) at rs32914632 near 138 Mb on chromosome 4 in the 6-d GST data (Figure 3 and Table 2), while retest GST had associations distal to this region that were highest (−log p 6.53 e−07) at rs32163108 near 150 Mb on chromosome 4 (Figure 3 and Table 2). While the day 6 association may overlap with the previously identified β-carboline-induced seizure QTL Bis1, congenic analyses performed by others in conjunction with our mapping results exclude the retest association from overlap with this locus (Zerr et al. 2000; B. Martin, personal communication). Based on this information, we designate the retest GST association as Epileptogenesis susceptibility factor 1 (Esf1); a novel seizure susceptibility locus that is dependent on prior seizure exposure to manifest its effects.
The D2 allele of the Szs1 locus influences Esf1 effects
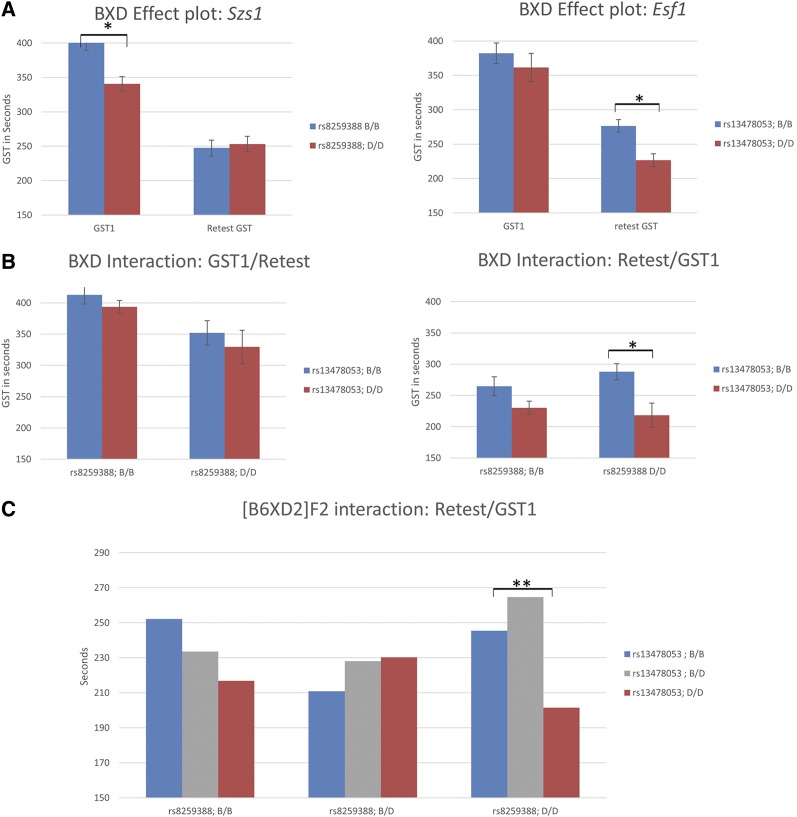
We considered the possibility that the temporal position of Szs1 and Esf1 effects in our model may indicate an interaction between these loci driven by repeated seizures. We tested for interdependency between the two genetic states by segregating BXD strains based on their genotype at Szs1 (i.e., B6 or D2 allele at rs8259388) to compare respective GSTs at either end of the seizure protocol (GST1 and retest GST). As expected, the average response between B6 and D2 Szs1 groupings was significantly different for GST1 (P = 0.001), but not for retest GST (Figure 6). Conversely, comparisons using Esf1 SNP rs13478053 to sort B6 vs. D2 alleles in BXDs showed significant differences in retest GST (P < 0.001), but not GST1 response. Notably, when the B6 and D2 Szs1 subsets were further divided based on their genotypes at Esf1, we only observed significant differences between the D2-Szs1/B6-Esf1 compared to the D2-Szs1/D2-Esf1 subset (Figure 6). One possible explanation for this outcome is that the D2-Szs1 haplotype potentiates the effects of Esf1 in mice undergoing repeated seizures. These effects were also observed in our [B6×D2] F2 cohort (Figure 6C) confirming the dependencies between the opposite ends of our seizure paradigm.
Figure 6.
Interaction between Szs1/GST1 and Esf1/retest GST. (A) BXD strains were sorted according to their genotype for rs8259388 within the Szs1 locus and rs13478053 within Esf1 to compare their effects on their respective phenotypes. Significant differences in response between the groups are indicated by an asterisk. (B) The Szs1 subgroups were further divided according to their Esf1 genotype to compare the average retest GST in these four groups. Significant differences (P < 0.05 marked by an asterisk) in retest GST were seen between B6 and D2 mice carrying the D2 allele of Szs1, but not the B6 allele. (C) Similar sorting for the two loci was performed in a [B6×D2] F2 intercross to reveal a similar pattern of significance between B6 and D2 homozygous animals, but not heterozygotes.
Genes tightly linked to Esf1 are influenced by cis-eQTL in BXD mice
Legacy data generated on several collections of recombinant inbred lines and genetic reference populations are available on the GeneNetwork systems genetics resource website (Williams and Mulligan 2012). Among these data, RNA expression profiles have compared gene expression levels between BXD strains for several brains regions (Chesler et al. 2005). Mapping expression differences as quantitative traits can reveal genetically controlled expression QTL (eQTL), a subset of which map to the same location as the differentially expressed gene (i.e., cis-eQTL) (Mortazavi et al. 2008; Wang et al. 2012). We exploited this resource as a first-pass evaluation of candidate loci in the vicinity of Esf1.
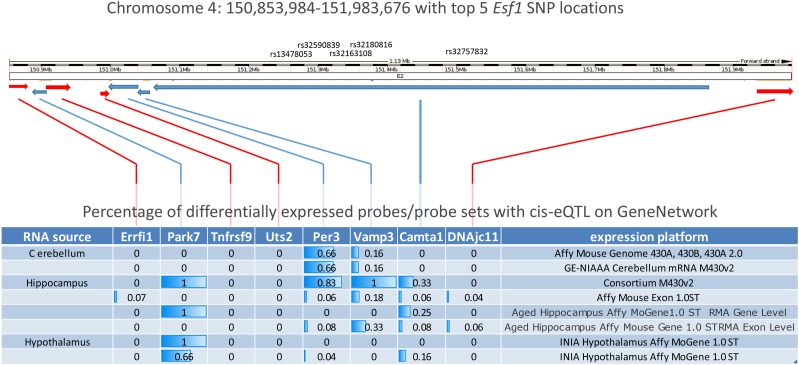
We interrogated mouse hippocampus expression data available on GeneNetwork comparing the relative expression among BXD strains subjected to the repeated-flurothyl seizure model. Expression differences between BXD strains were observed in genes across the Esf1 interval (Table 3). Among these expressed differences, multiple cis-eQTLs were detected. A cluster of probe sets with highly significant LRS (>25) were detected within four genes: Camta1, Period 3 (Per 3), Parkinson protein 7 (Park 7), and Vesicle Associated Membrane Protein 3 (Vamp3) (Figure 7 and Table 3). These cis-eQTLs attained the highest significance in the distal portion of the Camta1 gene directly adjacent to the region with our top SNP associations for Esf1 (Figure 7).
Table 3. eQTL whose expression correlates with retest GST around Esf1.
| SNP ID | Positiona | Transcript Influencedb | LRS | R2c |
|---|---|---|---|---|
| D4Mit343 | 142711337 | Vamp3(3) | 21 | −0.21 |
| rs13478031 | 144528745 | Errfi1(1) | 14 | 0.03 |
| rs3669806 | 148822719 | Nol9(5) | 9 | −0.15 |
| rs3697097 | 150397103 | Per3 (2) | 30 | 0.38 |
| rs3697097 | 150397103 | Camta1(7) | 52 | 0.61 |
| rs3697097 | 150397103 | Camta1(10) | 72 | 0.64 |
| rs4140148 | 150476099 | Park7(2) | 46 | 0.26 |
| rs6358921 | 150476099 | Vamp3(3) | 21 | −0.21 |
| rs6358921 | 150476099 | Vamp3(4) | 28 | −0.31 |
| rs3680006 | 151497797 | Tnfrsf25(7,8) | 10 | 0.18 |
| rs13478063 | 153325361 | Camta1(9) | 10 | 0.37 |
| rs32463773 | 156010835 | Camta1(13) | 13 | 0.22 |
| rs6279100 | 156183528 | Per3(6) | 10 | −0.07 |
Position relative to NCBI Build 38 genome assembly.
Gene symbol (exon containing probe/probe set).
Correlations with retest GST in bold P < 0.05; underlined bold P < 0.0001.
Figure 7.
Summary figure of Esf1 candidate loci cis-eQTL. Schematic of the genomic interval surrounding Esf1 including the approximate location of top SNPs from GWAS analysis (top) and the relative size and orientation of genes in the vicinity (red arrows indicate plus strand, blue arrows indicate minus strand) adapted from the Ensembl website (GRCm38). Table showing percentage of probes sets showing cis-eQTL/candidate gene from various datasets available on the GeneNetwork systems genetics resource. Note predominance of cis-eQTL on genes on the minus strand.
To better define the relationship between these cis-eQTL and Esf1, we correlated the expression values for each probe set with retest GST values. Most probe sets with LRS over 25 were significantly correlated (P < 0.05) with retest GST (Table 3). Exons from Vamp3 showed negative correlations with Esf1, while those for Camta1, Per3, and Park7 had positive correlations (Table 3). The most significant correlations (P < 0.0001) were observed in two Camta1 probes present in exons 7 and 10 of Camta1 (Table 3).
Similar cis-eQTLs were found in other expression datasets using distinct arrays and brain tissues (Figure 7). The percentage of probes within candidate genes showing cis-eQTL (selected based on a LRS score of 15 or greater within 2 Mb of a given locus) trended toward tissue-specific patterns. For example, while Camta1 and Park7 had cis-eQTL in hypothalamus (Figure 7, Geo accession GSE36674), Vamp3 and Per3 eQTL were more likely to be detected in cerebellum. Hippocampal cis-eQTLs were detected for all four loci in the region, further supporting a locally controlled difference in transcription that is differentially regulated between B6 and D2 animals.
We confirmed the presence of differential expression between B6 and D2 inbred strains using qPCR. We compared control cerebellum and hippocampus from age-matched B6 and D2 animals as well as similar sets that had undergone the full flurothyl induction protocol, collecting tissue 90 min after the last seizure. As shown in Table 4, significant differences in expression (P < 0.05) were observed between B6 and D2 hippocampus in the seizure-naive set for Camta1, Park7, and Per3, but not in cerebellar mRNA. In mice undergoing repeated-flurothyl seizures, there was an induction of Park7 and Vamp3 in B6 cerebellum that did not occur in D2 cerebellum. However, only Camta1 expression in hippocampus was significantly and differentially expressed between B6 and D2 mice after seizures.
Table 4. Quantitative PCR of expression levels in cis-eQTL genes in B6 and D2 brain tissues.
| Preseizure Exposure | Postseizure Exposure | ||||||
|---|---|---|---|---|---|---|---|
| Tissue | Gene | B6/D2 Ratio | Expression B6/D2a | P value | B6/D2 Ratio | Expression B6/D2a | P value |
| Cerebellum | Camta1 | 0.82 | 0.92(0.1)/1.18(1.1) | 0.9508 | 0.83 | 1.0(0.14)/1.2(0.17) | 0.7964 |
| Hippocampusb | Camta1 | 2.08 | 0.54(0.07)/0.28(0.02) | 0.0019 | 1.96 | 0.49(0.34)/0.25(0.08) | 0.0434 |
| Cerebellum | Park7 | 0.53 | 0.33(0.03)/0.62(0.16) | 0.9246 | 1.31 | 0.67(0.11)/0.51(0.19) | 0.1243 |
| Hippocampus | Park7 | 1.63 | 0.36(0.06)/0.19(0.03) | 0.0317 | 1.84 | 0.31(0.23)/0.53(0.28) | 0.0903 |
| Cerebellum | Per3 | 1.32 | 1.25(0.19)/0.95(0.12) | 0.1172 | 1.14 | 1.04(0.25)/0.91(0.16) | 0.344 |
| Hippocampus | Per3 | 2.10 | 0.86(0.15)/0.41(0.07) | 0.0196 | 0.55 | 0.6(0.07)/0.54(0.19) | 0.9797 |
| Cerebellum | Vamp3 | 0.46 | 0.51(0.02)/1.23(0.43) | 0.9051 | 1.40 | 1.25(0.37)/0.89(0.47) | 0.1967 |
| Hippocampus | Vamp3 | 1.33 | 0.36(0.03)/0.27(0.06) | 0.1632 | 0.58 | 0.38(0.04)/0.66(0.15) | 0.93 |
Expression levels quantified using relative standard curve approach normalized to β actin mRNA levels (SEM).
Data listed in bold indicates P < 0.05.
Discussion
The repeated-flurothyl seizure model is a novel approach to experimental epileptogenesis
This work demonstrates a novel preclinical approach to discover gene by environment interactions that mediate epileptogenesis. While rodent genetic studies have uncovered important seizure mechanisms among mammals (Hall 1947; Fletcher et al. 1996; Buono et al. 2004; Ferraro et al. 2004), ours is the first to follow genetic susceptibility over repeated seizures. Thus, unique modifiers dependent upon prior seizure activity can be discovered that are risk indicators or potential treatment targets for epilepsy.
A detailed genetic analysis of the Szs1 QTL locus has given insight into the molecular basis of seizure susceptibility (Ferraro et al. 2004). Congenic and transgenic rescue studies in mice have led to the election of a coding mutation within Kcnj10 as the strongest candidate for Szs1 differences in maximal electroshock threshold (Ferraro et al. 2004). KCNJ10 associations are also observed in individuals with epilepsy (Buono et al. 2004; Lenzen et al. 2005), indicating that similar epilepsy mechanisms are shared between rodents and humans. Additional work has shown that Kcnj10 function in glia is critical in K+ clearance, leading to increased neuronal hyper-excitability when conditionally deleted from astrocytes (Djukic et al. 2007). We show here that flurothyl-induced seizures are also influenced by Szs1, indicating a common connection between our approach and other experimental epilepsy models.
Dynamic QTL effects are observed across multiple flurothyl-induced seizures
While association of the Szs1 locus with day 1 GST was confirmed, Szs1 associations were not detected on subsequent seizure trials. Using the slope of daily GST reductions over eight trials as a measure of kindling, we detected highly significant associations within Szs1, 100 kb distal to Kcnj10. We are now investigating how seizure-dependent factor(s) could influence Szs1 effects, either by suppression of an existing Szs1 mutation, like the reported Kcnj10 mutation, or via alterations in other genes in this region. Multiple QTL affecting a variety of neurological traits in mice are present on distal chromosome 1 (Mozhui et al. 2008) that could be responsible for transient Szs1 effects on seizure susceptibility.
Reductions in astroglial Kir4.1 channels (formed by Kcnj10) have been observed in both human temporal lobe epilepsy and experimental epilepsy models (Schroder et al. 2000; Losi et al. 2012; Steinhäuser et al. 2012). Thus, the negative correlation between day 1 GST and kindling (r2 = −0.71) could be due to suppression of Kcnj10 activity in subsequent seizure trials. Reductions in Kcnj10 activity may be a common feature of repeated seizures that are mediated by uncharacterized mechanisms of epileptogenesis. In our model, suppressing both normal and mutant alleles of Kcnj10 as a means of seizure adaptation would consequently remove the Szs1 effects from associations after the initial seizure. The mechanisms responsible for these effects may be within the Szs1 interval or could be trans-acting factors that modify the effects of Szs1. While environmental confounds like animal age could contribute to seizure progression, all animals in our study were 6–7 wk old at the onset of the analysis. The presence of significantly different rates of kindling among HMDP strains suggests genetic factors mediate this activity within the HMDP. However, poor correlation between HMDP strain haplotypes within Szs1 and kindling in our data indicates that the uncharacterized kindling modifier and Szs1 are likely unlinked.
Novel locus on distal chromosome 4 influences seizures after repeated seizures
As repeated seizures are administered, seizure-related brain responses are invoked to stabilize neuronal function (González et al. 2015). Thus, detection of significant QTL effects are likely confounded by increased environmental variance during induction phase seizures after the initial seizure. Kindling-sensitive strains, like B6, show GST values that stabilize toward the second half of the induction phase (Papandrea et al. 2009). Once all strains reach this plateau, a second window of heritable effects can be revealed.
Significant SNP associations reappear on day 6 in the induction phases that correspond with the timing of the GST plateau effect seen in B6 mice (Papandrea et al. 2009). Among the potential candidates for day 6 GST associations is the calcium/calmodulin-dependent protein kinase II inhibitor (Camk2n), whose inhibition is reported to result in epileptiform activity (Murray et al. 2000) making it a viable candidate gene for modifying epileptogenesis. Also, calcium signaling is an important factor in epilepsy progression (Pal et al. 2001). Ongoing work will establish the parameters that influence such transient susceptibilities and their relationship to seizure progression.
Seizure-driven epileptogenesis is predicted to continue into the incubation phase of the repeated-flurothyl model due to a period of spontaneous seizures where B6 mice have as many as six seizures a day remitting to less than one seizure a day by the end of our model (Kadiyala et al. 2016). Interestingly, D2 mice also develop spontaneous seizures following flurothyl induction, however at lower frequencies than B6 mice, but that persist throughout the 28-day incubation period (Kadiyala and Ferland, 2017). When retest seizures are delivered, Esf1 is the predominant effect controlling seizure susceptibility with the top five associations all occurring in Camta1.
Genotype of Szs1 can influence the effects of Esf1
Complex networks of neuronal adaptation are activated by seizures (Kadiyala et al. 2015) that are further influenced by genetic background, leading to differential outcomes in epilepsy development. Based on our data, mutations in Kcnj10 segregating in the HMDP are associated with initial seizure effects that diminish over time, which may also be true in humans with epilepsy, raising the question of the nature of the association of Kcnj10 with epilepsy. A possibility is that the effect of Kcnj10 deficiency on seizure susceptibility is twofold. Its primary effect on hyper-excitability is mitigated by repeated seizures, but its effects on baseline K+ buffering in the interictal period remains abnormal. Such imbalances in K+ would be amplified in the context of repeated seizures leading to imbalances in downstream signaling cascades, in particular, Ca2+ signaling that has been shown to be altered by the presence of functional Kir4.1 channels (Härtel et al. 2007).
We show that strains carrying D2 alleles of Szs1, which is likely due in part to deficits in Kcnj10, has a significant impact on retest GST when these mice are further segregated based on their Esf1 genotype, while those carrying a B6 Szs1 haplotype have reduced effect on Esf1 outcome. Thus, differences in Szs1 could prime the ultimate outcomes that are observed in Esf1, possibly through an imbalance in steady-state K+ levels that exacerbate alternative mechanisms of epileptogenesis either within astrocytes or through effects on neuronal networks.
System genetics analyses point to coordinate deregulation between B6 and D2 genes near Esf1
Utilization of BXD strains was important to our project in two ways. First, it added increased power in our association analyses that would not have been possible with inbred strains alone (Bennett et al. 2010). The BXD strains also have a unique advantage in legacy data that is publicly available to perform correlation and system genetic studies to further characterize the genetic basis of epileptogenesis (Williams and Mulligan 2012). The presence of cis-eQTL around Esf1 was previously associated with other neuronal complex traits, including stress response (Wang et al. 2012). The effects were primarily attributed to differential expression of Per3. We also found differential baseline expression in Per3 in B6 and D2 mice (Table 4), depending on the tissue type being interrogated, but this differential expression was not observed in mice completing the repeated-flurothyl model and did not correlate as well with retest GST data as Camta1 expression differences.
Confirmation of the differential expression of cis-eQTL adjacent to Esf1 was also observed in the hippocampus of B6 and D2 parental strains by qPCR. While three of the four candidate loci showed significant differences in mRNA prior to seizures, only Camta1 maintained significant differences after completion of the repeated-flurothyl model. The differential expression of these loci was consistently higher in B6 mice indicating that a global regulatory control may impact this effect. Thus, our observation that the two SNPs rs369709 and rs3697097 that account for all of the cis-eQTL with LRS over 20 map to a region proximal to Esf1 could provide a good place to compare these genomes for transcriptional elements that may facilitate this kind of coordinated differential transcription.
Each of these potential candidates for Esf1 have plausible ways they could contribute to epileptogenesis. As mentioned above, Per3 has previously been implicated in neuronal stress response and other gene family members have been implicated in induced seizures in mice (Gachon et al. 2004). Park 7 protects against oxidative stress and may protect neurons from death (Kolisek et al. 2015) and has also been shown to be overexpressed in hippocampi of individuals with epilepsy (Persike et al. 2012). Vamp3 is a downstream target of MeCP2 that mediates SNARE-dependent exocytosis of glutamate (Kandratavicius et al. 2014). While these genes have reduced significance, based on our GWAS associations and system genetics data, they will need to be considered for their potential contributions to epileptogenesis going forward.
Camta1 is an excellent candidate for epileptogenesis susceptibility. In addition to having the five most significant SNPs associated with retest GST, and the best correlation with eQTLs in our system genetics data, it has several known functions that make it a good candidate from a biological perspective. First, calmodulin is a well-known mediator of postictal brain activity and has been implicated in brain adaptation to chemical (DeLorenzo 1986) and electrical kindling (Goldenring et al. 1986). Camta proteins are well-known mediators of the stress response in multiple species (Pandey et al. 2014; Doherty et al. 2009) and are well suited to mediate the stress response from repeated seizures. Increased Camta1 activity is a predictable consequence of inducing repeated seizures, the activity of which would be enhanced by reduced K+ buffering capacity that is an aspect of our model. Finally, mutations in CAMTA1 are associated with multiple neurological phenotypes, including seizure susceptibility in humans (Miller et al. 2011; Shinawi et al. 2015). Identifying the basis of this susceptibility will lead to improved diagnosis and novel treatment options for individuals with epilepsy.
Acknowledgments
We thank Rob Williams for helpful discussions during the progress of this study. The exceptional animal husbandry expertise of B.S.B., who has since passed away, was critical to the success of this project. This work was supported by a National Institutes of Health grant through the National Institute of Neurological Disorders and Stroke R01NS064283 grant to R.J.F. We are grateful to Veterinary Sciences at the Wadsworth Center and Albany Medical College that supported the completion of this work. The authors declare no competing financial interests.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Buono R. J., Lohoff F. W., Sander T., Sperling M. R., O’Connor M. J., et al. , 2004. Association between variation in the human KCNJ10 potassium ion channel gene and seizure susceptibility. Epilepsy Res. 58: 175–183. [DOI] [PubMed] [Google Scholar]
- Chaix Y., Ferraro T. N., Lapouble E., Martin B., 2007. Chemoconvulsant-induced seizure susceptibility: toward a common genetic basis? Epilepsia 48: 48–52. [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Lu L., Shou S., Qu Y., Gu J., et al. , 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37: 233–242. [DOI] [PubMed] [Google Scholar]
- DeLorenzo R. J., 1986. A molecular approach to the calcium signal in brain: relationship to synaptic modulation and seizure discharge. Adv. Neurol. 44: 435–464. [PubMed] [Google Scholar]
- Djukic B., Casper K. B., Philpot B. D., Chin L.-S., McCarthy K. D., 2007. Conditional knock-out of Kir4.1 leads to glial membrane depolarization, inhibition of potassium and glutamate uptake, and enhanced short-term synaptic potentiation. J. Neurosci. 27: 11354–11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F., 2009. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England M. J., Liverman C. T., Schultz A. M., Strawbridge L. M., 2012. Epilepsy across the spectrum: promoting health and understanding: a summary of the Institute of Medicine report. Epilepsy Behav. 25: 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom F. L., Woodbury D. M., 1988. Seizure susceptibility in DBA and C57 mice: the effects of various convulsants. Epilepsia 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Ferland, R. J. 2017 The repeated flurothyl seizure model in mice. Bio-protocol 7(11): e2309. DOI: 10.21769/BioProtoc.2309. [DOI] [PMC free article] [PubMed]
- Ferraro T. N., 2016. Barriers to the use of genetic information for the development of new epilepsy treatments. Expert Rev. Neurother. 16: 5–8. [DOI] [PubMed] [Google Scholar]
- Ferraro T. N., Golden G. T., Smith G. G., Schork N. J., St. Jean P., et al. , 1997. Mapping murine loci for seizure response to kainic acid. Mamm. Genome 8: 200–208. [DOI] [PubMed] [Google Scholar]
- Ferraro T. N., Golden G. T., Smith G. G., Martin J. F., Lohoff F. W., et al. , 2004. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm. Genome 15: 239–251. [DOI] [PubMed] [Google Scholar]
- Fletcher C. F., Lutz C. M., O’Sullivan T. N., Shaughnessy J. D., Hawkes R., et al. , 1996. Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell 87: 607–617. [DOI] [PubMed] [Google Scholar]
- Gachon F., Fonjallaz P., Damiola F., Gos P., Kodama T., et al. , 2004. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 18: 1397–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A., Rau C. D., Farber C. R., Bennett B. J., Orozco L. D., et al. , 2012. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm. Genome 23: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring J. R., Wasterlain C. G., Beate Oestreicher A., de Graan P. N. E., Farber D. B., et al. , 1986. Kindling induces a long-lasting change in the activity of a hippocampal membrane calmodulin-dependent protein kinase system. Brain Res. 377: 47–53. [DOI] [PubMed] [Google Scholar]
- González O. C., Krishnan G. P., Chauvette S., Timofeev I., Sejnowski T., et al. , 2015. Modeling of age-dependent epileptogenesis by differential homeostatic synaptic scaling. J. Neurosci. 35: 13448–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grone B. P., Baraban S. C., 2015. Animal models in epilepsy research: legacies and new directions. Nat. Neurosci. 18: 339–343. [DOI] [PubMed] [Google Scholar]
- Gupta S., Kwan P., Faught E., Tsong W., Forsythe A., et al. , 2016. Understanding the burden of idiopathic generalized epilepsy in the United States, Europe, and Brazil: an analysis from the National Health and Wellness Survey. Epilepsy Behav. 55: 146–156. [DOI] [PubMed] [Google Scholar]
- Hall C. S., 1947. Genetic differences in fatal audiogenic seizures between two inbred strains of house mice. J. Hered. 38: 2–6. [PubMed] [Google Scholar]
- Härtel K., Singaravelu K., Kaiser M., Neusch C., Hülsmann S., et al. , 2007. Calcium influx mediated by the inwardly rectifying K+ channel Kir4.1 (KCNJ10) at low external K+ concentration. Cell Calcium 42: 271–280. [DOI] [PubMed] [Google Scholar]
- International League Against Epilepsy Consortium on Complex Epilepsies , 2014. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 13: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala S. B., Papandrea D., Tuz K., Anderson T. M., Jayakumar S., et al. , 2015. Spatiotemporal differences in the c-fos pathway between C57BL/6J and DBA/2J mice following flurothyl-induced seizures: a dissociation of hippocampal Fos from seizure activity. Epilepsy Res. 109: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala S. B., Yannix J. Q., Nalwalk J. W., Papandrea D., Beyer B. S., et al. , 2016. Eight flurothyl-induced generalized seizures lead to the rapid evolution of spontaneous seizures in mice: a model of epileptogenesis with seizure remission. J. Neurosci. 36: 7485–7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiyala, S. B., and R. J. Ferland, 2017 Dissociation of spontaneous seizures and brainstem seizure thresholds in mice exposed to eight flurothyl-induced generalized seizures. Epilepsia Open 2: 48–58. [DOI] [PMC free article] [PubMed]
- Kandratavicius L., Alves Balista P., Lopes-Aguiar C., Ruggiero R. N., Umeoka E. H., et al. , 2014. Animal models of epilepsy: use and limitations. Neuropsychiatr. Dis. Treat. 10: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisek M., Montezano A. C., Sponder G., Anagnostopoulou A., Vormann J., et al. , 2015. PARK7/DJ-1 dysregulation by oxidative stress leads to magnesium deficiency: implications in degenerative and chronic diseases. Clin. Sci. 129: 1143–1150. [DOI] [PubMed] [Google Scholar]
- Kosobud A. E., Crabbe J. C., 1990. Genetic correlations among inbred strain sensitivities to convulsions induced by 9 convulsant drugs. Brain Res. 526: 8–16. [DOI] [PubMed] [Google Scholar]
- Lenzen K. P., Heils A., Lorenz S., Hempelmann A., Höfels S., et al. , 2005. Supportive evidence for an allelic association of the human KCNJ10 potassium channel gene with idiopathic generalized epilepsy. Epilepsy Res. 63: 113–118. [DOI] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–837. [DOI] [PubMed] [Google Scholar]
- Losi G., Cammarota M., Carmignoto G., 2012. The role of astroglia in the epileptic brain. Front. Pharmacol. 3: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, L. A., J. Gunstad, M. B. Spitznagel, J. Mccaffery, J. Mcgeary et al., 2011 CAMTA1 T polymorphism is associated with neuropsychological test performance in older adults with cardiovascular disease. Psychogeriatrics 11: 135–140. [DOI] [PubMed]
- Morgan A. P., Fu C.-P., Kao C.-Y., Welsh C. E., Didion J. P., et al. , 2015. The mouse universal genotyping array: from substrains to subspecies. G3 6: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Mozhui K., Ciobanu D. C., Schikorski T., Wang X., Lu L., et al. , 2008. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression (J. Flint, Ed.). PLoS Genet. 4: e1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K., Lu L., Armstrong W. E., Williams R. W., 2012. Sex-specific modulation of gene expression networks in murine hypothalamus. Front. Neurosci. 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. D., Isackson P. J., Eskin T. A., King M. A., Montesinos S. P., et al. , 2000. Altered mRNA expression for brain-derived neurotrophic factor and type II calcium/calmodulin-dependent protein kinase in the hippocampus of patients with intractable temporal lobe epilepsy. J. Comp. Neurol. 418: 411–422. [DOI] [PubMed] [Google Scholar]
- Neumann P. E., Collins R. L., 1991. Genetic dissection of susceptibility to audiogenic seizures in inbred mice. Proc. Natl. Acad. Sci. USA 88: 5408–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall R., Kempermann G., Peirce J., Lu L., Goldowitz D., et al. , 2009. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front. Neurosci. 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Sun D., Limbrick D., Rafiq A., DeLorenzo R. J., 2001. Epileptogenesis induces long-term alterations in intracellular calcium release and sequestration mechanisms in the hippocampal neuronal culture model of epilepsy. Cell Calcium 30: 285–296. [DOI] [PubMed] [Google Scholar]
- Pandey K., Dhoke R. R., Rathore Y. S., Nath S. K., Verma N., et al. , 2014. Low pH overrides the need of calcium ions for the shape-function relationship of calmodulin: resolving prevailing debates. J. Phys. Chem. B 118: 5059– 5074. [DOI] [PubMed] [Google Scholar]
- Papandrea D., Anderson T. M., Herron B. J., Ferland R. J., 2009a Dissociation of seizure traits in inbred strains of mice using the flurothyl kindling model of epileptogenesis. Exp. Neurol. 215: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandrea, D., W. S. Kukol, T. M. Anderson, B. J. Herron, and R. J. Ferland, 2009b Analysis of flurothyl-induced myoclonus in inbred strains of mice. Epil. Res. 87: 130–136. [DOI] [PMC free article] [PubMed]
- Persike D., Lima M., Amorim R., Cavalheiro E., Yacubian E., et al. , 2012. Hippocampal proteomic profile in temporal lobe epilepsy. J. Epilepsy Clin. Neurophysiol. 18: 53–56. [Google Scholar]
- Samoriski G. M., Applegate C. D., 1997. Repeated generalized seizures induce time-dependent changes in the behavioral seizure response independent of continued seizure induction. J. Neurosci. 17: 5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder W., Hinterkeuser S., Seifert G., Schramm J., Jabs R., et al. , 2000. Functional and molecular properties of human astrocytes in acute hippocampal slices obtained from patients with temporal lobe epilepsy. Epilepsia 41: S181–S184. [DOI] [PubMed] [Google Scholar]
- Shinawi M., Coorg R., Shimony J. S., Grange D. K., Al-Kateb H., 2015. Intragenic CAMTA1 deletions are associated with a spectrum of neurobehavioral phenotypes. Clin. Genet. 87: 478–482. [DOI] [PubMed] [Google Scholar]
- Steinhäuser C., Seifert G., Bedner P., 2012. Astrocyte dysfunction in temporal lobe epilepsy: K+ channels and gap junction coupling. Glia 60: 1192–1202. [DOI] [PubMed] [Google Scholar]
- Tse K., Puttachary S., Beamer E., Sills G., and T. Thippeswamy, 2014. Advantages of repeated low dose against single high dose of kainate in C57BL/6J mouse model of status epilepticus: behavioral and electroencephalographic studies. PLoS One 9: e96622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. G., Nelson S. B., 2004. Homeostatic plasticity in the developing nervous system. Nat. Rev. Neurosci. 5: 97–107. [DOI] [PubMed] [Google Scholar]
- Wang X., Mozhui K., Li Z., Mulligan M. K., Ingels J. F., et al. , 2012. A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Transl. Psychiatry 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. W., Mulligan M. K., 2012. Genetic and molecular network analysis of behavior. Int. Rev. Neurobiol. 104: 135–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr P., Martin B., Adelman J. P., 2000. The murine Bis1 seizure gene and the Kcnab2 gene encoding the beta2-subunit of the K+ channel are different. Neurogenetics 2: 231–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All seizure threshold data are available from the GeneNetwork website under accession numbers 18963–18981 (http://www.genenetwork.org/).