Abstract
Reproductive barriers are commonly observed in both animals and plants, in which they maintain species integrity and contribute to speciation. This report shows that a combination of loss-of-function alleles at two duplicated loci, DUPLICATED GAMETOPHYTIC STERILITY 1 (DGS1) on chromosome 4 and DGS2 on chromosome 7, causes pollen sterility in hybrid progeny derived from an interspecific cross between cultivated rice, Oryza sativa, and an Asian annual wild rice, O. nivara. Male gametes carrying the DGS1 allele from O. nivara (DGS1-nivaras) and the DGS2 allele from O. sativa (DGS2-T65s) were sterile, but female gametes carrying the same genotype were fertile. We isolated the causal gene, which encodes a protein homologous to DNA-dependent RNA polymerase (RNAP) III subunit C4 (RPC4). RPC4 facilitates the transcription of 5S rRNAs and tRNAs. The loss-of-function alleles at DGS1-nivaras and DGS2-T65s were caused by weak or nonexpression of RPC4 and an absence of RPC4, respectively. Phylogenetic analysis demonstrated that gene duplication of RPC4 at DGS1 and DGS2 was a recent event that occurred after divergence of the ancestral population of Oryza from other Poaceae or during diversification of AA-genome species.
Keywords: reproductive barrier, genetic drift, neutral gene, nonfunctionalization, hybrid sterility
Hybrid incompatibility is a phenomenon of maladaptive traits such as inviability, weakness, and sterility in hybrids between two diverged populations or species and in their progeny. Hybrid incompatibility is a common feature of a wide range of organisms (Coyne and Orr 2004). It plays a vital role in intrinsic postzygotic reproductive isolation, which conserves species integrity and uniformity and contributes to speciation by limiting gene flow within and between species (Lynch and Force 2000; Taylor et al. 2001).
The genetic basis of hybrid incompatibility has been analyzed in many plants and animals (Coyne and Orr 2004; Bomblies and Weigel 2007; Rieseberg and Blackman 2010). Epistasis of nuclear-encoded genes for hybrid incompatibility, known as Bateson–Dobzhansky–Muller (BDM) incompatibility, is a major genetic phenomenon in which interaction of alleles at different loci leads to maladaptive traits in hybrids derived from crosses between two diverged populations or species (Coyne and Orr 2004; Noor and Feder 2006). Models suggest that genes causing hybrid incompatibility are maintained in species with a compatible genetic background. Even though the BDM model is simple and universal, the interaction of independently evolved genes from two diverged populations leads to conflicts in a variety of biological processes, including molecular interaction related to DNA binding (Brideau et al. 2006), intercellular protein trafficking through plasmodesmata (Long et al. 2008), the endoplasmic reticulum stress response (Yang et al. 2012), and autoimmune responses caused by aberrant signal transduction systems (Bomblies and Weigel 2007; Yamamoto et al. 2010; Chen et al. 2014).
Gene duplication is considered to be a main driver of evolution because it supplies functional redundancy, which allows functional changes in one or more of the duplicate gene copies while keeping the original function of the ancestral gene, which may be essential for survival (Ohno 1970; Lynch and Conery 2000; Moore and Purugganan 2005). Duplicated genes are suggested to meet three functional fates: acquisition of a novel and beneficial function (neofunctionalization), specialization of an ancestral gene’s function across duplicates (subfunctionalization), and silencing or loss of function through degenerative mutations (nonfunctionalization). Although duplicate genes arise at a very high rate, the vast majority of duplicate genes rapidly undergo nonfunctionalization within a few million years (Lynch and Conery 2000). Since silencing or loss of function at unlinked gene duplicates are stochastic processes for young duplicate copies (designated A1A1A2A2), the probabilities of generating genotype A1A1a2a2 in one species and genotype a1a1A2A2 in the other species are assumed to be identical. These processes for gene duplications and subsequent losses at the reciprocal loci in each species [hereafter designated as gene duplication and reciprocal loss (GDRL)] result in positional variation of functional genes among diverged populations or species, and zygotes (a1a1a2a2) or gametes (a1a2) carrying the double null genes exhibit maladaptive phenotypes in hybrid progeny (Lynch and Force 2000). Hybrid incompatibility caused by the GDRL process can be considered as a kind of BDM incompatibility. Molecular cloning studies have revealed the zygotic type of BDM incompatibility caused by GDRL in a histidinol-phosphate aminotransferase gene, which results in lack of free histidine in hybrid embryos (Bikard et al. 2009); gametophytic BDM incompatibility caused by the GDRL process affecting nuclear-encoded mitochondrial ribosomal protein in pollen (Yamagata et al. 2010; Win et al. 2011); and GDRL leading to failure of pollen tube germination (Mizuta et al. 2010).
Eukaryotes synthesize RNA by multi-subunit protein complexes, the RNAPs: designated PolI for 45S preribosomal RNA, PolII for mRNA and noncoding RNA, and PolIII for tRNA and 5S rRNA (Vannini and Cramer 2012). PolI, PolII, and PolIII have similar structures with shared ancestral origin from 12 subunits (Rpo1–12) of archaean RNAP (Ream et al. 2009). In addition to the canonical PolII, plants have acquired two plant-specific multi-subunit RNAP homologs, PolIV and PolV, for RNA-directed DNA methylation and transcriptional silencing via neofunctionalization of gene duplicates for PolII components (Onodera et al. 2005; Haag and Pikaard 2011). Different angiosperm groups have experienced different lineage-specific gene duplication of several RNAP subunits, implying independent functional divergences of RNAP components (Wang and Ma. 2015). Neofunctionalization for RNAPs is likely to have resulted in acquisition of specific biological processes in plants. However, it has not been fully understood whether nonfunctionalization of gene duplicates for RNAP members drives diversification of plant species.
Here, we report the identification and cloning of the genes for F1 pollen sterility in an interspecific hybrid between the cultivated species Oryza sativa L. and the wild species O. nivara Sharma et Shastry. We show that the pollen sterility in the F1 hybrid is due to the epistatic interaction of two duplicated loci on chromosomes 4 and 7. Map-based cloning revealed that the two genes encode a putative RNAP III subunit C4 (RPC4), which is known to be involved in the transcription of 5S rRNAs and tRNAs.
Materials and Methods
Plant materials
Accessions of O. nivara Sharma et Shastry IRGC105715, originating from Cambodia, were kindly provided by the International Rice Research Institute, Manila, the Philippines. O. sativa L. ssp. japonica “Taichung 65” (T65) was crossed with pollen of IRGC105715 to produce F1 plants with T65 cytoplasm. Near-isogenic lines were developed by performing repeated backcrosses with T65 as the male parent (Supplemental Material, Figure S1 in File S1). For the linkage mapping of DGS1 and DGS2 (see Results), two kinds of BC4F3 plants were selected from the BC4F2 population: (1) plants heterozygous at RM16862 (linked to DGS1) on chromosome 4 and homozygous for the T65 allele at M41_STS (linked to DGS2) on chromosome 7 (later designated the DGS2-T65s allele), and (2) plants homozygous for the O. nivara allele at RM16862 (later designated the DGS1-nivaras allele) and heterozygous at M41_STS. These plants were grown to develop BC4F5 populations for high-resolution mapping. For genetic analysis of the epistatic interaction between DGS1 and DGS2, we used two BC4F3 populations derived from BC4F2 plants heterozygous for RM16862 and M41_STS.
Observation of mature pollen grains
Panicles at the preflowering stage were collected and fixed in solution containing 4% (w/v) paraformaldehyde, 0.25% (w/v) glutaraldehyde, 0.02% (v/v) Triton X-100, and 100 mM sodium phosphate (pH 7.5) at 4° for 24 hr. The panicles were rinsed in 100 mM sodium phosphate buffer and stored in the same buffer containing 0.1% (w/v) sodium azide (NaN3). Pollen grains were stained in hematoxylin solution according to Chang and Neuffer (1989) and observed under a light microscope.
Evaluation of pollen fertility
Spikelets at the flowering stage were fixed in 70% ethanol. Pollen grains from all six anthers of a single spikelet were released onto a glass slide and stained with 1% I2–KI solution. Approximately 200 grains were observed and evaluated for fertility under an Axioplan light microscope (Zeiss, Jena, Germany). Pollen germination was tested in vitro by releasing pollen grains from dehisced anthers immediately onto germination gel containing 15% sucrose, 0.01% H3BO3, 0.03% CaCl2, and 0.6% Gelrite (pH 7.2) (Wako, Osaka, Japan) dropped on a slide glass. The slide glasses were incubated at room temperature for 5 min, and pollen germination was observed under a light microscope. Images were processed in Photoshop software (Adobe, San Jose, CA).
Linkage mapping
A total of 111 simple sequence repeat (SSR) markers were used for genome-wide genotyping of the BC4F1 pollen-sterile plants to find regions of chromosomal introgression from O. nivara that were involved in pollen sterility in the T65 genetic background (Table S1 in File S2). Three pairs of primers for SSR markers WGS76, WGS1, and WG1 were newly designed in this study in SSRIT software (Temnykh et al. 2001). Five SSR markers (RM471 and RM1359 on chromosome 4 and RM6652, RM1353, and RM6081 on chromosome 7) were used for linkage analysis in the BC4F3 population (Table S1 in File S2). Total genomic DNA was extracted according to the method of Dellaporta et al. (1983) with minor modifications. Each 15 μl reaction mixture consisted of 50 mM KCl, 10 mM Tris (pH 9.0), 1.5 mM MgCl2, 200 μM dNTPs, 0.2 μM primers, 0.75 units of Taq polymerase (Takara, Otsu, Japan), and ∼10 ng genomic DNA template. PCR was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The cycling profile was an initial denaturation at 95° for 5 min; 35 cycles of 95° for 30 sec, 55° for 30 sec, 72° for 40 sec; and a final elongation step at 72° for 7 min. Amplified products were electrophoresed in 4% agarose gel in 0.5× TBE buffer. MAPMAKER/EXP v. 3.0 software (Lander et al. 1987) was used to construct a linkage map using Kosambi’s mapping function.
Map-based cloning
For the high-resolution mapping of DGS1 and DGS2, genotypes of recombinants were determined by additional SSR and sequence-tagged site (STS) markers shown in Table S2 in File S2. PCR primers for the STS markers M45_STS, M41_STS, and M23_STS were designed from the rice reference sequence of Nipponbare in SSRIT (Temnykh et al. 2001), and amplified PCR products from the T65 and O. nivara genomes were separated by electrophoresis in 4% agarose gel. For the BAC clone screening by PCR to investigate the genomic structures of T65 and O. nivara, RM16862 and RM16867 were used for DGS1 and M45_STS and M41_STS were used for DGS2. The obtained BAC clones GN0025A01, GN0028G14, and GN0031H20 (T65) and IRGC105715_0023B24 (O. nivara) were sequenced by next-generation sequencing using Roche/454 GS-FLX Titanium sequencing (Roche, Basel, Switzerland) and de novo assembly in GS De novo Assembler software (Roche). Shotgun sequencing by the Sanger sequencing method was conducted for the O. nivara BAC clone IRGC105715_0046I04.
A complementation test was conducted as described by Yamagata et al. (2010). In brief, EcoRI-digested genomic fragments containing predicted RPC4 genes were cloned into the Ti-plasmid binary vector pPZP2H-lac (Fuse et al. 2001), which was transformed into DGS1 semisterile plants (T+/Ns|Ts/Ts) and DGS2 semisterile plants (Ns/Ns|Ts/N+) via Agrobacterium-mediated transformation (The two semisterile plant types were designated by which of the two loci was heterozygous for fertile and sterile alleles. In each case, the other locus was homozygous for the sterile allele.). Complementation of pollen fertility by the transgenes was examined by the segregation ratios of genotypes at DGS1 and DGS2 using SSR markers tightly linked to DGS1 (marker RM471) and DGS2 (marker RM6574) in the T1 generation.
Copy numbers of the transgenes inserted in the genome of the T0 and T1 plants were estimated by quantitative PCR in a MX3000P QPCR system (Agilent Technologies, Santa Clara, CA) using QuantiTect SYBR Green PCR Kits (QIAGEN, Venlo, The Netherlands). The standard curve was based on the amplification of the CaMV 35S promoter region using primers 5′-CGT AAG GGA TGA CGC ACA ATC C-3′ and 5′-CGA GAT TCT TCG CCC TCC GA-3′. Amplification of a single-copy region of rice genomic chromosome 8 DNA using the primers 5′-GGA CTG GAC AGA TTG AGA GTG-3′ and 5′-AAC GCC GAA CAA GCC CTT ACA-3′ was used as the internal control. The thermal cycling profile was an initial denaturation at 95° for 15 min, followed by 35 cycles of 95° for 30 sec, 55° for 30 sec, and 72° for 30 sec.
Gene structure and expression analysis
To investigate the structure of the transcript of the causal gene(s) of DGS1 and DGS2, 5′- and 3′-rapid amplification of cDNA ends (RACE) was conducted using the SMARTer RACE cDNA synthesis kit (Clontech, Mountain View, CA). Total RNA was extracted from anthers using the mirVana miRNA isolation kit (Ambion, Carlsbad, CA). Amplification products were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and introduced into DH5α competent cells. The plasmids were sequenced with a BigDye Terminator v. 3.1 cycle sequencing kit and analyzed on a 3130xl Genetics Analyzer (Applied Biosystems). The sequencing data were aligned to the genomic sequences of T65 and O. nivara from BAC clones in Sequencher software (Gene Codes, Ann Arbor, MI).
For expression analysis of DGS1 and DGS2, 20 ng/μl of total RNA was used for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis using the forward primer 5′-ATG TCT CTC CGG GTT CAA ATT GC-3′ and the reverse primer 5′-TTA CGC TTC CAT CTT GTC GAA AGA ATC C-3′. The SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA) was used for real-time reactions as described previously (Nguyen et al. 2010). Rice Ub-CEP52-1 encoding ubiquitin-fused 60S ribosomal protein L40-1 (Os03g0234200), amplified by the forward primer 5′-CTG TCA ACT GCC GCA AGA AG-3′ and the reverse primer 5′-GGC GAG TGA CGC TCT AGT TC-3′, was used as an internal control for the qRT-PCR.
Promoter–GUS assay
To analyze the spatial expression patterns of RPC4, a 2.4 kb PCR fragment upstream of the coding sequence was amplified from T65 BAC clone GN0028G14 using the forward primer 5′-CAC CGG TAG GGG AAG GAC ATG A-3′ and the reverse primer 5′-ACA AGA AAA GAA GCA CAA ATC CTG CG-3′ and cloned into the pENTR/D-TOPO vector (Invitrogen). Plasmids were sequenced to confirm that no mutation had occurred during the PCR amplification. The cloned endogenous promoter sequence was fused to a promoterless GUS gene derived from the pGWB3 Gateway-system destination vector (Nakagawa et al. 2007) by using LR Clonase (Invitrogen). The final construct was transformed into T65 using Agrobacterium-mediated transformation. Spikelets of the T0 plants at the mature stage of pollen development were collected and stained for in situ determination of GUS activity, as described by Yamagata et al. (2010).
Data availability
Strains are available upon request. File S1 contains all of the supplemental figures and methods. All genotype data are presented in the supplemental tables (File S2). Sequence data have been deposited in the DNA Data Bank of Japan for RPC4 from the DGS1-T65+ allele (AB758279); RPC4 from the DGS1-nivaras allele (AB758280); RPC4a (AB758281), RPC4b (AB758282), RPC4c (AB758283), RPC4d (AB758284), and RPC4e (AB758285) from the DGS2-nivara+ allele; and BAC sequences of GN0028G14 for DGS1-T65+ (LC060740), IRGC105715_0023B24 for DGS1-nivaras (LC060741), GN0031H20 for DGS2-T65s (LC060743), and IRGC105715_0046I04 for DGS2-nivara+ (LC060742).
Results
Identification of genetic factors on chromosomes 4 and 7
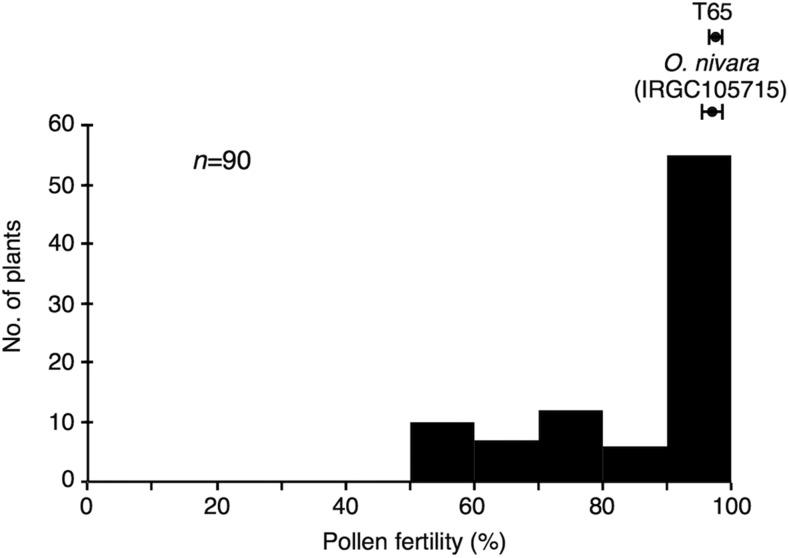
In developing introgression lines of O. nivara genomic segments in a T65 genetic background, we produced a BC4F1 population by several generations of backcrossing with T65 as the male parent. In the progeny of a single BC4F2 population (“BC4F2 42” in Figure S1 in File S1), we observed a kind of trimodal distribution with pollen fertility ranging from 50 to 100% (Figure 1) in the two BC4F3 populations derived from two BC4F2 plants heterozygous for markers on chromosomes 4 (at RM471 and RM1359) and 7 (at RM6652, RM1353, and RM6081) (Figure S1 in File S1). We assumed that this pollen sterility was governed by epistatic interaction of duplicated sterile genes at two segregating loci, known as the gametophytic type of BDM incompatibility model, in which pollen sterility levels of 50, 75, and 100% are expected to segregate in a 2:3:7 ratio (Figure S2 in File S1), because we have seen a similar pollen fertility distribution in the hybrid pollen sterility governed by the duplicated loci S27 and S28 in O. sativa and O. glumaepatula hybrid (Yamagata et al. 2010). The observed frequency distribution of pollen fertility in the BC4F3 populations was similar to the distribution predicted by the model (Figure 1 and Figure S2 in File S1). Genotyping at RM471 on chromosome 4 and RM6081 on chromosome 7 showed that genotypes in the BC4F3 populations fit the expected ratio for the gametophytic type of BDM incompatibility model of 1:2:1:1:3:2:0:1:1, which assumes that pollen grains carrying the O. nivara allele at RM471 and the T65 allele at RM6081 are sterile (Figure S2 in File S1, Table S3 in File S2 and Table 1) [hereafter, T65 and O. nivara alleles are indicated by T and N, respectively, and the letters to the left and right of the vertical bar indicate genotypes at RM471 (DGS1) and RM6081 (DGS2), respectively; sterile alleles, i.e., the O. nivara allele at RM471 (DGS1) and the T65 allele at RM6081 (DGS2), are indicated with a superscript of s; and normal alleles, i.e., the T65 allele at DGS1 and the O. nivara allele at DGS2, are indicated with a superscript of +]. In particular, no plants homozygous for O. nivara at RM471 and homozygous for T65 at RM6081 (genotype Ns/Ns|Ts/Ts; Table 1) were observed among 90 individuals of this BC4F3 population, proving further evidence that pollen carrying Ns|Ts was sterile and therefore not transmitted to the progeny.
Figure 1.
Frequency distribution of pollen fertility in the BC4F3 populations derived from a cross between T65 (O. sativa) and IRGC105715 (O. nivara). The BC4F1 parents were heterozygous for markers on chromosomes 4 (at RM471 and RM1359) and 7 (at RM6652, RM1353, and RM6081) (Figure S1 in File S1). The pollen fertility of each parent is indicated at the top; error bars indicate SEs (n = 3). No., number.
Table 1. Epistatic interaction observed between genotypes at RM6081 and RM471 fitted to a gametophytic BDM incompatibility model in BC4F3 populations derived from a cross between T65 (O. sativa) and IRGC105715 (O. nivara).
| Genotype | Expected Ratioa | Number of Individuals | χ2b | ||
|---|---|---|---|---|---|
| RM471 (DGS1) | RM6081 (DGS2) | Expected | Observed | ||
| T/Tc | T/T | 1/12 | 7.5 | 9 | 0.3 |
| T/N | 2/12 | 15 | 14 | 0.07 | |
| N/N | 1/12 | 7.5 | 9 | 0.3 | |
| T/N | T/T | 1/12 | 7.5 | 10 | 1.7 |
| T/N | 3/12 | 22.5 | 18 | 0.9 | |
| N/N | 2/12 | 15 | 15 | 7.5 | |
| N/N | T/T | 0/12 | 0 | 0 | — |
| T/N | 1/12 | 7.5 | 7 | 0.03 | |
| N/N | 1/12 | 7.5 | 8 | 0.03 | |
| Total | 1 | 90 | 90 | 2.46 | |
| P | 0.92 | ||||
Expected ratio based on the hypothesis that DGS1 and DGS2 segregate independently, and that pollen of genotype N|T is sterile. See Figure S2 in File S1 for further explanation.
Each value in the column contains rounding error.
T/T, homozygous for T65 allele; T/N, heterozygous; and N/N, homozygous for O. nivara allele at the indicated marker locus.
Identification of DGS1 and DGS2
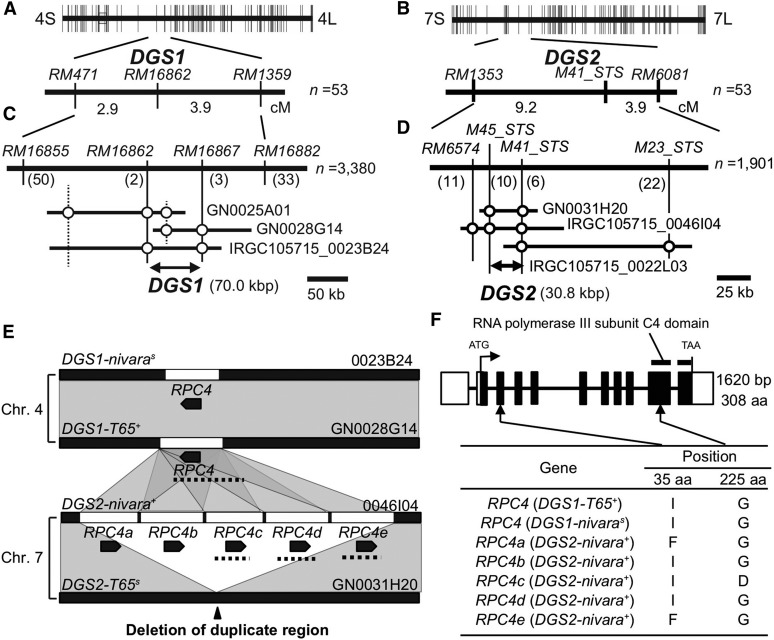
We conducted linkage analysis to identify the postulated genetic factors on chromosomes 4 and 7. For linkage mapping of the genetic factor on chromosome 4, we selected the BC4F2 individual that was heterozygous on chromosome 4 and homozygous for the T65 allele on chromosome 7 (T+/Ns|Ts/Ts) to produce a BC4F3 population segregating only for the segment on chromosome 4. Pollen-fertile and semisterile plants segregated in a ratio of 24:29 in the BC4F3 population (Table S4 in File S2 and Table 2). All homozygotes for the T65 allele at RM16862 (T+/T+|Ts/Ts) were fertile and all heterozygotes (T+/Ns|Ts/Ts) were pollen semisterile. As in previous analyses, no homozygotes for the O. nivara allele on chromosome 4 (Ns/Ns|Ts/Ts) were observed. These data suggest that sterility in this population was governed by a single gametophytically-acting gene tightly linked to RM16862 on chromosome 4, which induced selective abortion or sterility of pollen grains carrying the O. nivara allele when the chromosomal segment on chromosome 7 was fixed for the T65 allele. We designated this locus on chromosome 4 as DGS1. Linkage analysis showed that DGS1 cosegregated with RM16862 and was located between RM471 and RM1359 with map distances of 2.9 and 3.9 cM, respectively (Figure 2A). The observed segregation distortion of genotypes in the BC4F3 population would be the expected result of gametophytic sterility of pollen grains carrying the DGS1-nivaras allele and the DGS2-T65s allele (pollen genotype Ns|Ts) in the semisterile BC4F2 plants.
Table 2. Linkage analysis of pollen fertility in a BC4F3 population derived from a BC4F2 plant heterozygous at RM16862 (chromosome 4) and homozygous for the T65 allele at M41_STS (chromosome 7).
| Pollen Fertility | Genotypes at RM16862a | ||
|---|---|---|---|
| T/T | T/N | N/N | |
| Fertile | 24 | 0 | 0 |
| Semisterile | 0 | 29 | 0 |
T/T, homozygous for T65 allele; T/N, heterozygous; and N/N, homozygous for O. nivara allele.
Figure 2.
Linkage mapping and map-based cloning of DGS1 and DGS2. (A and B) Linkage mapping of (A) DGS1 and (B) DGS2. (C and D) High-resolution mapping of (C) DGS1 and (D) DGS2. The number in parentheses beneath each marker represents the number of recombinants between that marker and the DGS gene. (E) Diagrams of the BAC clones in the DGS1 and DGS2 regions of T65 and O. nivara. Segmental duplications (white boxes) were found in the DGS1-T65+, DGS1-nivaras, and DGS2-nivara+ alleles, but not in the DGS2-T65s allele. Black pentagons represent the predicted RPC4 genes. Dotted lines represent the EcoRI fragments used for the transgenic complementation tests. Homologous genomic regions are connected by translucent gray areas. Black arrowhead indicates site where segmental duplication was not found or deleted in DGS2-T65s. (F) Structure of RPC4. White boxes represent 5′- and 3′-untranslated regions. Black boxes indicate coding sequences within exons. Arrows indicate the positions of amino acid polymorphisms among the copies of RPC4. Horizontal bars indicate position of the RNA polymerase III subunit C4 domain. BAC, bacterial artificial chromosome; Chr., chromosome.
For linkage mapping of the genetic factor on chromosome 7, we took a similar approach. We developed three BC4F3 populations from three BC4F2 plants, which were homozygous for O. nivara at RM471 on chromosome 4 and heterozygous at RM6081 on chromosome 7 (Ns/Ns|Ts/N+) in the BC4F2 42 population (Figure S1 in File S1), to obtain a population segregating only for the segment on chromosome 7. In the BC4F3 population, fertile and semisterile plants segregated in a 25:28 ratio (Table S5 in File S2 and Table 3). All of the fertile plants were homozygous for the O. nivara allele at M41_STS (Ns/Ns|N+/N+), and all of the semisterile plants were heterozygous (Ns/Ns|Ts/N+); no homozygotes for the T65 allele (Ns/Ns|Ts/Ts) were observed. As with DGS1, we considered that the semisterility and segregation ratios were caused by gametophytic sterility of pollen grains carrying the T65 allele of a locus tightly linked to M41_STS (Ns|Ts genotype). We named this locus DGS2. DGS2 was mapped between RM1353 (9.2 cM) and RM6081 (3.9 cM) (Figure 2B).
Table 3. Linkage analysis of pollen fertility in BC4F3 populations derived from BC4F2 plants homozygous for the O. nivara allele at RM16862 (chromosome 4) and heterozygous at M41_STS (chromosome 7).
| Pollen Fertility | Genotype at M41_STSa | ||
|---|---|---|---|
| T/T | T/N | N/N | |
| Fertile | 0 | 0 | 25 |
| Semisterile | 0 | 28 | 0 |
T/T, homozygous for T65 allele; T/N, heterozygous; and N/N, homozygous for O. nivara allele.
Characterization of sterility caused by DGS1 and DGS2
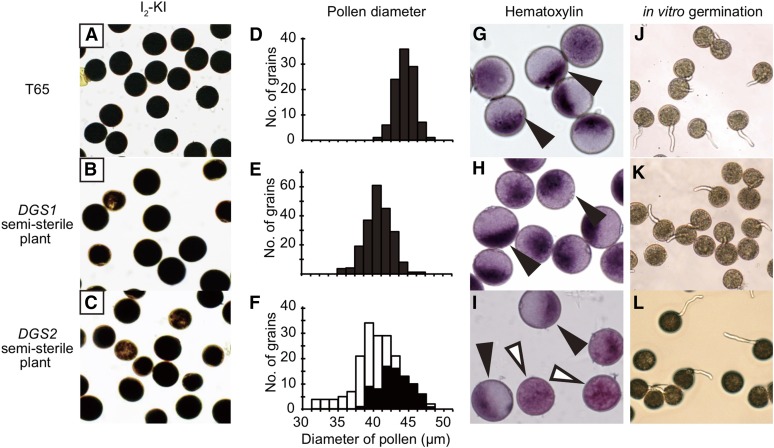
We investigated abnormalities induced by the epistatic interaction between DGS1 and DGS2 in postmeiotic development by observing the phenotypes of T65, BC4F5 plants carrying the T+/Ns|Ts/Ts genotype (DGS1 semisterile plants), and BC4F5 plants carrying the Ns/Ns|Ts/N+ genotype (DGS2 semisterile plants). In each of the two semisterile plant types, 50% pollen sterility was expected because of the sterility of pollen grains carrying the Ns|Ts genotype. No abnormalities were observed in male gametophytes during the tetrad, unicellular, bicellular, or tricellular stages among T65, DGS1 semisterile plants, or DGS2 semisterile plants (data not shown). At the mature pollen stage, which occurs a few days before anthesis, almost all pollen grains stained well with I2–KI (Figure 3, A–C). Smaller pollen grains began to appear, but it was technically difficult to count their frequency because the size distribution was continuous, so two categories could not be clearly distinguished (Figure 3, D–F). At 1 hr before flowering (preflowering stage), most of pollen grains had begun glycosylation of starch in T65 and the DGS1 semisterile plants (Figure 3, G–H), whereas only half of the pollen grains (91 glycosylated/191 grains in total) in the DGS2 semisterile plants showed glycosylation (Figure 3I). At the flowering stage, 44.3 ± 1.6% of pollen grains from the DGS1 semisterile plants and 44.4 ± 3.5% of those from the DGS2 semisterile plants germinated in vitro, whereas 80.3 ± 4.5% of the T65 pollen grains germinated (Figure 3, J–L and Figure 4). Thus, half of the pollen grains from the DGS1 and DGS2 semisterile plants appeared to have lost their fertilization ability. These data demonstrate that for the DGS1 and DGS2 semisterile plants, the pollen germination and glycosylation stages, respectively, were the critical stages at which fertile and sterile pollen grains became evident.
Figure 3.
Characterization of sterility caused by DGS1 and DGS2. (A–C) Mature pollen stained with I2–KI. (D–F) Frequency distribution of pollen grain diameter at the preflowering stage. Black and white bars indicate numbers of glycosylated and nonglycosylated pollen grains, respectively. (G–I) Pollen grains stained by hematoxylin at the preflowering stage. Black arrowheads, glycosylated grains; white arrowheads, nonglycosylated grains. (J–L) In vitro pollen germination test. No., number.
Figure 4.
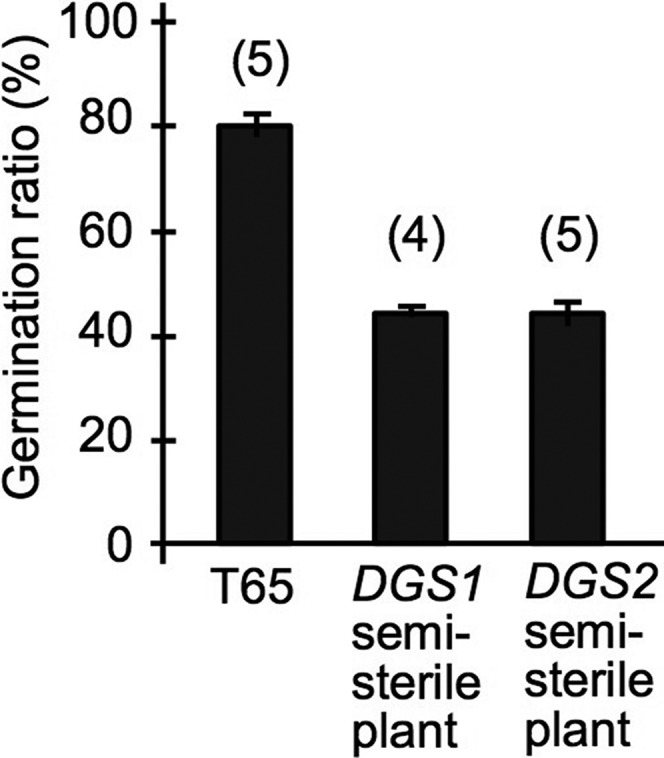
Pollen germination in vitro. Numbers in parentheses indicate numbers of plant replicates. Error bars indicate SEs in replications shown in parenthesis.
Map-based cloning
We conducted high-resolution mapping of DGS1 using 3380 plants from BC4F5 populations derived from DGS1 semisterile plants (Figure 2C). DGS1 was mapped to a 70 kb genomic region between RM16862 and RM16867 in the reference sequence of Nipponbare, with two recombinants between RM16862 and DGS1 and three between RM16867 and DGS1 (Table S6 in File S2). In the high-resolution mapping of DGS2 using 1901 plants of BC4F5 populations derived from DGS2 semisterile plants, we located DGS2 within a 30.8 kb genomic region of Nipponbare between M45_STS and M41_STS (Figure 2D). Ten recombinants were obtained between M45_STS and DGS2, and six between M41_STS and DGS2 (Table S7 in File S2).
To determine the genomic structure around the DGS1 and DGS2 regions, we performed BAC screening for the candidate region of each gene (Figure 2, C and D). For the DGS1 region, the T65 BAC clones GN0025A01 and GN0028G14 and the O. nivara clone IRGC105715_0023B24 were isolated from BAC libraries by using DNA markers RM16855, RM16862, RM16867, and RM16882. For the BAC clones around the DGS2 region, T65 clone GN0031H20 and O. nivara clones IRGC105715_0046I04 and IRGC105715_0022L03 were identified by using DNA markers RM6574, M45_STS, M41_STS, and M23_STS. Sequencing of one of the O. nivara BAC clones at DGS2, IRGC105715_0046I04, revealed five tandem copies of a duplicated segment (14,747, 15,902, 14,462, 15,942, and 15,925 bp) in the “finishing-phase” assembly (Figure 2E). Interestingly, these duplicated segments of ∼14–15 kb showed homology to BAC clones covering the regions responsible for DGS1-T65+ as one copy (13,677 bp) and DGS1-nivaras as one copy (15,968 bp), whereas the duplicated segment was not found (or had been deleted) in BAC clones from the sterile allele DGS2-T65s (Figure 2E, black arrowhead). We assumed a working hypothesis that the sterility could possibly be explained by the GDRL model at the DGS1 and DGS2 loci, which was caused by loss of function of a duplicated gene in the interchromosomal duplicated segment at DGS1-nivaras and absence of the duplicated gene at DGS2-T65s. In the segmental duplication on chromosome 4, only one gene, Os04g0394500, was predicted in the Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp). The MSU Rice Genome Annotation Project Database Release 7 (MSU7; http://rice.plantbiology.msu.edu/) also predicted RPC4 at the same genomic region of Os04g0394500 as LOC_Os04g32350. We conducted 5′- and 3′-RACE reactions to determine the cDNA structure of the RPC4 homolog of DGS1-T65+ (Figure 2F). The obtained coding sequence of DGS1-T65+ has a different structure from those of LOC_Os04g32350 and Os04g0394500 in the second, third, and seventh exons (Figure S3 in File S1). One RNA polymerase III subunit C4 domain (Pfam accession number PF05132) was detected by a Pfam domain search (Finn et al. 2014) in the C-terminal region of all three deduced amino acid sequences (Figure 2F and Figure S3 in File S1).
Consistent with the BAC sequencing results, the DGS1-T65+ region contains one copy of the RPC4 gene, whereas the DGS2-nivara+ region has five tandem copies of RPC4 (here designated RPC4a–e; Figure 2, E and F), whose deduced protein sequences are identical except at the 35th and 225th amino acid residues. At the 35th amino acid residue, the fertile allele DGS1-T65+, the sterile allele DGS1-nivaras, and three of the five tandem copies in DGS2-nivara+ possess an isoleucine (I) residue. At the 225th amino acid residue, all of the sequences except one of the five tandem copies in DGS2-nivara+ have a glycine (G) residue. Thus, the observed amino acid substitutions among the deduced proteins were not clearly associated with the allelic functionalities (i.e., fertile or sterile).
To confirm the functionality of the predicted RPC4 copies, we prepared genomic constructs containing individual RPC4 genes from the BAC clones of the DGS1-T65+ and DGS2-nivara+ alleles (Figure 2E) and transformed them into DGS1 semisterile plants (Table 4) and DGS2 semisterile plants (Table 5). For the transformation of RPC4 copies from DGS2-nivara+, we used the RPC4c, RPC4d (identical to RPC4b), and RPC4e (identical to RPC4a) copies (Figure 2F). Since sterile pollen grains were predicted to carry sterile alleles at both loci, i.e., DGS1-nivaras and DGS2-T65s, we hypothesized that introducing a copy of an RPC4 gene from either of the functional alleles (DGS1-T65+ or DGS2-nivara+) would rescue the fertility of pollen grains containing it and that Ns/Ns|Ts/Ts plants would segregate in the T1 generation.
Table 4. Segregation of genotypes in T1 generation derived by transformation of RPC4 genes derived from normal alleles into DGS1 semisterile plants.
| Transgene (Allele) | Segregation at RM16862 in T1 Generation | Pa | |||
|---|---|---|---|---|---|
| T+/T+|Ts/Ts | T+/Ns|Ts/Ts | Ns/Ns|Ts/Ts | Total | ||
| RPC4 (DGS1-T65+) | 19 | 39 | 10 | 68 | 8.0 × 10−4 |
| RPC4c (DGS2-nivara+) | 18 | 47 | 15 | 80 | 6.7 × 10−5 |
| RPC4d (DGS2-nivara+) | 22 | 33 | 18 | 73 | 1.1 × 10−6 |
| RPC4e (DGS2-nivara+) | 6 | 16 | 6 | 28 | 5.5 × 10−4 |
| Vector control | 45 | 48 | 1 | 94 | — |
Probabilities were calculated by Fisher’s exact test. We hypothesized that plants carrying the Ns/Ns|Ts/Ts genotype segregate with a probability of 1/94.
Table 5. Segregation of genotypes in T1 generation derived by transformation of RPC4 genes derived from normal alleles into DGS2 semisterile plants.
| Transgene (Allele) | Segregation at M41_STS in T1 Generation | |||
|---|---|---|---|---|
| Ns/Ns|Ts/Ts | Ns/Ns|N+Ts | Ns/Ns|N+N+ | Total | |
| RPC4 (DGS1-T65+) | 8 | 21 | 12 | 41 |
| RPC4c (DGS2-nivara+) | 3 | 17 | 6 | 26 |
| RPC4d (DGS2-nivara+) | 6 | 15 | 7 | 28 |
| RPC4e (DGS2-nivara+) | 17 | 37 | 15 | 69 |
| Vector control | 0 | 26 | 22 | 48 |
In the T1 population derived from transformation of an empty vector into DGS1 semisterile plants, T+/T+|Ts/Ts, T+/Ns|Ts/Ts, and Ns/Ns|Ts/Ts plants segregated in a 45:48:1 ratio at SSR marker RM16862. The single plant with an Ns/Ns|Ts/Ts genotype was assumed to be a recombinant between DGS1 and RM16862. Ns/Ns|Ts/Ts plants were recovered at significantly higher frequencies in the T1 progeny derived from transformation of the RPC4 genomic segment of DGS1-T65+ into DGS1 semisterile plants than in the vector control plants (Table 4). To examine the functional redundancy of RPC4 copies at DGS1-T65+ and DGS2-nivara+, we also transformed genomic segments containing RPC4c, RPC4d, and RPC4e of the DGS2-nivara+ allele into DGS1 semisterile plants (Table 4). Similarly, we obtained many Ns/Ns|Ts/Ts plants in all T1 progeny, at a frequency significantly higher than in the vector control. Likewise, the T1 generation of DGS2 semisterile plants transformed with RPC4 genes showed recovery of Ns/Ns|Ts/Ts plants, whereas the T1 generation produced by transformation with the empty vector had no Ns/Ns|Ts/Ts plants (Table 5). The observed genotypic frequency in the T1 progeny fit the expected ratio of genotypes depending on the numbers of introduced transgenes in the T0 generation as determined by quantitative PCR (Table S8 and Table S9 in File S2). These data demonstrate that DGS1 and DGS2 are duplicate loci encoding a protein homologous to RPC4.
The sterile allele of DGS2-T65s was found to be a null allele because RPC4 was absent (Figure 2E). The other sterile allele, DGS1-nivaras, is suggested to be a loss-of-function allele caused by a deficiency or absence of expression during male gametogenesis. Within the 3000-bp genomic sequence upstream from the initial codon of the RPC4 genes, the RPC4 gene of DGS1-nivaras has 2 nt substitutions—one from G to A at −2373 bp (G−2373A) and one from C to T at −2234 bp from the initial codon (C−2234T)—that distinguish it from all of the fertile alleles (Table S10 in File S2). But we could not conclude that these nucleotide substitutions were essential mutations for the sterility of pollen grains carrying genotype Ns|Ts.
Duplication of RPC4 homologs in rice
Another putative RPC4 homolog, LOC_Os01g66580, was found on rice chromosome 1 by similarity search using BLAST (Altschul et al. 1990). The LOC_Os01g66580 sequence in the T65 genome was identical to that in the rice reference sequence of Nipponbare. To examine the divergence of RPC4 homologs found in the rice genome, a maximum-likelihood inference tree of the RPC4 domain was constructed using the predicted amino acid sequences of RPC4 homologs from rice and other plants (Figure S4 in File S1). The analysis suggests that the RPC4 homologs of Poales or Poaceae species form two monophyletic clades (designated Poaceae1 and Poaceae2) that separated from those of dicots. The RPC4 homologs encoded by DGS1-T65+ and DGS2-nivara+ were classified into the Poaceae1 clade and the LOC_Os01g66580 protein was classified into the Poaceae2 clade. The RPC4 homologs found in the Zingiberales (banana) and Arecales (African oil palm), which represent other commelinid species, were similar to the ones in the Poaceae1 clade but not to those in the Poaceae2 clade, suggesting that the gene duplication leading to the RPC4 homologs that distinguish the Poaceae1 and Poaceae2 groups occurred in an ancestral population of the Poales or Poaceae, and that RPC4 members in the Poaceae1 clade probably retain the canonical RPC4 function in commelinids. The rice RPC4 members encoded by DGS1-T65+ and DGS2-nivara+ formed one monophyletic group in the Poaceae1 clade. This result suggests that interchromosomal segmental duplication (ISD) between chromosomes 4 and 7 and tandem duplication of RPC4 members at DGS2-nivara+ occurred after divergence of rice from other Poaceae species.
Spatial and temporal expression of DGS1 and DGS2
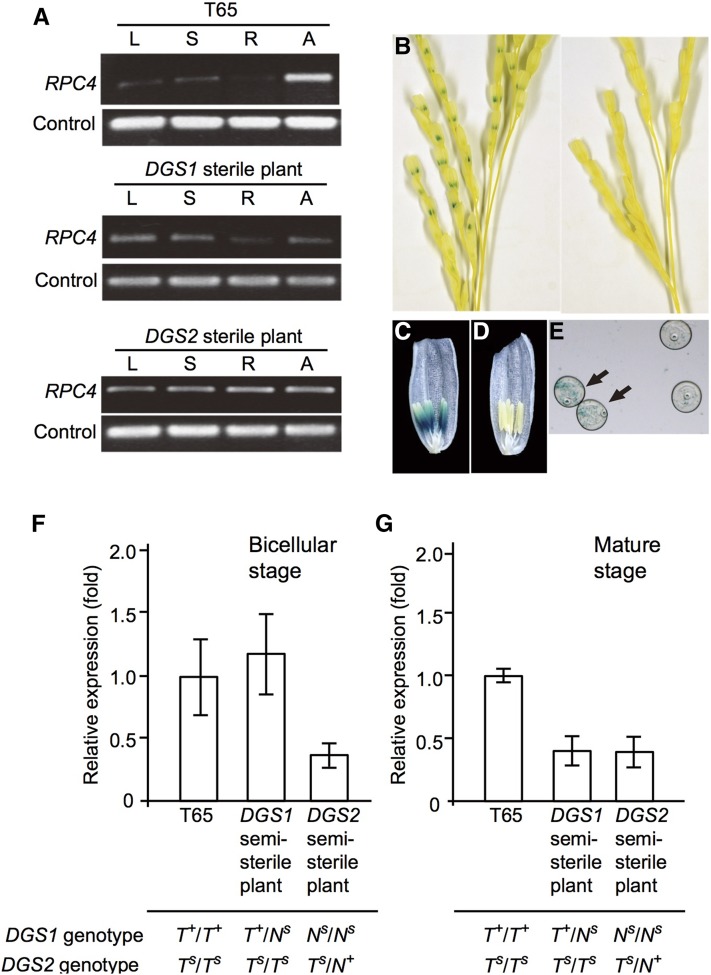
Expression analysis of DGS1 and DGS2 was conducted in the leaf, stem, roots, and anthers of T65 and of DGS1 and DGS2 semisterile plants. Because the RNA transcripts derived from different DGS genes are almost identical, the expression of those transcripts was analyzed as a whole. Expression of RPC4 was observed in various tissues, but it was strongest in the anther of T65 (Figure 5A). The expression of GUS driven by the RPC4 promoter derived from the DGS1-T65+ allele was strong in pollen grains, but was not detected in the anther wall or anywhere else (Figure 5, B–E).
Figure 5.
Expression of DGS1 and DGS2. (A) RT-PCR expression analysis of RPC4 in different plant and tissue types. L, leaf blade; S, leaf sheath; R, roots; A, anther containing pollen at the bicellular stage. Expression was normalized to that of Ub-CEP52-1 (Os03g0234200). (B) Promoter–GUS assay of a T0 plant transformed with an RPC4 promoter–GUS construct (left) and T65 (right). (C and D) Promoter–GUS assay of the floret of a T0 plant (C) and T65 (D). (E) Promoter–GUS assay of pollen grains from the floret of a T0 plant. Arrows indicate blue GUS signal, which segregated in the pollen as expected. (F and G) Relative expression of RPC4 in anthers of T65, DGS1 semisterile plants, and DGS2 semisterile plants, at the (F) bicellular and (G) mature pollen stage. Error bars indicate SEs of three biological replicates (plants). RT-PCR, reverse-transcription polymerase chain reaction.
By using qRT-PCR, we estimated the RPC4 transcript levels in anthers of T65, DGS1 semisterile plants, and DGS2 semisterile plants (Figure 5, F–G). In the DGS1 semisterile plants, the RPC4 transcript level was reduced by half at the mature stage compared with the T65 control (Figure 5G). T65 is homozygous for DGS1-T65+, encoding functional RPC4, and homozygous for DGS2-T65s, from which the RPC4 gene is completely absent (Figure 2E). T65 produces all normal pollen grains carrying the T+|Ts genotype, whereas the DGS1 semisterile plants produce normal pollen grains carrying T+|Ts and sterile pollen grains carrying Ns|Ts. Therefore, we considered that the reduction in RPC4 transcript level was probably caused by the 50% reduction in the number of normal pollen grains carrying T+|Ts in the DGS1 semisterile plants (T+/Ns|Ts/Ts) compared with T65 (T+/T+|Ts/Ts), and that DGS1-nivaras was expressed weakly or not at all at the mature stage (Figure 5G). The DGS2 semisterile plants (Ns/Ns|Ts/N+ genotype) also showed half the amount of RPC4 transcripts compared with T65. These results demonstrate that RPC4 of DGS2-nivara+ and RPC4 of DGS1-T65+ express at similar levels because, as previously described, the sterile alleles at each locus appear to contribute little if any RPC4 transcript at the mature stage.
At the bicellular stage, the amount of RPC4 transcript in the DGS2 semisterile plants was approximately one-third of that in the T65 control, whereas the amounts of RPC4 transcripts in the DGS1 semisterile plants and T65 were comparable (Figure 5F). In Arabidopsis, it is suggested that male gametophytes have two sources of PolIII subunits: PolIII machinery synthesized by the diploid meiocyte (sporophyte) and persisting in mature pollen, and machinery synthesized in the developing pollen (gametophyte) (Onodera et al. 2008). As one possibility, RPC4 transcripts synthesized in meiocytes might contribute substantially to the total RPC4 transcripts in pollen in the DGS1 semisterile plants at the bicellular stage, since pollen carrying the sterile genotype Ns|Ts did not show any abnormal phenotype. The PolIII machinery synthesized in zygotic meiocytes seems to be enough for pollen maturation and pollen tube germination, because pollen grains carrying the loss-of-function allele for the PolIII subunit complete pollen maturation and pollen tube germination in Arabidopsis mutants. However, the mutant Arabidopsis pollen did not complete pollen tube elongation because de novo synthesis of PolIII machinery is required for late stages of pollen tube elongation (Onodera et al. 2008). We speculate that the timing of starvation of PolIII machinery synthesized in the meiocyte is critical for Ns|Ts pollen grains prior to their developmental failure. The sterile pollen grains produced by DGS1 semisterile plants accomplish starch glycosylation at flowering but fail to germinate, whereas those produced by DGS2 semisterile plants fail starch glycosylation as early as the preflowering stage (Figure 3, G–L). Although the evidence at this point is indirect, we assume that DGS1 semisterile plants maintain the same RPC4 transcript levels as T65 up through the bicellular stage and, thus, achieve further development of pollen grains carrying the sterile genotype Ns|Ts than do DGS2 semisterile plants. However, all Ns|Ts pollen grains eventually fail in pollen tube germination owing to little or no de novo RPC4 transcription in pollen and starvation of RPC4 transcripts synthesized in meiocytes of both DGS1 and DGS2 semisterile plants.
Discussion
Duplication and loss-of-function of RPC4 caused hybrid pollen sterility
We revealed that a combination of loss-of-function alleles of RPC4 in male gametophytes in hybrids caused hybrid pollen sterility in an interspecific rice hybrid between O. sativa ssp. japonica-type cultivar T65 and wild O. nivara. Pollen sterility was observed in pollen grains carrying two sterile alleles, DGS1-nivaras and DGS2-T65s (pollen genotype Ns|Ts). The DGS2-T65s allele did not possess an RPC4 copy (Figure 2). For the absence of the RPC4 copy at DGS2-T65s, two hypotheses were suggested; (1) the RPC4 copy of the Poaceae1 group was originally located at DGS1 on chromosome 4 in genus Oryza and an ISD did not occur on the ancestral lineage of T65 at DGS2 on chromosome 7; or (2) the DGS2-T65s allele is the result of an ISD followed by a deletion. It is possible that these hypotheses can be tested by diversity analysis in O. sativa and O. rufipogon gene pools to investigate the presence or absence of the ISD at DGS2. On the other hand, the RPC4 gene in DGS1-nivaras seemed to contribute little to no RPC4 transcript in pollen grains carrying the Ns|Ts genotype (Figure 5, F and G). Two nucleotide substitutions, G−2373A and C−2234T, found in a region 3000 bp upstream from the deduced initial codon of the RPC4 homolog in DGS1-nivaras, are suggested to be causal variations for nonfunctionalization of the RPC4 homolog in DGS1-nivaras (Table S10 in File S2). Using genome editing technology, substitution of the “G” nucleotide at position −2373 in functional RPC4 alleles with the “A” found in DGS1-nivaras would confirm whether this substitution is required for pollen sterility.
Male-biased sterility due to lack of an RPC4 homolog in rice
Since RNAPs are vital and fundamental transcriptional machineries in all eukaryotes, a deficit of functional RNAP components would lead to life cycle failure. In this study, gametes carrying the Ns|Ts genotype showed male-biased sterility: male gametophytes carrying both sterile alleles (Ns|Ts) failed in either pollen maturation (DGS2 semisterile plants) or pollen tube germination (DGS1 semisterile plants), but there was no obvious abnormality on the female side. Sex-biased sterility in RNAP subunit-deficient mutants has also been observed in Arabidopsis thaliana. In mutant lines deficient in the second largest subunits of RNAP I (PolI), II (PolII), and III (PolIII) (designated NRPA2, NRPB2, and NRPC2, respectively), the female gametes carrying defective alleles nrpa2, nrpb2, or nrpc2 completely aborted and male gametes with the defective alleles partially retained fertilization ability (Onodera et al. 2008). The female-biased sterility in these mutants was suggested to be caused by different characteristics of RNAP synthesis in male and female gametes: the female gamete is autonomous from sporophytic tissues with regard to gene expression and its RNAP is synthesized in the haploid female gametes de novo, whereas male gametophytes contain both RNAP machinery that is synthesized in the diploid meiocyte and partitioned into four microspores and machinery newly synthesized in haploid pollen during pollen maturation.
To explain the male-biased sterility observed in this study, we suggest two hypotheses. One is subfunctionalization of DGS1/DGS2 and LOC_Os01g66580 with respect to male vs. female gametogenesis. If de novo synthesis of RPC4 is indispensable in female gametogenesis in rice, perhaps the RPC4 homolog LOC_Os01g66580 (chromosome 1) is active in female gametes, thus preserving female fertility. The other hypothesis suggests that the G−2373A nucleotide substitution at DGS1-nivaras results in loss of transcriptional control of RPC4 in male gametes only. Analysis of a mutant deficient for LOC_Os01g66580 would provide better understanding of sex-biased sterility in RPC4 in rice.
Evolution of transcriptional machinery via gene duplication
Our phylogenetic analysis demonstrated that divergence of the RPC4 homologs DGS1/DGS2 (Poaceae1) and LOC_Os01g66580 (Poaceae2) occurred in an ancestral population of the Poales or Poaceae (Figure S4A in File S1), and that the RPC4 duplication giving rise to DGS1 and DGS2 occurred in an ancestral lineage of O. sativa and O. nivara. Although duplicated genes rapidly lose their function in general, the RPC4 members of the Poaceae2 group have been well conserved in all Poaceae members used in the analysis. RPC4 members in Poaceae2 may have the diverged role in RNA synthesis whereas the Poaceae1 group sequences retain the canonical RPC4 function.
The ancestral lineage of the Poaceae family, such as the BEP clade (rice and purple false brome) and the PACMAD clade (sorghum, maize, foxtail millet, and switchgrass), experienced at least two instances of whole-genome duplication (WGD; paleopolyploidization), called σ and ρ, following the divergence of commelinids (Paterson et al. 2004; Wang et al. 2005; Tang et al. 2010). It has been estimated that the most recent paleopolyploidization, ρ, occurred 70 MYA and that σ paleopolyploidization occurred after the Poaceae–Arecaceae split 120–83 MYA, before divergence of the major Poaceae crop species (Paterson et al. 2004, 2009). Arecaceae (African oil palm), Zingiberales (banana), and Asparagales (Crocus) species do not have RPC4 members in Poaceae2 (Figure S4 in File S1). The genus Oryza diverged 8–14 MYA (Guo and Ge 2005) and diversification of the AA-genome species occurred over the past 2 MY (Zhu and Ge 2005). Thus, we estimate that the gene duplication leading to DGS1/DGS2 and LOC_Os01g66580 occurred after the Poaceae–Arecaceae split 120–83 MYA, and that duplication of RPC4 leading to DGS1 and DGS2 occurred > 2–14 MYA. Tang et al. (2010) found homeologous chromosomal regions on chromosomes 1 and 4 that originated from the σ paleopolyploidization. However, we do not have sufficient evidence to conclude that the RPC4 homologs DGS1/DGS2 and LOC_Os01g66580 were derived from WGD. The other possibility is that the gene duplication leading to DGS1/DGS2 (Poaceae1) and LOC_Os01g66580 (Poaceae2) resulted from segmental genomic duplication independent from the σ and ρ paleopolyploidization in the ancestral population of the Poaceae.
Segmental duplication and WGD are primary sources of gene redundancy, consequent massive gene losses, and evolutionary novelty (Ohno 1970; Lynch and Conery 2000; Moore and Purugganan 2005). WGD events are found to have independently and sporadically occurred in many angiosperm lineages (James and Lyons 2015). The number of RPC4 copies observed in this study was fewer than expected based on the number of WGD events experienced in each lineage (Figure S4B in File S1). For example, we found only two RPC4 copies at present in the banana (Zingiberales) genome, despite the fact that this genome experienced three successive rounds of WGD after divergence from the Poales. Arabidopsis underwent three rounds of WGD (α, β, and γ) (Bowers et al. 2003), but A. thaliana has only two RPC4 copies, NPBC14a and NPBC14b, the products of which were proven to have the ability to bind to the PolIII complex in Arabidopsis (Ream et al. 2015). The phylogenetic analysis showed that the duplication of NRPC14a and NPRC14b in the Brassicaceae probably originated from a duplication event independent of that related to RPC4 of the Poaceae1 and Poaceae2 groups. These results suggest that duplicated genes may have been eliminated from gene pools in intermediate evolutionary steps, and positional variations of RPC4 homologs on chromosomes may have occurred during diploidization after WGDs in several evolutionary lineages, leading to reproductive isolation barriers. In plants, WGD or polyploidization has long been considered a major force of speciation (Stebbins 1950; Soltis et al. 2014). Other RNAP components, as well as other duplicated ubiquitous genes indispensable for the life cycle in eukaryotes, may represent potential sources of postzygotic reproductive isolation without any acquisition of functional novelty. In crop species including rice, high-throughput and comprehensive investigation of sequence variants among many varieties and related wild species has actively progressed. This massively accumulated information reveals the numbers, positions, and birth-and-death dating of duplicated genes, and consequently the crop-lineage-specific landscape of genome complexity and plasticity in the history of monocotyledons.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043943/-/DC1.
Acknowledgments
This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, QTL-5002 to A.Y.), from the Ministry of Education, Culture, Sports, Science and Technology of Japan [Grant-in-Aid for Scientific Research (A) grant number 24248002 and National Bioresource Project (Rice) to A.Y. and Grant-in-Aid for Scientific Research (B) grant number 16KT0034 to Y.Y.], and from and the Japanese Society for Promotion of Science (P10401 to G.N.N.).
Footnotes
Communicating editor: A. H. Paterson
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 1215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bikard D., Patel D., Mette L. C., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Weigel D., 2007. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393. [DOI] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J., Paterson A. H., 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Brideau N., Flores H., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Chang M. T., Neuffer M. G., 1989. Maize microsporogenesis. Genome 32: 232–244. [Google Scholar]
- Chen C., Chen H., Lin Y. S., Shen J. B., Shan J. X., et al. , 2014. A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5: 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Dellaporta S., Wood J., Hicks J., 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Report. 1: 19–21. [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T., Sasaki T., Yano M., 2001. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18: 219–222. [Google Scholar]
- Guo Y. L., Ge S., 2005. Molecular phylogeny of Oryzeae (Poaceae) based on DNA sequences from chloroplast, mitochondrial, and nuclear genomes. Am. J. Bot. 92: 1548–1558. [DOI] [PubMed] [Google Scholar]
- Haag J. R., Pikaard C. S., 2011. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. [DOI] [PubMed] [Google Scholar]
- James, S., and E. Lyons, 2015 Plant paleopolyploidy. Figshare. Available at: https://figshare.com/articles/Plant_Paleopolyploidy/1538627.
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., et al. , 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Long Y., Zhao L., Niu B., Su J., Wu H., et al. , 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 105: 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A. G., 2000. The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156: 590–605. [DOI] [PubMed] [Google Scholar]
- Mann C., Micouin J. Y., Chiannilkulchai N., Treich I., Buhler J. M., et al. , 1992. RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol. Cell. Biol. 12: 4314–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta Y., Harushima Y., Kurata N., 2010. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 107: 20417–20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. C., Purugganan M. D., 2005. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., et al. , 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nguyen G. N., Hailstones D. L., Wilkes M., Sutton B. G., 2010. Role of carbohydrate metabolism in drought-induced male sterility in rice anthers. J. Agron. Crop Sci. 196: 346–357. [Google Scholar]
- Noor M. A. F., Feder J. L., 2006. Speciation genetics: evolving approaches. Nat. Rev. Genet. 7: 851–861. [DOI] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer, Berlin, Heidelberg, Germany. [Google Scholar]
- Onodera Y., Haag J. R., Ream T., Costa Nunes P., Pontes O., et al. , 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Onodera Y., Nakagawa K., Haag J. R., Pikaard D., Mikami T., et al. , 2008. Sex-biased lethality or transmission of defective transcription machinery in Arabidopsis. Genetics 180: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Chapman B. A., 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Ream T. S., Haag J. R., Wierzbicki A. T., Nicora C. D., Norbeck A., et al. , 2009. Subunit compositions of the RNA-silencing enzymes pol IV and pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream T. S., Haag J. R., Pontvianne F., Nicora C. D., Norbeck A. D., et al. , 2015. Subunit compositions of Arabidopsis RNA polymerases I and III reveal Pol I- and Pol III-specific forms of the AC40 subunit and alternative forms of the C53 subunit. Nucleic Acids Res. 43: 4163–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Blackman B. K., 2010. Speciation genes in plants. Ann. Bot. 6: 439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Visger C. J., Soltis P. S., 2014. The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 101: 1057–1078. [DOI] [PubMed] [Google Scholar]
- Stebbins G. L., 1950. Variation and Evolution in Plants. Oxford University Press, London, UK. [Google Scholar]
- Tang H. J. E., Bowers X., Wang A. H., Paterson A. H., 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 107: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Van de Peer Y., Meyer A., 2001. Genome duplication, divergent resolution and speciation. Trends Genet. 17: 299–301. [DOI] [PubMed] [Google Scholar]
- Temnykh S., DeClerck G., Lukashova A., 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Cramer P., 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45: 439–446. [DOI] [PubMed] [Google Scholar]
- Wang X., Shi X., Hao B., Ge S., Luo J., 2005. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 165: 937–946. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ma H., 2015. Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits. New Phytol. 207: 1198–1212. [DOI] [PubMed] [Google Scholar]
- Win K. T., Yamagata Y., Miyazaki Y., Doi K., Yasui H., et al. , 2011. Independent evolution of a new allele of F1 pollen sterility gene S27 encoding mitochondrial ribosomal protein L27 in Oryza nivara. Theor. Appl. Genet. 122: 385–394. [DOI] [PubMed] [Google Scholar]
- Yamagata Y., Yamamoto E., Aya K., Win K. T., Doi K., et al. , 2010. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. USA 107: 1494–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Takashi T., Morinaka Y., Lin S., Wu J., et al. , 2010. Gain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol. Genet. Genomics 283: 305–315. [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao X., Cheng K., Du H., Ouyang Y., et al. , 2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Ge S., 2005. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 167: 249–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. File S1 contains all of the supplemental figures and methods. All genotype data are presented in the supplemental tables (File S2). Sequence data have been deposited in the DNA Data Bank of Japan for RPC4 from the DGS1-T65+ allele (AB758279); RPC4 from the DGS1-nivaras allele (AB758280); RPC4a (AB758281), RPC4b (AB758282), RPC4c (AB758283), RPC4d (AB758284), and RPC4e (AB758285) from the DGS2-nivara+ allele; and BAC sequences of GN0028G14 for DGS1-T65+ (LC060740), IRGC105715_0023B24 for DGS1-nivaras (LC060741), GN0031H20 for DGS2-T65s (LC060743), and IRGC105715_0046I04 for DGS2-nivara+ (LC060742).