Abstract
Spongy degeneration with cerebellar ataxia (SDCA) is a genetically heterogeneous neurodegenerative disorder with autosomal recessive inheritance in Malinois dogs, one of the four varieties of the Belgian Shepherd breed. Using a combined linkage and homozygosity mapping approach we identified an ∼10.6 Mb critical interval on chromosome 5 in a Malinois family with four puppies affected by cerebellar dysfunction. Visual inspection of the 10.6 Mb interval in whole-genome sequencing data from one affected puppy revealed a 227 bp SINE insertion into the ATP1B2 gene encoding the β2 subunit of the Na+/K+-ATPase holoenzyme (ATP1B2:c.130_131insLT796559.1:g.50_276). The SINE insertion caused aberrant RNA splicing. Immunohistochemistry suggested a reduction of ATP1B2 protein expression in the central nervous system of affected puppies. Atp1b2 knockout mice had previously been reported to show clinical and neurohistopathological findings similar to the affected Malinois puppies. Therefore, we consider ATP1B2:c.130_131ins227 the most likely candidate causative variant for a second subtype of SDCA in Malinois dogs, which we propose to term spongy degeneration with cerebellar ataxia subtype 2 (SDCA2). Our study further elucidates the genetic and phenotypic complexity underlying cerebellar dysfunction in Malinois dogs and provides the basis for a genetic test to eradicate one specific neurodegenerative disease from the breeding population in Malinois and the other varieties of the Belgian Shepherd breed. ATP1B2 thus represents another candidate gene for human inherited cerebellar ataxias, and SDCA2-affected Malinois puppies may serve as a naturally occurring animal model for this disorder.
Keywords: Canis familiaris, canine, Malinois, Na+/K+-ATPase, β2 subunit, adhesion molecule on glia, AMOG, astrocytes, brain, central nervous system, epilepsy, KCNJ10, cerebellar dysfunction
Inherited (cerebellar) ataxia in humans represents a broad group of clinically, pathologically, and genetically heterogeneous neurodegenerative disorders characterized by progressive degeneration of cerebellum and, to a variable degree, of extracerebellar structures (Manto and Marmolino 2009; Hersheson et al. 2012; Jayadev and Bird 2013). Inherited ataxias are divided in autosomal recessive, autosomal dominant, X-linked, and mitochondrial ataxias. Autosomal recessive and autosomal dominant inheritance patterns represent the most prevalent inherited ataxias with ∼20 and 40 identified causative genetic variants so far, respectively (Washington University Neuromuscular Disease Center Web site: http://neuromuscular.wustl.edu; Mancuso et al. 2014; Klockgether and Paulson 2011; Jayadev and Bird 2013). Cerebellar ataxia, the main clinical feature of these disorders, becomes manifest as imbalance and lack of coordination. Ataxia may be the sole sign of cerebellar dysfunction or, more frequently, be accompanied by a wide spectrum of additional neurological manifestations (Hersheson et al. 2012; Mancuso et al. 2014). Disparate pathophysiological mechanisms have been described for inherited cerebellar ataxias, however some recurrent components emerge. These include accumulation of protein aggregates, impaired ion channel functions, defects in the DNA-repair system, and mitochondrial dysfunction (Manto and Marmolino 2009; Klockgether and Paulson 2011; Sandford and Burmeister 2014).
In veterinary medicine, similar to human medicine, cerebellar ataxias are described as a heterogeneous group of neurodegenerative disorders with variability in disease onset, severity and histopathological lesions (Urkasemsin et al. 2010; Urkasemsin and Olby 2014). However, to date, a genetic basis has been described for only some autosomal recessive inherited cerebellar ataxias in the dog (Supplemental Material, Table S1, Online Mendelian Inheritance in Animals Web site: http://omia.angis.org.au). Currently, there are no consensus criteria for the classification of canine neurodegenerative diseases, and denominations of entities are mainly based on clinical and/or neuropathological features. The increasing knowledge of the genetic defects underlying these disorders is expected to facilitate the implementation of a neurodegeneration classification scheme in animals and the study of pathogenetic mechanisms (Urkasemsin and Olby 2014).
We studied ataxias in the Belgian Shepherd breed and recently reported a candidate causative variant in the KCNJ10 gene for spongy degeneration with cerebellar ataxia, subtype 1 (SDCA1). This study revealed an unexpected genetic heterogeneity in clinically comparable cases, suggesting that more than one type of cerebellar ataxia is present in Belgian Shepherd dogs (Kleiter et al. 2011; Mauri et al. 2017). The KCNJ10 variant was also identified in an independent study (Stee et al. 2016; Van Poucke et al. 2017).
The aim of the present study was to identify the presumed causative genetic defect of a second form of SDCA in Belgian Shepherd dogs, which we propose to term SDCA2.
Materials and Methods
Ethics statement
All animal experiments were performed according to the local regulations. All dogs in this study were examined with the consent of their owners. The collection of blood samples was approved by the Cantonal Committee for Animal Experiments (Canton of Bern; permit 75/16).
Breed nomenclature
The Federation Cynologique Internationale (FCI) describes the Malinois, together with the Groenendael, the Laekenois, and the Tervueren, as a variety of the Belgian Shepherd dog breed. The American Kennel Club, however, officially recognizes the Belgian Malinois, the Belgian Sheepdog (FCI: Groenendael), the Belgian Laekenois (FCI: Laekenois), and the Belgian Tervuren (FCI: Tervueren) as four distinct breeds. In this paper all references to the breed nomenclature correspond to the FCI standards.
Animal selection
We used the same animals as in our previous study (Mauri et al. 2017). We investigated six related Malinois families in which 12 puppies showed clinical signs of cerebellar dysfunction (Figure 1). We also examined seven Malinois puppies with reported cerebellar signs, for which no relatives were available. In addition to these individuals, we genotyped 230 other Malinois, 25 Groenendael, two Laekenois, and 35 Tervueren dogs whose blood samples were donated to the biobank of the Institute of Genetics at the University of Bern. Furthermore, we analyzed 503 samples from 87 genetically diverse dog breeds.
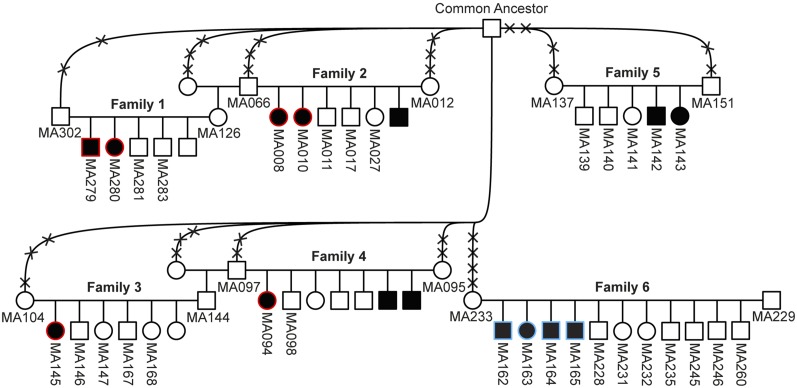
Figure 1.
Pedigree of Malinois dogs used for genetic mapping of the disease loci, modified from Mauri et al. (2017). Filled symbols represent animals with cerebellar dysfunction. Numbers indicate dogs from which samples were available. Six dogs affected by SDCA1 (KCNJ10:c.986T>C) are indicated by red contours. Four affected siblings from family 6, which did not carry the previously identified KCNJ10 variant, are indicated by blue contours and were selected for homozygosity mapping in this study. The affected animals MA142 and MA143 from family 5 seem to have yet another genetic form of cerebellar dysfunction (see Results and Discussion). Crosses intersecting the connection lines to the common ancestor represent the numbers of generations (e.g., MA302 is a great-grandson of the common ancestor).
Neuropathology and immunohistochemistry
Two Malinois puppies from family 5 and three from family 6 with signs of cerebellar dysfunction were necropsied (MA142, MA143, MA162, MA164, MA165, Figure 1). Brain and spinal cord samples from these five puppies were collected and fixed in 4% buffered formaldehyde solution, embedded in paraffin, and sectioned at 2–5 µm. Eye samples were available from puppy MA162 and processed in the same manner. Sections were stained with hematoxylin and eosin and examined by light microscopy. Furthermore, we performed immunohistochemistry (IHC) with a polyclonal rabbit antibody raised against a peptide corresponding to amino acids 115–141 of the human ATP1B2 protein. This epitope is 100% identical between human and dog. IHC was performed on paraffin-embedded cerebellar and brain stem sections from the SDCA2-affected puppies MA162, MA164, MA165; and 13 control dogs, which consisted of nine dogs that did not suffer from cerebellar ataxia and four Malinois puppies that were affected by SDCA1 and homozygous for the KCNJ10:c.986T>C variant. To this end, sections were deparaffinized, and antigen heat retrieval was performed by boiling sections in pH 9 buffer (Dako Target Retrieval Solution, pH 9) in a laboratory microwave (20 min at 95°) following a peroxidase block and a blocking step with 10% normal goat serum. Tissue sections were incubated overnight at 4° with the primary antibodies (Thermo Fisher Scientific PA5-26279, 1:50 dilution of the 0.5 mg/ml stock solution) and the reaction was visualized with the Dako REAL Detection System according to the manufacturer’s instructions. The manufacturer’s documentation for the primary antibody showed a Western Blot in which only a single specific band of ∼34 kDa was detected.
DNA extraction and genotyping
Genomic DNA was isolated from EDTA blood samples with the Maxwell RSC Whole Blood DNA Kit, which were used with the Maxwell RSC Instrument (Promega). Genotyping was done on Illumina CanineHD Chips containing 173,662 genome-wide SNPs by GeneSeek/Neogen. Genotypes were stored in a BC/Gene database version 3.5 (BC/Platforms).
Linkage and homozygosity mapping
Linkage analysis was performed with Illumina CanineHD SNP Chip genotypes from 20 dogs belonging to family 5 and 6 (Figure 1). We analyzed the data set for parametric linkage under a fully penetrant, recessive model of inheritance with the Merlin software (Abecasis et al. 2002). PLINK v1.07 (Purcell et al. 2007) was used as described (Wiedmer et al. 2016) to search for extended intervals of homozygosity with shared alleles across selected affected animals.
Reference sequences
The dog CanFam 3.1 genome assembly and NCBI annotation release 103 was used for all analyses. All references to the canine ATP1B2 gene correspond to the accessions XM_546597.5 (mRNA) and XP_546597.2 (protein). XP_546597.2 has the same length as the human protein (NP_001669.3; 290 amino acids) and 286 out of 290 amino acids (99%) are identical between dog and human.
Whole-genome resequencing
A PCR-free fragment library was prepared from one affected Malinois dog (MA163) with a 400 bp insert size. We sequenced the library to roughly 32× coverage on an Illumina HiSeq3000 instrument using 2 × 150 bp paired-end reads. The reads were mapped to the dog reference genome assembly CanFam3.1 as previously described (Mauri et al. 2017). The sequence data were deposited under the study accession number PRJEB16012 at the European Nucleotide Archive (ENA). The sample accession number is SAMEA104032048. We also used 146 control genomes, which were either publicly available (Bai et al. 2015) or produced during other projects in our group (Table S2).
Single nucleotide and small indel variants were individually identified using GATK HaplotypeCaller in gVCF mode, and subsequently genotyped per chromosome and genotyped across all samples simultaneously (Van der Auwera et al. 2013). We filtered the obtained data with the variant filtration module of GATK and used the ENSEMBL annotation CanFam 3.1 (version 72) to predict the functional effects of the called variants together with SnpEff software (Cingolani et al. 2012). The resulting sequence alignments of MA163 were visually inspected and screened for structural variants with the Integrated Genomics Viewer (IGV) software (Robinson et al. 2011).
PCR and Sanger sequencing
Sanger sequencing was used to confirm the variant identified from whole-genome resequencing. For these experiments we amplified PCR products from genomic DNA using AmpliTaq Gold 360 Master Mix (Life Technologies). The PCR primers used for the genotyping of the ATP1B2:c.130_131insLT796559.1:g.50_276 variant were GAACCCCCTGACTCCATTTC (forward primer) and GGAGCAGTTAAAGGCTGGTG (reverse primer). PCR products were directly sequenced on an ABI 3730 capillary sequencer (Life Technologies) after treatment with exonuclease I and shrimp alkaline phosphatase. Sanger sequence data were analyzed with Sequencher 5.1 (GeneCodes). The sequence of the PCR product with the insertion allele was submitted to the ENA under accession number LT796559.1.
Fragment length analysis
To genotype a large number of samples we used fragment length analyses by assessing the PCR product sizes on the Fragment Analyzer Automated CE (capillary gel electrophoresis) System [Advanced Analytical Technologies (AATI)]. We used the PROSize analytical software (AATI) to visually inspect the obtained gel images and classify the dogs as homozygous for the SINE (short interspersed nuclear elements) insertion (ins/ins, single band of ∼425 bp), heterozygous (wt/ins, two bands of ∼198 and ∼425 bp), or homozygous wild type (wt/wt, single band of ∼198 bp).
RNA isolation and reverse transcription-PCR
Total RNA was purified from the skin of one affected Malinois puppy (MA133) with the RNeasy Fibrous Tissue Mini Kit according to the manufacturer’s recommendations (QIAGEN). Blood total RNA from one control dog (SY045) was isolated as described (Wiedmer et al. 2016). The RNA samples were treated with RNase-free DNase to remove contamination with genomic DNA. The SuperScript IV Reverse Transcriptase kit was used to generate cDNA according to the manufacturer’s recommendations (Thermo Fisher Scientific). Primers for reverse transcription PCR (RT-PCR) were designed in exon 1 and at the boundary of exons 3 and 4 of the ATP1B2 gene (forward primer: GTGGTTGAGGAGTGGAAGGA; reverse primer: TGGAATCGTTGTAAGGCTCCAA). 30 cycles of PCR were performed with AmpliTaq Gold 360 Master Mix (Life Technologies). RT-PCR products were analyzed with the Fragment Analyzer and preparatively separated using the DNA Size Selection System PippinHT (Sage Science) according to the manufacturer’s recommendations. The resulting isolated fragments were sequenced separately as described above.
Data availability
File S1 is a video showing the clinical phenotype of the four affected Malinois siblings belonging to family 6 at 4 wk of age (MA162–165). File S2 depicts the sequence context of the 227 bp SINE insertion into exon 2 of the ATP1B2 gene. File S3 illustrates the aberrant ATP1B2 exon 2 splicing patterns in dogs with the SINE insertion and the predicted protein sequences resulting from the mutant transcripts. Figure S1 contains a screenshot of the IGV software in the region of the visually identified ATP1B2:c.130_131ins227 variant. Table S1 lists the genetic basis of inherited canine cerebellar disorders which have been reported in the literature. Table S2 shows the ENA accession numbers of the whole-genome sequencing data that was used. Table S3 contains genome regions ≥1 Mb that showed positive LOD scores in the linkage analysis. Table S4 presents the homozygous genome regions with shared alleles among the four analyzed affected Malinois puppies from family 6. Table S5 illustrates ATP1B2:c.130_131ins227 genotypes of 503 control dogs from 87 diverse dog breeds.
Results
Clinical presentation
Clinical signs in the puppies from family 5 and 6 had a similar time of onset of 4 wk of age and were mainly associated with cerebellar dysfunction. We predominantly observed generalized ataxic gait in all puppies. Due to the inability to ambulate, five out of six affected puppies were euthanized on welfare grounds by the sixth week of age. One puppy from family 6 died at 6.5 wk of age during a seizure.
Upon more detailed investigation, only the four affected Malinois puppies from family 6, but not the affected puppies from family 5, additionally had seizures, showed pacing as well as circling, and were diagnosed with central blindness. Moreover, they had a very rapid progression of clinical signs (File S1).
Neuropathological findings
Neuropathological analysis was hindered in two puppies due to autolysis of the CNS (MA164, MA165). Histopathological changes in the three affected puppies from family 6 were characterized by bilateral-symmetric vacuolation of the neuropil, targeting the cerebellar nuclei (Figure 2); the ventral horn gray matter of the spinal cord, in particular at the level of the cervical intumescence; and the brain stem. In the spinal cord, vacuolation was associated with neuronal necrosis and severe gliosis. Additionally, in the puppy MA162, neuronal necrosis and diffuse presence of hypertrophic astrocytes with vesicular nuclei, reminiscent of Alzheimer type II cells, were observed in the hippocampus, caudate nucleus, and diffusely in the cortex. Histopathological eye abnormalities were not noticed.
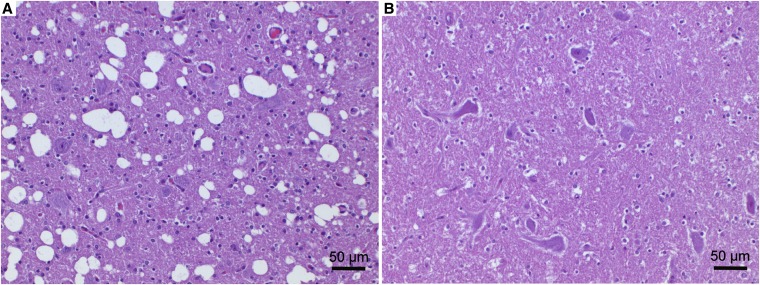
Figure 2.
Histopathology of a cerebellar nucleus. (A) Malinois dog MA162 with spongy degeneration and (B) nonaffected control dog. The affected Malinois puppy (A) showed a prominent vacuolation of the neuropil with large numbers of clearly defined and empty vacuoles of varying size and gliosis. Hematoxylin and eosin stain.
Neuropathological lesions noted in family 5 differed from those observed in family 6. The affected puppies in family 5 displayed gliosis in the cerebellar nuclei, in selected medullary nuclei, and in the spinal cord gray matter. However, CNS vacuolation was not present.
Genetic mapping of the causative variant in family 6
As the genetic analyses were performed independently and at the same time as the clinical and neuropathological investigations, we initially considered the possibility that the affected puppies in families 5 and 6 shared the same genetic defect. To map the causative locus we investigated these two Malinois families with a total of six puppies with cerebellar dysfunction (Figure 1). A combined linkage analysis revealed a single linked segment on chromosome 5 reaching a statistically significant LOD score of 3.657. However, when we performed linkage analysis separately for each of the two families, we noted that family 6 only presented a single linked segment on chromosome 5 (LOD score of 2.680), whereas family 5 showed linked segments on 12 different chromosomes (maximal LOD score of 0.977, Table S3).
To fine map the region of interest, we then analyzed the six affected Malinois puppies belonging to family 5 and 6 for extended regions of homozygosity with simultaneous allele sharing. This initial homozygosity-mapping approach did not reveal any shared segment on chromosome 5 between all six investigated puppies. As the histopathological findings and the clinical presentation had already suggested phenotypic differences between these two families, we then subsequently performed the homozygosity mapping for each family separately. The analyses showed that only the four affected puppies from family 6 had a homozygous genome region on chromosome 5 (Table S4). By intersecting the linked segment and the homozygous interval from the four cases of family 6, we could define an exact critical interval of 10,564,105 bp at Chr5:29,906,132–40,470,236. Moreover, upon inspection of the SNP-chip genotypes of the isolated Malinois cases with unknown relationships to our families, we identified one additional puppy, which also carried the disease-associated haplotype in the homozygous state (MA133). We therefore assumed that this puppy was affected by the same genetic disease as the four cases in family 6.
Identification of the causative variant
A total of 255 genes were annotated in the 10.6 Mb critical interval on chromosome 5. To acquire a comprehensive overview of all variants in this region we resequenced the whole genome of one affected Malinois puppy (MA163) and called single nucleotide as well as indel variants with respect to the reference genome of a presumably nonaffected Boxer. The genotype of the affected Malinois puppy was further compared with 146 dog genomes from various breeds that had been sequenced in the course of other studies (Table S2). We hypothesized that the causative variant should be completely absent from all dog breeds in the sample set except the Belgian Shepherd breed. After applying this filter, 37 disease-associated variants remained. However, none of these variants was predicted to change an encoded protein by our automated bioinformatic analysis (Table 1).
Table 1. Variants detected by whole-genome resequencing of one affected Malinois puppy (MA163).
| Filtering Step | Number of Variants |
|---|---|
| Variants in the whole genomea | 1,889,727 |
| Variants in the critical 10.6 Mb interval on chromosome 5 | 75,231 |
| Variants in the critical interval that were absent from 146 other dog genomes | 37 |
| Protein-changing variants in the whole genomea | 7936 |
| Protein-changing variants in the 10.6 Mb critical interval on chromosome 5 | 817 |
| Protein-changing variants in the critical interval, absent from 146 other dog genomes | 0 |
The sequences were compared to the reference genome (CanFam 3.1) from a Boxer. Protein-changing variants were classified based on the ENSEMBL annotation (version 72).
Therefore, we considered the possibility that the disease-associated variant was a structural variant, which would not have been detected by our automated variant calling pipeline. Thus, we visually inspected the 10.6 Mb critical interval on chromosome 5 and identified a candidate structural variant in the ATP1B2 gene (Figure S1). The structural variant arose from a 227 bp SINE insertion within exon 2 of the ATP1B2 gene, including a 15 bp duplication flanking the insertion site, and can be described as ATP1B2:c.130_131insLT796559.1:g.50_276 or in abbreviated form as ATP1B2:c.130_131ins227 or Chr5:32,551,064_32,551,065ins227 (Figure 3 and Figure S1).
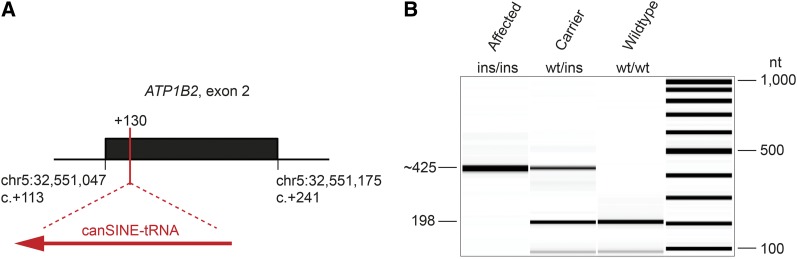
Figure 3.
SINE insertion in exon 2 of the ATP1B2 gene (ATP1B2:c.130_131ins227). (A) A 227 bp SINE insertion was found in homozygous state in five Malinois puppies affected by SDCA2 after position 130 of the ATP1B2 coding sequence. The SINE belonged to the SINEC2A1_CF family derived from an endogenous tRNA gene. 15 nt flanking the insertion site were duplicated. (B) Experimental genotyping of the SINE insertion by fragment size analysis. We amplified exon 2 of ATP1B2 and flanking intron segments by PCR and separated the products of dogs with the three different genotypes by capillary gel electrophoresis.
The presence of this structural variant in homozygous state was confirmed by Sanger sequencing in the four affected Malinois puppies belonging to family 6 and in the isolated case (MA133). The two available parents (MA229, MA233) were heterozygous for this insertion as expected for obligate carriers. Furthermore, we genotyped this variant by fragment length analysis in 251 other Malinois, 25 Groenendael, two Laekenois, 35 Tervueren, and 503 dogs of genetically diverse other breeds. The variant was not found outside the Belgian Shepherd population, but it also occurred in a heterozygous state in 38 Malinois, one Groenendael, and seven Tervueren dogs. In the two cases from family 5 and in the five remaining isolated cases, the variant was also absent (Table 2 and Table S5).
Table 2. Association of the SINE insertion with cerebellar dysfunction.
| Genotype ATP1B2:c.130_131ins227 | wt/wt | wt/ins | ins/ins |
|---|---|---|---|
| Malinois cases (family 6 and MA133) | — | — | 5 |
| Malinois cases (families 1–5 and six isolated puppies)a | 13 | 1 | — |
| Malinois controls | 199 | 38 | — |
| Groenendael controls | 24 | 1 | — |
| Laekenois controls | 2 | — | — |
| Tervueren controls | 28 | 7 | — |
| Control dogs from other breedsb | 503 | — | — |
Six of these Malinois puppies, which belonged to family 1–4, and one isolated case, MA152, were previously reported to be affected by SDCA1 caused by the KCNJ10:c.986T>C variant (Mauri et al. 2017).
These dogs were specifically genotyped by fragment length analysis for the ATP1B2:c.130_131ins227 variant. The genome sequences of 146 independent control dogs were also homozygous wt/wt. Therefore, the number of control dogs totals 948.
Analyses of the ATP1B2 transcript
Unfortunately, no suitable brain RNA samples were available for this study. To examine the effect of the 227 bp SINE insertion in exon 2 of the ATP1B2 gene on the transcript, we therefore isolated total skin and blood RNA from one affected Malinois puppy (MA133) and an unaffected control dog (SY045), respectively. In the affected Malinois puppy we identified at least three distinct transcript isoforms due to altered splicing. After separation and sequencing, none of the experimentally obtained RT-PCR products had the expected size. Sequencing of the products demonstrated either the complete skipping of exon 2 or the activation of two new cryptic splice sites generating aberrant exon lengths. All three mutant transcripts maintained the original reading frame (Figure 4 and File S3).
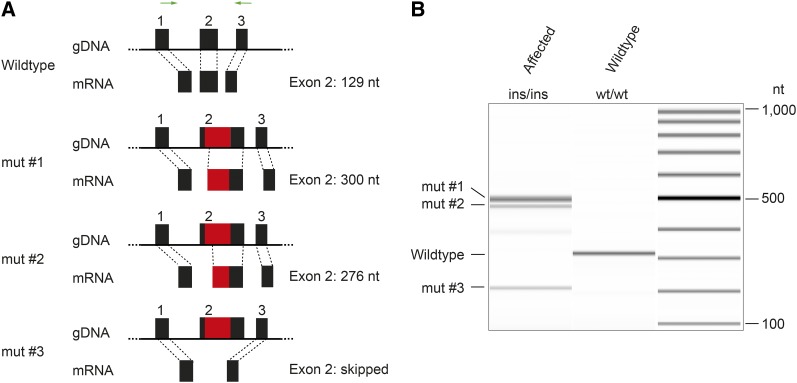
Figure 4.
Effect of the SINE insertion on ATP1B2 transcripts. (A) Schematic representation of exons 1–3 of the ATP1B2 gene. The 227 bp SINE insertion in exon 2 is displayed in red. Different mutant transcripts were identified and three of them characterized (mut #1–3). In mut #1 and #2 the SINE insertion leads to the utilization of novel splice acceptors, in which parts of the normal exon 2 were replaced with mutant sequences. In mut #3, exon 2 was skipped. RT-PCR primers in exon 1 and at the boundary of exons 3 and 4 of the ATP1B2 gene are indicated with green arrows (B) RT-PCR products of an affected and a control dog. All products were sequenced to confirm their identities (File S3).
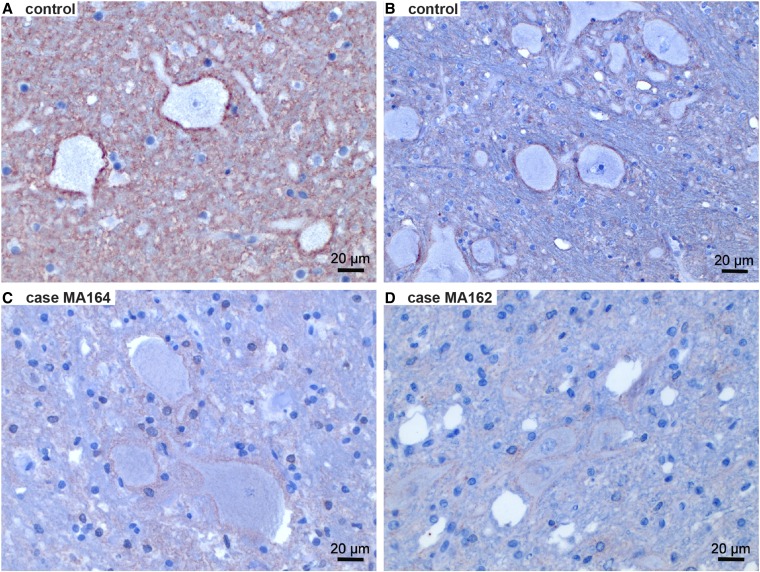
IHC
To assess the ATP1B2 protein expression we performed IHC on cerebellar and brain stem tissues with anti-ATP1B2 polyclonal antibodies (Figure 5). In the control dogs, the generated IHC signal was consistently present, albeit with varying intensity. The signal was mostly seen around neurons and in a glial-like pattern, with extensions similar to astrocytic processes, in the whole cerebellar and brain-stem parenchyma, especially in the gray matter. The obtained IHC sections from MA162 and MA164 showed a weaker IHC signal around neurons (Figure 5). In the affected Malinois puppy MA165, no ATP2B1 expression could be detected by IHC.
Figure 5.
IHC for the ATP2B1 protein. (A and B) Two control dogs showed a clear perineuronal expression of ATP2B1. However, the intensity of the IHC signal was variable between dogs. (C and D) In two of the affected Malinois puppies, perineuronal expression was present, but appeared to be weaker than in the control dogs. In a third affected puppy, no ATP2B1 expression was observed.
Discussion
In this study, we identified a structural variant in exon 2 of the ATP1B2 gene as candidate causative genetic variant for SDCA in the Belgian Shepherd breed. While we only observed affected puppies in the Malinois variety, the proposed pathogenic variant also segregates in other varieties of the Belgian Shepherd breed. We suggest to call this particular phenotype spongy degeneration with cerebellar ataxia, subtype 2 (SDCA2).
To the best of our knowledge, so far, no ATP1B2 variants have been described in human patients with neurologic symptoms. However, Atp1b2 knockout mice were reported with similar clinical signs as the affected Malinois puppies. Atp1b2−/− mice showed rapidly worsening motor impairment and spongy degeneration of the CNS, resembling the neurohistopathological findings observed in the Malinois puppies but with a different topographical distribution (Magyar et al. 1994). Our hypothesis is also supported by a study where the Na+/K+-ATPase was inhibited in vivo by subdural injections of ouabain in guinea pigs. The inhibition of the Na+/K+-ATPase holoenzyme caused seizures and, on histopathology, an evident spongy degeneration in the CNS (Calandriello et al. 1995).
The Na+/K+-ATPase is a ubiquitously expressed multi-subunit protein in the plasma membrane. It is the principal regulator of intracellular homeostasis in every animal cell. The holoenzyme consists of α, β, and auxiliary γ subunits. It is responsible for the active Na+ extrusion (three ions) and K+ uptake (two ions) necessary to generate and maintain the cellular transmembrane ionic gradients that are essential for the activity of secondary transporters such as voltage-gated Na+ and K+ channels, the Na+/Ca2+ exchanger, and neurotransmitter uptake transporters (Mobasheri et al. 2000; Tokhtaeva et al. 2009; Friedrich, et al. 2016). The α subunit is the main component of the Na+/K+-ATPase and is also defined as a catalytic subunit. It is responsible for ion transport. The β subunit is essential for correct folding, assembly, and targeting of the holoenzyme to the plasma membrane as well as for holoenzyme function by regulating Na+ affinity (Habiba et al. 2000; Mobasheri et al. 2000; Geering 2008; Tokhtaeva et al. 2009). Furthermore the β subunit may play a role in cell adhesion and CNS development (Antonicek et al. 1987; Müller-Husmann et al. 1993; Lecuona et al. 1996; Boer et al. 2010). In contrast to the α and β subunits, the γ subunit is not essential for the function of the Na+/K+-ATPase. If present, it acts as an ion transport regulator. To date, four different α, four β, and seven γ subunit isoforms were identified (Geering 2001; Hilbers et al. 2016).
The ATP1B2 gene encodes the β2 subunit isoform, first discovered as adhesion molecule on glia (AMOG) (Antonicek et al. 1987; Gloor et al. 1990). The β2 subunit isoform is predominantly expressed in the brain, especially in the cerebellum, and it preferentially binds to α2, which is mainly found in astrocytes after completion of development. The main task of the α2β2 Na+/K+-ATPase holoenzyme in astrocytes is to restore extracellular K+ homeostasis following neuronal depolarization. A failure of K+ buffering and clearance would result in high extracellular K+ and consequently to sustained glial and neuronal hyperexcitability compromising neuronal firing, synaptic transmission, and neurotransmitter reuptake (Tokhtaeva et al. 2012; Friedrich et al. 2016; Hilbers et al. 2016; Larsen et al. 2016).
Disturbances in the K+ homeostasis in the CNS are often associated with neurological disorders such as cerebellar dysfunction or epilepsy (Hirose 2006; Larsen et al. 2014). Recently, we genetically characterized a hereditary cerebellar ataxia in the Belgian Shepherd breed caused by a pathologic variant in KCNJ10, encoding the astrocytic Kir4.1 potassium channel (Mauri et al. 2017). Both KCNJ10 and the α2β2 Na+/K+-ATPase seem to play a pivotal role for K+ homeostasis in the CNS, especially in the cerebellum. They are not interchangeable, but serve temporally distinct roles, with KCNJ10 acting during and α2β2 Na+/K+-ATPase working after neuronal depolarization (Larsen et al. 2014). In addition to an impaired K+ clearance after neuronal depolarization, dysfunction of the α2β2 Na+/K+-ATPase might also lead to extracellular accumulation of glutamate and/or to an increase in intracellular Ca2+ and/or to cell swelling, which all might contribute to the observed histopathological changes (Friedrich et al. 2016; Prontera et al. 2017).
The central blindness in the SDCA2-affected puppies is most likely explained by necrotic changes involving the visual cortex. However, the α3β2 Na+/K+-ATPase holoenzyme is also associated with retinoschisin on the photoreceptor and bipolar cells of the eye, and the retinoschisin–α3β2 Na+/K+-ATPase complex is necessary for maintenance of retinal cell organization as well as photoreceptor-bipolar synaptic structure. Variants in the retinoschisin gene, RS1, cause splitting of retinal cell layers and loss in central vision. The phenotype is defined as X-linked juvenile retinoschisis (XLRS) (MIM#312700; Molday et al. 2007, 2012; Friedrich et al. 2011). However, we did not observe any histopathological changes of the retina in an SDCA2-affected puppy and thus think that blindness in these dogs is caused by the central lesions.
Our RNA experiments demonstrated altered splicing of the ATB1B2 transcript in skin RNA from one affected puppy. It has to be cautioned that the splicing might be different in the CNS. However, given the sequence context of the SINE insertion, it seems impossible that the insertion allele could give rise to the expression of the wild-type mRNA.
Our IHC findings suggest a reduction in ATP1B2 protein expression in the CNS of SDCA2-affected Malinois. However, IHC signal could still be identified, possibly indicating the expression of mutant ATP1B2 protein. The antibody that was used was directed against a peptide corresponding to amino acids 115–141 located in the extracellular domain of the wild-type protein. The three characterized aberrant transcripts all maintained the original reading frame and encoded this epitope. Thus, translation of any of these mutant transcripts might have led to the expression of a detectable mutant protein (File S3). Further investigation is needed to assess if any of these mutant ATP1B2 proteins might retain some residual functional activity. For the mut #3 protein, this seems highly unlikely as it is lacking the entire predicted transmembrane domain. The mut #1 and mut #2 proteins contain insertions of 59 and 51 amino acids into the cytoplasmic domain respectively, and it is at least questionable whether they can exert the same functions as the normal β2 subunit of the Na+/K+-ATPase.
In humans, ATP1B2 variants have not been reported to date. However, severe neurological disorders have been associated with variants in the ATP1A2 and ATP1A3 genes encoding the α2 and α3 subunit isoforms. ATP1A2 variants are responsible for hemiplegic migraine type 2 (FHM2, MIM#602481; Prontera et al. 2017) and alternating hemiplegia of childhood 1 (AHC1, MIM#104290; Swoboda et al. 2004). ATP1A3 defects cause rapid-onset dystonia Parkinsonism (RDP, DYT12, MIM#128235), as well as alternating hemiplegia of childhood 2 (AHC2, MIM#614820) and CAPOS syndrome (cerebellar ataxia, areflexia, pes cavus, optic atrophy, and sensorineural hearing loss syndrome, MIM#601338; Heinzen et al. 2014; Holm and Lykke-Hartmann 2016).
To conclude, we identified a SINE insertion in exon 2 of the ATP1B2 gene (ATP1B2:c.130_131insLT796559.1:g.50_276), leading to altered splicing and an impaired ATP1B2 protein expression in the CNS, as most likely causative for SDCA2 in the Belgian Shepherd breed. Cerebellar dysfunction in this breed is heterogeneous and, together with the previously reported KCNJ10:c.986T>C variant, we still cannot explain all cases affected by similar clinical signs. Further investigation is needed to resolve the genetic and phenotypic complexity underlying cerebellar dysfunction in Malinois dogs. Our findings encourage genetic testing of Belgian Shepherd dogs so that the nonintentional breeding of affected puppies with SDCA2 can be avoided in the future. Moreover, our data imply ATP1B2 as an additional candidate gene for human inherited cerebellar ataxias of unknown etiology. Affected puppies represent a spontaneous animal model for hereditary ataxia.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043018/-/DC1.
Acknowledgments
We are grateful to all dog owners and breeders who donated samples and shared pedigree data and other valuable information regarding their dogs. We acknowledge collaborators of the Dog Biomedical Variant Database Consortium, which also includes authors V.J. and T.L.: Gus Aguirre, Catherine André, Danika Bannasch, Doreen Becker, Cord Drögemüller, Oliver Forman, Eva Furrow, Urs Giger, Christophe Hitte, Marjo Hytönen, Hannes Lohi, Cathryn Mellersh, Jim Mickelson, Leonardo Murgiano, Anita Oberbauer, Jeffrey Schoenebeck, Sheila Schmutz, and Claire Wade for sharing dog genome sequence data from control dogs. We thank Nathalie Besuchet Schmutz, Muriel Fragnière, Camille Monney, and Marianne Wyss for expert technical assistance, the Next Generation Sequencing Platform of the University of Bern for performing the whole-genome sequencing experiments, and the Interfaculty Bioinformatics Unit of the University of Bern for providing computational infrastructure. This study was supported by a grant from the Albert Heim Foundation.
Footnotes
Communicating editor: D. J. de Koning
Literature Cited
- Abecasis G. R., Cherny S. S., Cookson W. O., Cardon L. R., 2002. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30: 97–101. [DOI] [PubMed] [Google Scholar]
- Antonicek H., Persohn E., Schachner M., 1987. Biochemical and functional characterization of a novel neuron-glia adhesion molecule that is involved in neuronal migration. J. Cell Biol. 104: 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B., Zhao W.-M., Tang B.-X., Wang Y.-Q., Wang L., et al. , 2015. DoGSD: the dog and wolf genome SNP database. Nucleic Acids Res. 43: D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer K., Spliet W. G. M., van Rijen P. C., Jansen F. E., Aronica E., 2010. Expression patterns of AMOG in developing human cortex and malformations of cortical development. Epilepsy Res. 91: 84–93. [DOI] [PubMed] [Google Scholar]
- Calandriello L., Curini R., Pennisi E. M., Palladini G., 1995. Spongy state (status Spongiosus) and inhibition of Na,K-ATPase: a pathogenetic theory. Med. Hypotheses 44: 173–178. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T., Tavraz N. N., Junghans C., 2016. ATP1A2 mutations in migraine: seeing through the facets of an ion pump onto the neurobiology of disease. Front. Physiol. 7: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich U., Stöhr H., Hilfinger D., Loenhardt T., Schachner M., et al. , 2011. The Na/K-ATPase is obligatory for membrane anchorage of retinoschisin, the protein involved in the pathogenesis of X–linked juvenile retinoschisis. Hum. Mol. Genet. 20: 1132–1142. [DOI] [PubMed] [Google Scholar]
- Geering K., 2001. The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33: 425–438. [DOI] [PubMed] [Google Scholar]
- Geering K., 2008. Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17: 526–532. [DOI] [PubMed] [Google Scholar]
- Gloor S., Antonicek H., Sweadner K. J., Pagliusi S., Frank R., et al. , 1990. The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na,K-ATPase. J. Cell Biol. 110: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiba A., Blanco G., Mercer R. W., 2000. Expression, activity and distribution of Na,K-ATPase subunits during in vitro neuronal induction. Brain Res. 875(1–2): 1–13. [DOI] [PubMed] [Google Scholar]
- Heinzen E. L., Arzimanoglou A., Brashear A., Clapcote S. J., Gurrieri F., et al. , 2014. Distinct neurological disorders with ATP1A3 mutations. Lancet Neurol. 13: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersheson J., Haworth A., Houlden H., 2012. The inherited ataxias: genetic heterogeneity, mutation databases, and future directions in research and clinical diagnostics. Hum. Mutat. 33: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Hilbers F., Kopec W., Isaksen T. J., Holm T. H., Lykke-Hartmann K., et al. , 2016. Tuning of the Na,K-ATPase by the beta subunit. Sci. Rep. 6: 20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., 2006. A new paradigm of channelopathy in epilepsy syndromes: intracellular trafficking abnormality of channel molecules. Epilepsy Res. 70: S206–S217. [DOI] [PubMed] [Google Scholar]
- Holm T. H., Lykke-Hartmann K., 2016. Insights into the pathology of the α3 Na(+)/K(+)-ATPase ion pump in neurological disorders. Front. Physiol. 7: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev S., Bird T. D., 2013. Hereditary ataxias: overview. Genet. Med. 15: 673–683. [DOI] [PubMed] [Google Scholar]
- Kleiter M., Högler S., Kneissl S., Url A., Leschnik M., 2011. Spongy degeneration with cerebellar ataxia in Malinois puppies: a hereditary autosomal recessive disorder? J. Vet. Intern. Med. 25: 490–496. [DOI] [PubMed] [Google Scholar]
- Klockgether T., Paulson H., 2011. Milestones in ataxia. Mov. Disord. 26: 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B. R., Assentoft M., Cotrina M. L., Hua S. Z., Nedergaard M., et al. , 2014. Contributions of the Na+/K+-ATPase, NKCC1, and Kir4.1 to hippocampal K+ clearance and volume responses. Glia 62: 608–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B. R., Stoica A., MacAulay N., 2016. Managing brain extracellular K+ during neuronal activity: the physiological role of the Na+/K+-ATPase subunit isoforms. Front. Physiol. 7: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuona E., Luquín S., Avila J., García-Segura L. M., Martín-Vasallo P., 1996. Expression of the β1 and β2(AMOG) subunits of the Na,K-ATPase in neural tissues: cellular and developmental distribution patterns. Brain Res. Bull. 40: 167–174. [DOI] [PubMed] [Google Scholar]
- Magyar J. P., Bartsch U., Wang Z. Q., Howells N., Aguzzi A., et al. , 1994. Degeneration of neural cells in the central nervous system of mice deficient in the gene for the adhesion molecule on glia, the Beta 2 subunit of murine Na,K-ATPase. J. Cell Biol. 127: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M., Orsucci D., Siciliano G., Bonuccelli U., 2014. The genetics of ataxia: through the labyrinth of the Minotaur, looking for Ariadne’s thread. J. Neurol. 261: S528–S541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manto M., Marmolino D., 2009. Cerebellar ataxias. Curr. Opin. Neurol. 22: 419–429. [DOI] [PubMed] [Google Scholar]
- Mauri N., Kleiter M., Leschnik M., Högler S., Dietschi E., et al. , 2017. A missense variant in KCNJ10 in Belgian Shepherd dogs affected by spongy degeneration with cerebellar ataxia (SDCA1). G3 (Bethesda) 7: 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobasheri A., Avila J., Cózar-Castellano I., Brownleader M. D., Trevan M., et al. , 2000. Na+, K+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Biosci. Rep. 20: 51–91. [DOI] [PubMed] [Google Scholar]
- Molday L. L., Wu W. W. H., Molday R. S., 2007. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J. Biol. Chem. 282: 32792–32801. [DOI] [PubMed] [Google Scholar]
- Molday R. S., Kellner U., Weber B. H. F., 2012. X-linked juvenile retinoschisis: clinical diagnosis, genetic analysis, and molecular mechanisms. Prog. Retin. Eye Res. 31: 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Husmann G., Gloor S., Schachner M., 1993. Functional characterization of beta isoforms of murine Na,K-ATPase. The adhesion molecule on glia (AMOG/Beta 2), but not Beta 1, promotes neurite outgrowth. J. Biol. Chem. 268: 26260–26267. [PubMed] [Google Scholar]
- Prontera P., Sarchielli P., Caproni S., Bedetti C., Cupini L. M., et al. , 2017. Epilepsy in hemiplegic migraine: genetic mutations and clinical implications. Cephalalgia DOI: 10.1177/0333102416686347 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandford E., Burmeister M., 2014. Genes and genetic testing in hereditary ataxias. Genes 5: 586–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stee K., Van Poucke M., Bhatti S., Vanhaesebrouck A., Bosseler L., et al. , 2016. The novel homozygous KCNJ10 c.986T>C (p.Leu329-Pro) variant is pathogenic for the SeSAME/EAST homologue in Malinois dogs (conference abstract). J. Vet. Intern. Med. 30: 1934. Proceedings 29th Symposium ESVN-ECVN Edinburgh, United Kingdom 16th–17th September 2016. [Google Scholar]
- Swoboda K. J., Kanavakis E., Xaidara A., Johnson J. E., Leppert M. F., et al. , 2004. Alternating hemiplegia of childhood or familial hemiplegic migraine? A novel ATP1A2 mutation. Ann. Neurol. 55: 884–887. [DOI] [PubMed] [Google Scholar]
- Tokhtaeva E., Sachs G., Vagin O., 2009. Assembly with the Na,K-ATPase α1 subunit is required for export of β1 and β2 subunits from the endoplasmic reticulum. Biochemistry 48: 11421–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E., Clifford R. J., Kaplan J. H., Sachs G., Vagin O., 2012. Subunit isoform selectivity in assembly of Na,K-ATPase α-β heterodimers. J. Biol. Chem. 287: 26115–26125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urkasemsin G., Olby N. J., 2014. Canine hereditary ataxia. Vet. Clin. North Am. Small Anim. Pract. 44: 1075–1089. [DOI] [PubMed] [Google Scholar]
- Urkasemsin G., Linder K. E., Bell J. S., de Lahunta A., Olby N. J., 2010. Hereditary cerebellar degeneration in Scottish Terriers. J. Vet. Intern. Med. 24: 565–570. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., et al. , 2013. From fastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poucke M., Stee K., Bhatti S. F., Vanhaesebrouck A., Bosseler L., et al. , 2017. The novel homozygous KCNJ10 c.986T>C (p.(Leu329Pro)) variant is pathogenic for the SeSAME/EAST homologue in Malinois dogs. Eur. J. Hum. Genet. 25: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmer M., Oevermann A., Borer-Germann S. E., Gorgas D., Shelton G. D., et al. , 2016. A RAB3GAP1 SINE insertion in Alaskan Huskies with polyneuropathy, ocular abnormalities, and neuronal vacuolation (POANV) resembling human warburg micro syndrome 1 (WARBM1). G3 (Bethesda) 6: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 is a video showing the clinical phenotype of the four affected Malinois siblings belonging to family 6 at 4 wk of age (MA162–165). File S2 depicts the sequence context of the 227 bp SINE insertion into exon 2 of the ATP1B2 gene. File S3 illustrates the aberrant ATP1B2 exon 2 splicing patterns in dogs with the SINE insertion and the predicted protein sequences resulting from the mutant transcripts. Figure S1 contains a screenshot of the IGV software in the region of the visually identified ATP1B2:c.130_131ins227 variant. Table S1 lists the genetic basis of inherited canine cerebellar disorders which have been reported in the literature. Table S2 shows the ENA accession numbers of the whole-genome sequencing data that was used. Table S3 contains genome regions ≥1 Mb that showed positive LOD scores in the linkage analysis. Table S4 presents the homozygous genome regions with shared alleles among the four analyzed affected Malinois puppies from family 6. Table S5 illustrates ATP1B2:c.130_131ins227 genotypes of 503 control dogs from 87 diverse dog breeds.