Abstract
Eukaryote genomes are replete with repetitive DNAs. This class includes tandemly repeated satellite DNAs (satDNA) which are among the most abundant, fast evolving (yet poorly studied) genomic components. Here, we used high-throughput sequencing data from three cactophilic Drosophila species, D. buzzatii, D. seriema, and D. mojavensis, to access and study their whole satDNA landscape. In total, the RepeatExplorer software identified five satDNAs, three previously described (pBuM, DBC-150 and CDSTR198) and two novel ones (CDSTR138 and CDSTR130). Only pBuM is shared among all three species. The satDNA repeat length falls within only two classes, between 130 and 200 bp or between 340 and 390 bp. FISH on metaphase and polytene chromosomes revealed the presence of satDNA arrays in at least one of the following genomic compartments: centromeric, telomeric, subtelomeric, or dispersed along euchromatin. The chromosomal distribution ranges from a single chromosome to almost all chromosomes of the complement. Fiber-FISH and sequence analysis of contigs revealed interspersion between pBuM and CDSTR130 in the microchromosomes of D. mojavensis. Phylogenetic analyses showed that the pBuM satDNA underwent concerted evolution at both interspecific and intraspecific levels. Based on RNA-seq data, we found transcription activity for pBuM (in D. mojavensis) and CDSTR198 (in D. buzzatii) in all five analyzed developmental stages, most notably in pupae and adult males. Our data revealed that cactophilic Drosophila present the lowest amount of satDNAs (1.9–2.9%) within the Drosophila genus reported so far. We discuss how our findings on the satDNA location, abundance, organization, and transcription activity may be related to functional aspects.
Keywords: satellite DNA, cactophilic Drosophila, centromeres, telomeres, concerted evolution
The genomes of many organisms are replete with highly repetitive (>1000 copies) tandemly repeated DNA sequences, commonly known as satellite DNAs (satDNAs) (Tautz 1993). Long and homogeneous arrays made of satDNA repeats are located in the heterochromatin (Charlesworth et al. 1994; Plohl 2012; Beridze 2013; Khost et al. 2017), but recent studies also revealed the presence of short arrays dispersed along the euchromatin (Brajković et al. 2012; Kuhn et al. 2012; Larracuente 2014; Pavlek et al. 2015). SatDNAs do not have the ability to transpose by themselves as transposable elements (TEs) do. However, there are some reported examples showing that TEs may act as a substrate for satDNA emergence and mobility (Dias et al. 2015; Meštrović et al. 2015; Satović et al. 2016).
The whole collection of satDNAs makes up large portions (usually >30%) of animal and plant genomes (reviewed by Plohl et al. (2014)). Although satDNAs do not code for proteins, they may play important cellular roles, including participation in chromatin packaging (Blattes et al. 2006; Feliciello et al. 2015), centromere formation/maintenance (Rošić et al. 2014; Aldrup-MacDonald et al. 2016), and gene regulation (Menon et al. 2014; Feliciello et al. 2015; Urrego et al. 2017).
Despite their abundance, diversity and contribution to genomic architecture and function, our knowledge about several features of satDNAs is still limited. In the past decades, satDNAs have been mostly studied from a small sample of cloned repeats obtained by biased experimental approaches (usually by restriction digestion and/or PCR), isolated from one or few species. Experimental strategies for the identification of satDNAs were expensive, time-consuming, and insufficient for the identification of the whole collection of satDNAs from any chosen genome.
Next-generation sequencing technologies have provided a revolution in the number of species with sequenced genomes, while new and efficient bioinformatic tools have been specifically developed toward genome-wide identification of repetitive DNAs. Consequently, we now have new tools and strategies to access the whole collection of satDNAs from a given genome. For example, software tools known as RepeatExplorer have been successfully used for genome-wide characterization of repetitive DNAs from several animal and plant genomes, including those sequenced with >1× coverage (Barghini et al. 2014; Marques et al. 2015; Ruiz-Ruano et al. 2016; Zhang et al. 2017). This algorithm directly uses short next-generation sequencing reads as rough material for the identification of repeats. Together with the results from similarity searches and abundance, the repeat families can be identified and classified.
Within the genus Drosophila, most studies on satDNA were conducted in D. melanogaster and in a few closely related species from the melanogaster group (e.g., Strachan et al. 1985; Kuhn et al. 2012; Larracuente 2014; Jagannathan et al. 2017). The study of satDNAs of species distantly related to D. melanogaster are expected to broaden the understanding of this major fraction of the eukaryote genome. In this context, the repleta group is of particular interest. It contains at least 100 species that breed in cactuses in North and South America (Oliveira et al. 2012). Species from the repleta group are separated from the melanogaster group by >40 MY (Powell 1997). Intense vertical studies in some species of this group revealed several aspects related to chromosome and genome evolution that have broad interest (e.g., Cáceres et al. 1999; Negre et al. 2005; Kuhn et al. 2009; Guillén et al. 2015).
At present, three repleta group species have available sequenced genomes: D. mojavensis (Drosophila 12 Genomes Consortium 2007), D. buzzatii (Guillén et al. 2015), and D. seriema (Dias G.B., M. Svartman and G.C.S. Kuhn, unpublished data). D. buzzatii and D. seriema belong to the buzzatii cluster, a monophyletic group of South American origin that contains seven species morphologically very similar and came from a radiation process dated at 6 MYA (Manfrin and Sene 2006; Oliveira et al. 2012). D. mojavensis lives in the deserts and dry tropical forests of the southwestern United States and Mexico (Reed et al. 2007). The time since the split between D. buzzatii and D. mojavensis has been estimated at 11 MYA (Oliveira et al. 2012; Guillén et al. 2015).
Previous studies in D. buzzatii and D. seriema conducted before the genomic era allowed the identification of three satDNA families. The first family, named pBuM, can be divided into two subfamilies according to its primary structure and size of the repeat units (Kuhn and Sene 2005). The pBuM-1 subfamily is comprised of alpha repeat units of ∼190 bp, whereas the pBuM-2 subfamily consists of 370-bp composite repeat units called alpha/beta, each one consisting of an alpha (∼190 bp) followed by a beta sequence (∼180 bp) of unknown origin. DNA hybridization data revealed pBuM-1 to be the major repeat variant present in D. buzzatii but pBuM-2 as the major repeat variant in D. seriema.
The second family, named DBC-150, consists of 150-bp long repeat units. This family is abundant in D. seriema but virtually absent in D. buzzatii (Kuhn et al. 2007). Finally, the third satDNA family, named SSS139, with 139-bp-long repeat units is abundant in D. seriema but absent in D. buzzatii (Franco et al. 2008). There is no significant sequence similarity among pBuM, DBC-150, and SSS139 satDNA repeats, suggesting that these families have independent evolutionary origins.
Three sequencing platforms (Sanger, 454, and Illumina) (Guillén et al. 2015) have been used to sequence the D. buzzatii genome, which became publicly available in 2015 (http://dbuz.uab.cat). In a preliminary approach, we used the Tandem Repeats Finder (TRF) software (version 4.04) (Benson 1999) to search for satDNAs with repeats longer than 50 bp in the D. buzzatii contigs. The two most abundant tandem repeat families identified were pBuM-1 (alpha repeats) and a novel family that we named CDSTR198, with 198-bp-long repeat units (Guillén et al. 2015). However, in D. melanogaster and D. virilis, for example, several abundant satDNA families showed repeat units <10-bp long (Gall et al. 1971; Lohe et al. 1993). Therefore, a new satDNA screen is necessary in the D. buzzatii sequenced genome in order to look for the presence of small-size satDNA repeat motifs.
There are no detailed studies involving satDNAs in D. mojavensis. Melters et al. (2013) developed a bioinformatic pipeline to identify the most abundant tandem repeats from 282 selected sequenced genomes from animal and plant species, including some Drosophila species. A satDNA with 183-bp-long repeat units was identified as the most abundant satDNA of D. mojavensis. Most recently, we showed that this satDNA actually belongs to the pBuM-1 satDNA subfamily (alpha repeats), previously described in D. buzzatii (Guillén et al. 2015).
Our group has recently sequenced the genome of D. seriema using the MiSeq platform (Dias et al., unpublished data). The availability of three sequenced genomes (D. buzzatii, D. seriema, and D. mojavensis) provides an unprecedented opportunity to study the satDNA collection from each species and to compare them in a scale never possible before. We combined bioinformatic, phylogenetic, and molecular cytogenetic tools to study the satDNA fraction from these three cactophilic Drosophila species. The resulting data are discussed in the context of satDNA genomic distribution, evolution, and potential functional roles.
Materials and Methods
Genomic data
The Illumina sequence reads from D. buzzatii, D. mojavensis, and D. seriema used for identification of satDNAs were obtained from three different sources. D. buzzatii reads (76× coverage) were generated by the Prof. Alfredo Ruiz group at Universitat Autònoma de Barcelona and were used for the genome assembly of D. buzzatii (Guillén et al. 2015). All D. buzzatii Illumina reads used on this paper were downloaded directly from the Drosophila buzzatii genome project webpage (http://dbuz.uab.cat). These data are publicly available for download on the FTP section: http://dbuz.uab.cat/ftp.php. We used D. mojavensis (SRX2932915) sequence reads (20× coverage) generated by Prof. Bernardo de Carvalho (Universidade Federal do Rio de Janeiro, Brazil), and D. seriema (ERX2037878) sequence reads (20× coverage) were generated by our group (Dias et al., unpublished data).
Identification of satellite DNAs
Similarity-based clustering, repeat identification, and classification were performed using RepeatExplorer (Novák et al. 2013) with whole-genome shotgun Illumina reads from D. buzzatii, D. mojavensis, and D. seriema. Initially, files containing all sequence reads from each species were uploaded (trimmed at 100 bp). The clustering analysis used RepeatExplorer default parameters. Clusters containing possible tandemly repeated satDNA families were identified based on the resultant graph-based clustering and then manually checked for the presence of tandem repeats using the TRF software (version 4.04) (Benson 1999). Genomic proportion was calculated from the number of reads present in each cluster divided by the total number of reads. We searched for clusters with high graph density, which is a typical characteristic of satDNAs families (Novák et al. 2013). The Dotlet software (Junier and Pagni 2000) was also used to generate a scrutinized description of full length copies of each satDNA family.
Sequence and phylogenetic analysis
Multiple satDNA sequences were aligned with the Muscle algorithm (Edgar 2004) of the MEGA5.05 software (Tamura et al. 2011), with manual optimization when necessary. MEGA5.05 was also used for the analysis of nucleotide composition and variability. Phylogenetic trees were constructed with the Neighbor Joining algorithm (Saitou and Nei 1987) of the MEGA program 5:05 (Tamura et al. 2011). The genetic distance between sequences was calculated using the “Tamura-Nei model” (Tamura and Nei 1993) after an analysis of best substitution model for the data on MEGA 5.05 (Tamura et al. 2011). Statistical evaluation of each branch of the tree was performed using analysis “bootstrap” (1000 replicates).
Samples, DNA extractions, PCR amplifications, cloning, and sequencing
For our experimental data we used DNA from the same sequenced strains: D. buzzatii (strain: ST01), D. seriema (strain: D73C3B), and D. mojavensis (strain: CI 12 IB -4 g8). DNA extraction of 30–50 adult flies was performed with the Wizard Genomic DNA Purification kit (Promega). PCR reactions consisted of an initial denaturation step of 94° for 3 min, followed by 30 cycles of 94° for 60 sec, 55° for 60 sec, and 72° for 60 sec and then a final extension at 72° for 10 min. The primers used for satDNA amplification are listed in Supplemental Material, Table S1 in File S1. PCR products were excised from 1% agarose gels and purified with the Wizard SV Gel and PCR Clean-up System kit (Promega). After cloning with the pGEM-T-Easy cloning kit (Promega), recombinant plasmids were sequenced on the ABI3130 platform (Myleus Biotechnology).
In situ hybridization experiments
Chromosome preparations, DNA fibers obtention, single and double-color FISH, and Fiber-FISH experiments were conducted as described in Kuhn et al. (2008). The probes labeled with digoxigenin-11-dUTP were detected with antidigoxigenin FITC (Roche) and probes labeled with biotin-14-dATP were detected with NeutrAvidin-rhodamine (Roche). Chromosomes were stained with DAPI (4′, 6-diamidino-2-phenylindole, dihydrochloride salt). The preparations were analyzed under an epifluorescence Zeiss Axiophot 2 microscope equipped with a CCD camera and the images were obtained using the AxioVision software (Zeiss). To determine the size of the DNA fibers, hybridization signals were measured according to the protocol described by Schwarzacher and Heslop-Harrison (2000).
Transcription analysis
Total RNA-seq data of D. mojavensis and D. buzzatii (st-1 strain) were those obtained by Guillén et al. (2015). Briefly, RNA samples were extracted from 10 to 20 individuals from each of the four development stages (embryos, third-stage larvae, pupae, adult females and males), enriched for mRNA by poly-A tail selection and sequenced by Illumina, generating ∼100 bp reads [see Guillén et al. (2015) for details]. All reads were aligned against consensus sequences representing the pBuM and CDSTR198 families from D. buzzatii and pBuM and CDSTR130 from D. mojavensis with the Bowtie2 software (Langmead and Salzberg 2012) incorporated into the usegalaxy.org server (Afgan et al. 2016). The mapped reads were normalized by the RPKM method (reads per kilobase per million mapped reads; Mortazavi et al. 2008).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Cactophilic Drosophila repetitive DNAs: general aspects
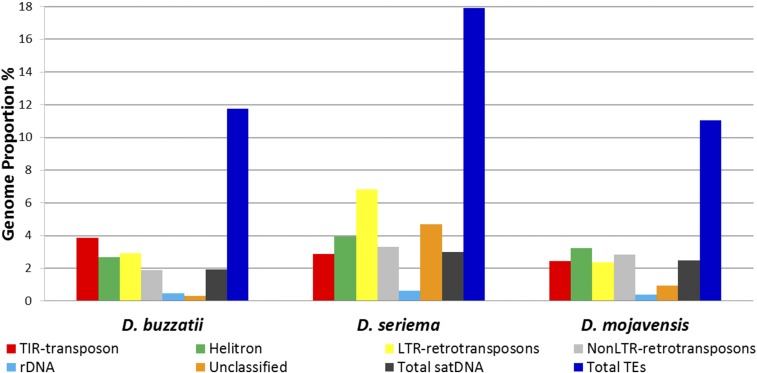
The RepeatExplorer graphic representation containing all identified repetitive DNA clusters in D. buzzatii, D. seriema, and D. mojavensis and their genome proportion (%) is shown in Figures S1–S3 in File S1. Most clusters making >0.01% of the genome could be classified into established groups of repetitive elements, such as TEs, satDNAs, or rDNA sequences (Figure 1 and Tables S2–S4 in File S1).
Figure 1.
Estimated repetitive DNA abundance in three cactophilic Drosophila species.
The satDNA genomic contribution is similar in the three species: ∼1.9% in D. buzzatii, ∼2.9% in D. seriema, and ∼2.5% in D. mojavensis. The genomic contribution of the classified TEs is on average 5.4× higher: ∼12% in D. buzzatii, ∼18% in D. seriema, and ∼11% in D. mojavensis. Rius et al. (2016) have recently estimated the TE content of D. buzzatii and D. mojavensis using the same genomic sequences used in this work, but with a different methodology, and found that TEs represent ∼11% of the D. buzzatii and ∼15% of the D. mojavensis genomes.
The genomic contribution of the different TE orders [TIR-transposons, Helitrons, long terminal repeat (LTR) retrotransposons, and non-LTR retrotransposons] differs among the three species (Figure 1). TIR-transposons are the most abundant TEs in the D. buzzatii genome (3.85%); in D. seriema, LTR retrotransposons (6.8%) are the most abundant and in D. mojavensis, Helitrons are the most abundant TE elements (3.25%). Conversely, Rius et al. (2016) described Helitrons as the most abundant TEs in the D. buzzatii and D. mojavensis genomes. Interestingly, the genomic contribution of LTR retrotransposons in D. seriema (6.8%) is at least two times higher than in D. buzzatii (2.9%) or in D. mojavensis (2.4%). The contribution of unclassified repetitive elements is also considerably higher in D. seriema (18%) than in the other two species (11% and 12%). These results suggest a recent burst of repetitive elements in D. seriema.
Satellite DNA landscape in the three cactophilic Drosophila species
We identified only two previously described satDNA families in D. buzzatii. The pBuM-1 satDNA (Kuhn and Sene 2005) with 189-bp-long alpha repeats is the most abundant, representing 1.7%. The second is CDSTR198 (Guillén et al. 2015), with 198-bp-long repeats and representing 0.2% of the genome. These genomic contributions revealed by RepeatExplorer are higher than those obtained by our first contig-based approach, most notably for pBuM-1 (0.04% for pBuM-1 and 0.03% for CDSTR198; Guillén et al. 2015). The organization of satDNAs, made of several tandem repeats with high DNA sequence similarity, imposes a huge limitation for assembly computer programs. Consequently, it is very likely that the bulk of pBuM and CDSTR198 satDNA repeats of D. buzzatii were omitted from the contigs used in our previous approach. Accordingly, although still low (see discussion below), we consider the values obtained in the present work as the most reliable ones.
We detected four satDNAs in D. seriema. The pBuM-2 satDNA with ∼340- to 390-bp-long alpha/beta repeat units (Kuhn and Sene 2005) is the most abundant, representing 1.93% of the genome. The second satDNA is DBC-150 (Kuhn et al. 2007), with ∼110- to 150-bp-long repeat units and representing 0.8% of the genome. The third satDNA is a novel one and was named CDSTR138, with 138-bp-long repeat units and representing 0.23% of the genome. The fourth satDNA is CDSTR198, which is shared with D. buzzatii, but represents only 0.02% of the D. seriema genome.
The SSS139 satDNA, with 139-bp-long repetition units was previously described in D. seriema (Franco et al. 2008). In the RepeatExplorer output, we found sequences homologous to SSS139 in the 10th most abundant repeat cluster, representing 0.5% of the genome. However, detailed sequence analysis revealed that this cluster is not made of tandem repeats. Instead, most sequences correspond to an ∼30-bp SSS139 inverted fragment interrupted by a region variable both in size and identity, followed by an ∼120-bp SSS139 sequence in direct orientation. Interestingly, these variable regions or the SSS139 sequences themselves showed no similarity to any TE or satDNA family previously described. Therefore, further studies will be necessary for elucidating the nature of the SSS139 repetitive elements.
We found two satDNAs in D. mojavensis. The most abundant is a novel one, which we named CDSTR130, with 130-bp-long repeat units and representing 1.63% of the genome. It is worth noting, however, that RepBase identified these sequences as a LTR BEL3_DM-I element described in D. mojavensis (Jurka 2012). This LTR has been characterized from D. mojavensis scaffold 5562 (nucleotide positions 8682–13,043 bp). However, the scrutinized analysis of 100 BEL3-DM insertions on the D. mojavensis genome showed that the 130-bp tandem repeats are not part of the LTR, but only flank the element in the scaffold 5562 (Figure 2). The identification of CDSTR130 as a satDNA highlights the importance of manual curation of the automated output provided by RepeatExplorer. It also explains why Melters et al. (2013) did not identify CDSTR130 as the most abundant tandem repeat family in the D. mojavensis genome.
Figure 2.
Schematic representation of the BEL3-DM-I transposable element present on RepBase, which is flanked by CDSTR130 satDNA arrays. Blue arrows represent the undescribed 185-bp-long terminal repeat of the BEL3-DM element.
The second most abundant satDNA identified in D. mojavensis is the pBuM-1 variant from the pBuM family (shared with D. buzzatii and D. seriema), with 185-bp-long repeats and representing 0.86% of the genome. This satDNA has been previously identified as the most abundant tandem repeat family of D. mojavensis by Melters et al. (2013).
The main features of the satDNAs identified above are summarized in Table 1 and a list containing consensus sequences from all the new satellites described in the present work can be seen in Figure S4 in File S1.
Table 1. Main features of satellite DNA families present on D. buzzatii, D. seriema, and D. mojavensis genomes.
| satDNA Family | Monomer Size | GC Content (%) | Copy Number (Analyzed) | Genomic Contribution (%) | Variability (%) | |
|---|---|---|---|---|---|---|
| D. buzzatii | pBuM | 189 | 29 | 379 | 1.71 | 12.1 |
| CDSTR198 | 198 | 34 | 79 | 0.23 | 13.1 | |
| D. seriema | pBuM-2 | 370 | 23.9 | 30a | 1.93 | 1.9a |
| DBC-150 | 150 | 55.9 | 5b | 0.81 | 11.3b | |
| CDSTR138 | 138 | 31.2 | 386 | 0.22 | 12.7 | |
| CDSTR198 | 198 | 34.8 | 67 | 0.02 | 15.5 | |
| D. mojavensis | CDSTR130 | 130 | 26.2 | 929 | 1.63 | 13.7 |
| pBuM | 185 | 26.5 | 600 | 0.86 | 4.1 |
Data from Kuhn et al. (2008).
Data from Kuhn et al. (2007).
Cactophilic Drosophila species present the lowest satDNA content within the genus
In most analyzed Drosophila species, the satDNA proportion fall within the range of between 15 and 40% (Bosco et al. 2007; Craddock et al. 2016). We found that the pBuM and CDSTR130 satDNAs represent only 2.5% of the D. mojavensis genome. Our result, obtained from the analyses of sequence reads using RepeatExplorer, was very close to the 2% satDNA contribution estimated by Bosco et al. (2007) using flow cytometry. In addition, we also found low amounts of satDNAs in the genomes of the other two cactophilic Drosophila: 1.9% for D. buzzatii and 2.9% for D. seriema. The additional 1% of the D. seriema in relation to D. buzzatii is probably represented by sequences located in the microchromosome of D. seriema, which is larger than that of D. buzzatii and also contains a higher amount of satellites (pBuM-2 and DBC-150) when compared to the other chromosomes (Figure 9; Kuhn et al. 2007, 2009). Our data revealed that cactophilic Drosophila present the lowest amount of satDNAs within the Drosophila genus reported so far. On the other hand, the estimated contribution of repetitive DNAs (satDNA+TE+unclassified repeats) in the three cactophilic Drosophila (14–27%) is not atypical for the genus (Drosophila 12 Genomes Consortium 2007; Craddock et al. 2016). Future studies focusing on satDNAs of more populations and species of the repleta group are expected to shed light on whether the low satDNA content in cactophilic Drosophila is a result of selective constraints or historical events.
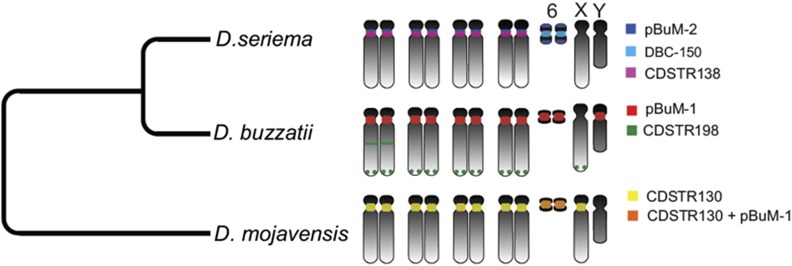
Figure 9.
Representative ideogram showing the chromosomal localization of all satDNAs identified in D. buzzatii, D. seriema, and D. mojavensis.
Preferential satDNA repeat lengths in cactophilic Drosophila
SatDNA repeats in the three studied cactophilic Drosophila have lengths of 130–200 bp or between 340 and 390 bp. To confirm this result, we ran RepeatExplorer with sequence reads from D. melanogaster where satDNA repeats <10 bp are abundant. RepeatExplorer correctly identified them as the most abundant repetitive DNAs of D. melanogaster (Table S5 in File S1). Therefore, we concluded that the preferential lengths for satDNA repeats in the three cactophilic Drosophila are not an artifact generated by RepeatExplorer.
Interestingly, satDNA repeats described before the genomic era in many plant and animal species (including Arabidopsis, maize, humans, and many insect species) typically show basic repeat units 150–180 or 300–360 bp long (Henikoff et al. 2001; Heslop-Harrison et al. 2003). Similar repeat-length patterns have been confirmed with recent genome-wide analysis of tandem repeats in other organisms. For example, Pavlek et al. (2015) showed that the most abundant tandem repeat families in the beetle Tribolium castaneum present repeat lengths either ∼170 bp or ∼340 bp long. It is difficult to explain such preferential repeat lengths by chance. On the other hand, it is striking that these two peak units closely correspond to the length of DNA wrapped around one or two nucleosomes.
It has been hypothesized that satDNA length could play a critical role in DNA packaging by favoring nucleosome positioning (or phasing) that in turn leads to condensation of certain genomic regions, such as the heterochromatin (Fitzgerald et al. 1994; Henikoff et al. 2001). Accordingly, the preferential lengths observed in the satDNA from cactophilic Drosophila could be selectively constrained by a possible role in chromatin packaging.
Satellite DNA candidates for centromeric function
The centromeres of most plant and animal species are composed of long arrays of tandemly repeated satellite DNAs (Plohl et al. 2014). There is increasing evidence to support a role for satDNA in centromeric function by providing motifs for centromeric-protein binding, e.g., CENP-B box in alphoid human satDNA (Ohzeki et al. 2002), and/or by producing RNA transcripts that are necessary for centromere/kinetochore assembly (Gent and Dawe 2012; Rošić et al. 2014). On the other hand, centromeric satDNAs may differ greatly even between closely related species. In fact, there are several examples supporting the observation that satDNA is one of the most rapidly evolving components of the genomes. Therefore, the identification of the most likely candidate for centromere function in a species is a task that in most cases has to be performed on a case-by-case basis.
Based on data collected from several animal and plant genomes, Melters et al. (2013) suggested that the most abundant tandem repeat of a genome would also be the most likely candidate for centromeric location and function. To test this hypothesis, we investigated by FISH the chromosomal location of all satDNAs identified in the three cactophilic Drosophila sampled in the present study.
All three species share the same basic karyotype (2n = 12) consisting of four pairs of telocentric autosomes, one pair of microchromosomes, and one pair of sex chromosomes (Baimal et al. 1983; Kuhn et al. 1996; Ruiz et al. 1990). Heterochromatin is located in the centromeric region of all four telocentric chromosomes, along the whole microchromosomes and Y chromosome and covering approximately one third of the proximal region of the X chromosome.
We identified the pBuM-1 alpha repeats as the most abundant satDNA of D. buzzatii. In a previous study, Kuhn et al. (2008) showed by FISH on mitotic chromosomes that pBuM-1 alpha repeats are located in the centromeric heterochromatin of all chromosomes except the X. In order to further investigate the chromosomal location of pBuM, we also hybridized a pBuM-1 probe to the polytene chromosomes. In these chromosomes, the centromeric heterochromatin is underreplicated and forms a dense central mass in the chromocenter – a region where the centromeres of all chromosomes bundle together. We observed that the pBuM-1 repeats are restricted to the chromocenter region (Figure 3A), therefore confirming their centromeric location. The second most abundant satDNA in D. buzzatii is CDSTR198, which was mapped by FISH in terminal and interstitial locations on metaphase chromosomes (these results are detailed below). Therefore, the most abundant satDNA of D. buzzatii, i.e., pBuM, is the one showing centromeric location in most chromosomes.
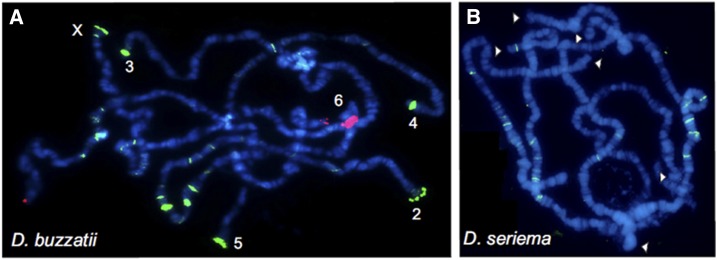
Figure 3.
FISH on polytene chromosomes of (A) D. buzzatii and (B) D. seriema using satDNA probes for pBuM (red) and CDSTR198 (green) (arrowheads indicate telomeric regions).
In D. seriema, the most abundant satDNA identified was pBuM-2 and the second most abundant was DBC-150. Previous studies showed that pBuM-2 is located on the centromeric regions of chromosomes 2, 3, 4, and 5 and on the telomeric regions of chromosome 6 (Kuhn et al. 2008). DBC-150 was found exclusively on the centromeric region of chromosome 6 (Kuhn et al. 2007). CDSTR138, the new satDNA described herein, is the third most abundant tandem repeat of this species and was mapped by FISH at the centromeric region of chromosomes 2, 3, 4, and 5 in mitotic chromosomes (Figure 4C). The centromeric location was also confirmed after FISH on polytene chromosomes, where no hybridization signals were observed outside the chromocenter (Figure 3A). The fourth identified satDNA in D. seriema, CDSTR198, showed no hybridization signal after FISH on mitotic chromosomes, confirming that it has very low copy number in this species (in contrast to D. buzzatii). However, we detected a few CDSTR198 repeats in the euchromatin after FISH on polytene chromosomes (Figure 3B; see below). Therefore, all three most abundant satDNAs of D. seriema are part of the centromeric region of most chromosomes.
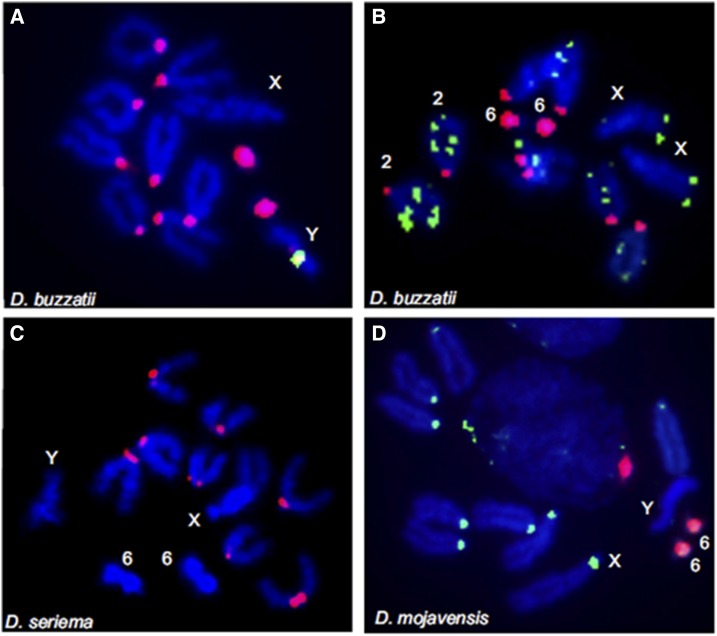
Figure 4.
FISH on mitotic chromosomes using satellite DNA probes. (A) pBuM-1a (red) and pBuM-1b (green) satDNA probes on D. buzzatii; (B) pBuM-1a (red) and CDSTR198 (green) probes on D. buzzatii; (C) CDSTR138 (red) on D. seriema; (D) CDSTR130 (green) and pBuM (red) probes on D. mojavensis.
CDSTR130 was identified as the most abundant satDNA in D. mojavensis; FISH on mitotic chromosomes showed that CDSTR130 repeats are located at the centromeric region of all autosomes and the X chromosome (Figure 4D). The second most abundant satDNA is pBuM-1, which covered the microchromosome (chromosome 6) almost entirely (Figure 4D). Therefore, both pBuM-1 and CDSTR130 are abundant in chromosome 6. However, given the size and dot-like morphology of this chromosome in this species, it is not possible to determine which one shows centromeric location. The analysis of the polytene chromosomes showed that the two satDNAs colocalize in the chromocenter region (Figure S5 in File S1).
Based on the collection and chromosome distribution of the satDNAs discussed herein, the centromeric regions of the X chromosome of D. buzzatii, of the X and Y of D. seriema, or of the Y of D. mojavensis are not composed of satDNAs. Some centromeres described in plants and animals are composed of TEs (reviewed by Plohl et al. 2014). In Drosophila, DINE-1 elements (helitrons) are one of the most abundant types of TEs (Yang and Barbash 2008). Kuhn and Heslop-Harrison (2011) and Dias et al. (2015) showed by FISH on mitotic chromosomes that these elements are highly enriched in the sex chromosomes (including the centromeric regions) in the three analyzed species from the repleta and virilis groups. It is possible that these DINE-1 elements are the main components of the centromeres of the sex chromosomes of cactophilic Drosophila species.
According to RepeatExplorer, the genomic proportion of satDNA in D. mojavensis (CDSTR130 + pBuM) is 2.5% (Table 1). This value is very close to the 2% satDNA contribution estimated by Bosco et al. (2007) using flow cytometry in the same species. According to the authors, if we split the ∼2% satDNA evenly among the D. mojavensis chromosomes that would result in ∼430 kb for each centromere. As noted by the authors, this value is also very close to what is considered as the minimum amount of centromeric DNA (420 kb) needed to fulfill centromeric function in Drosophila (Sun et al. 1997). In this context, Bosco et al. (2007) emphasized that it would be valuable to identify the centromeric satDNA of D. mojavensis and other Drosophila species to investigate whether they agree with the ∼420 kb limit observed in D. melanogaster.
In the present work, we found that pBuM and CDSTR130 are the main centromeric components of D. buzzatii and D. mojavensis. According to previous estimates, the male genome size of D. buzzatii and D. mojavensis is ∼170 Mb (Gregory and Johnston 2008; Romero-Soriano et al. 2016). Accordingly, we calculated that the bulk of centromeric satDNA in D. buzzatii is 2.9 Mb and in D. mojavensis, 2.8 Mb. If we split these values equally between the number of centromeres (= 6), each centromere will have ∼480 kb of centromeric DNA in D. buzzatii and ∼460 kb in D. mojavensis. This suggests cactophilic Drosophila have centromeric sizes roughly 470 kb on average, a value close to the suggested limit of 420 kb necessary for a functional centromere in Drosophila (Sun et al. 1997).
New insights on pBuM distribution and evolution
According to previous data on the distribution of pBuM-1 alpha and pBuM-2 alpha/beta repeats in the phylogeny of Drosophila species from the buzzatii cluster (repleta group), it was proposed that the ancestral state of the pBuM satDNA family consisted of alpha tandem repetition units ∼190 bp long. The alpha/beta repeats would have been originated subsequently from an insertion of a nonhomologous sequence of 180 bp (beta) in an alpha array, resulting in a composite alpha/beta repeat unit that also became abundant and tandemly organized (Kuhn and Sene 2005).
We found only alpha repeats in the genome of D. mojavensis, which is consistent with the hypothesis that alpha repeats represent the ancestral state of the pBuM family. According to current estimates, the split between the buzzatii and mojavensis clusters occurred ∼11 MYA (Oliveira et al. 2012; Guillén et al. 2015), which would be the minimum age for the origin of the pBuM family.
In D. seriema, we detected only pBuM-2 repeats, which agrees with previous DNA hybridization data (Kuhn and Sene 2005) suggesting that pBuM-2 is the only pBuM subfamily present in this species. The split between D. buzzatii and D. seriema was estimated to have happened ∼3 MYA (Franco et al. 2010). Therefore, in the last 3 MY, it seems that there was a complete turnover from pBuM-1 to pBuM-2 repeats in the genome of D. seriema.
According to our FISH experiments on mitotic and polytene chromosomes, pBuM repeats are restricted to the heterochromatic regions. However, BLAST on the assembled genome (Freeze 1 Scaffolds) of D. buzzatii revealed fragments of pBuM-1 repeats on three scaffolds (1, 88, and 90) that were mapped to the euchromatin from chromosomes 2, 5, and X [see Guillén et al. (2015) for exact location of scaffolds]. The three observed pBuM-1 euchromatic loci contain either a partial pBuM-1 repeat (<189 bp) or at most two partial pBuM-1 tandem repeats (<300 bp), and such small sizes were probably the reason they were undetected in our FISH experiments. The analysis of flanking sequences did not show evidence that these euchromatic pBuM-1 sequences could be integral parts of TEs and the mechanism(s) responsible for their presence on euchromatin are currently unknown.
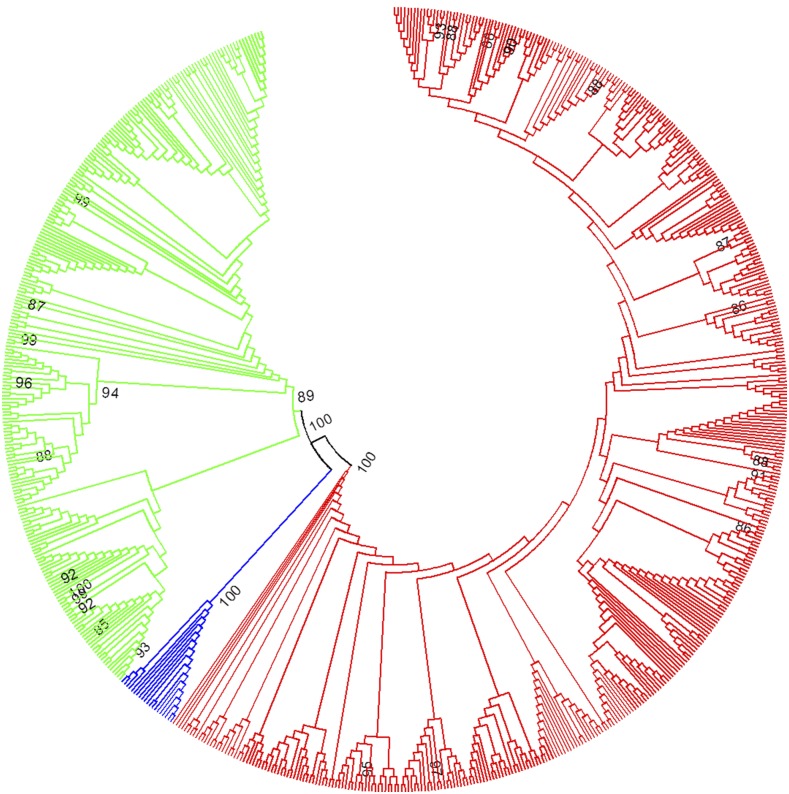
Previous phylogenetic analyses of pBuM repeats in D. buzzatii and D. seriema showed that these repeats have been evolving according to the concerted evolution model (Kuhn and Sene 2005). In other words, repeats within each species are more similar to each other than to repeats between species. In order to test whether pBuM also evolved in concert in D. mojavensis, we constructed a NJ tree with all pBuM repeats extracted from D. buzzatii, D. seriema, and D. mojavensis (Figure 5). The NJ tree revealed pBuM repeats from each species allocated in species-specific branches, indicating that pBuM has been evolving in a concerted manner in the last 11 MY.
Figure 5.
NJ tree containing a sample of pBuM repeats extracted from the sequenced genomes of Drosophila buzzatii (green), D. seriema (blue), and D. mojavensis (red). The tree was estimated using the T93 substitution model with 1000 bootstrap replicas.
The presence of pBuM in the nonrecombining Y allowed independent homogenization
In a previous report, the analysis of 63 pBuM-1 alpha repeats from D. buzzatii revealed very low levels of interrepeat variability (4.2% on average), indicating that, despite multiple chromosomal location, pBuM arrays have been efficiently homogenized at the intraspecific level (Kuhn et al. 1999). However, one repeat (Juan/4) showed atypical levels of nucleotide divergence in comparison to the remaining repeats (22% on average). Kuhn et al. (1999) suggested that this repeat may belong to another, less abundant, pBuM subfamily.
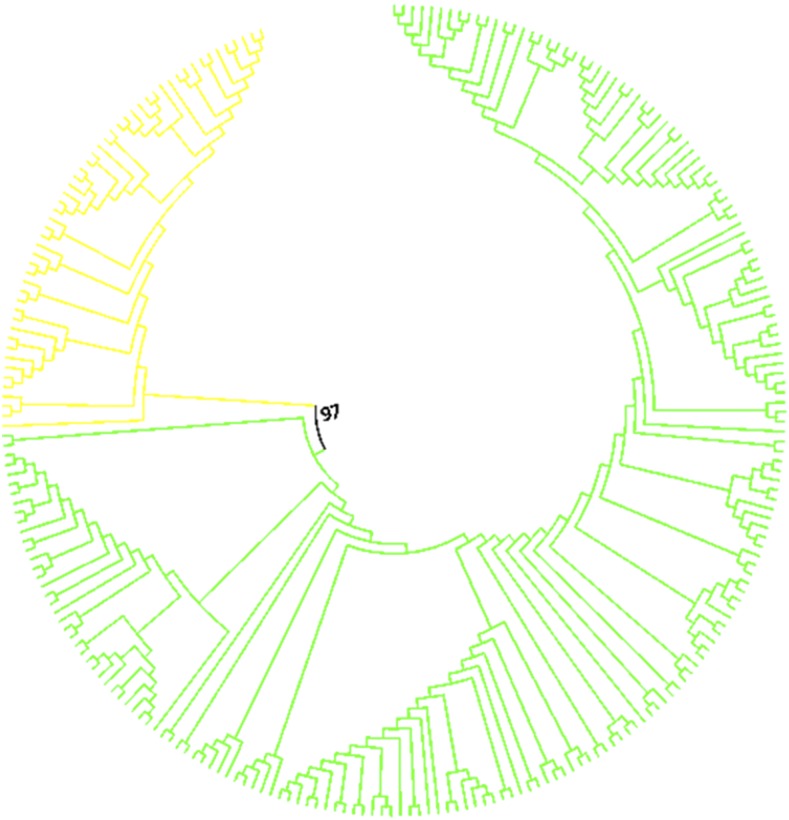
In the present work, we retrieved a sample of 247 pBuM-1 repeats from the sequenced genome of D. buzzatii and used them to construct a NJ tree. The resulting tree split the repeats into two main branches (Figure 6). The major one, containing 194 repeats, contains the “typical” pBuM-1 repeats, described in Kuhn et al. (1999). The second minor branch, with 53 repeats, contains “Juan/4-like” pBuM-1 repeats. Between the two groups, the nucleotide difference is 24.2%.
Figure 6.
NJ tree of pBuM satDNA repeats retrieved from the D. buzzatii assembled genome and previously described in Kuhn et al. (1999). Colored branches evidence Y chromosome-specific arrays (yellow) when compared to autosomal arrays (green). The tree was estimated using the T93 substitution model with 1000 bootstrap replicas.
These data are consistent with the hypothesis of two pBuM subfamilies being present in the D. buzzatii genome. Herein, we will name them as pBuM-1a (typical) and pBuM-1b (“Juan/4-like”). All the data generated so far about pBuM from D. buzzatii (including chromosomal location) concern the typical pBuM-1a repeat variant. There are several diagnostic nucleotide substitutions that allow discrimination between pBuM repeats from these two subfamilies. Such a situation allowed us to design oligonucleotides to specifically amplify pBuM-1b repeats by PCR for probe preparation. We then performed double-FISH with pBuM-1a and pBuM-1b on D. buzzatii mitotic chromosomes. The pBuM-1a probe showed the same multichromosomal distribution as described before. However, the pBuM-1b probe hybridized specifically to the Y chromosome (Figure 4A).
According to the model of concerted evolution, intraspecific homogenization of repeats occurs by recombination events such as unequal crossing over and gene conversion (Dover 1982; Dover and Tautz 1986). There is also some evidence suggesting that different arrays on the same or in different chromosomes may experience independent homogenization for arrays- or chromosomal-specific repeat variants (i.e., intragenomic concerted evolution) (Kuhn et al. 2012; Larracuente 2014; Khost et al. 2017). In this context, it is expected that arrays with tandem repeats on nonrecombining chromosomes, such as the Y, would be specially subjected to independent homogenization. This is most likely the reason for the existence of a different pBuM subfamily (pBuM-1b) on the Y chromosome of D. buzzatii. Furthermore, empirical and experimental data showed that low recombination is expected to increase interrepeat variability (Stephan and Cho 1994; Navajas-Pérez et al. 2006; Kuhn et al. 2007). In fact, pBuM-1a repeats had a nucleotide difference of 12%, while the pBuM-1b repeats (restricted to the Y chromosome) showed a higher variability of 17%.
The CDSTR198 satDNA shows terminal and dispersed distribution
The CDSTR198 satDNA was found in D. buzzatii and D. seriema, but with marked quantitative differences (0.23% in D. buzzatii and 0.02% in D. seriema). FISH on D. buzzatii mitotic chromosomes revealed that this satDNA is located in the terminal regions of chromosomes 2, 3, 4, 5, and X but also spread along euchromatic regions (Figure 4A). FISH on polytene chromosomes of the same species revealed strong hybridization signals in the telomeric regions of chromosomes 2, 5, and X, and in subtelomeric regions of chromosomes 3 and 4 (Figure 3A). Moreover, we detected the presence of CDSTR198 repeats along euchromatic regions of all chromosomes, except on the microchromosome. We found the highest number of CDSTR198 euchromatic signals concentrated in chromosomes 2 and 5 (Figure 3A). Similar results were also obtained by an overall analysis of 37 CDSTR198 euchromatic arrays present in the D. buzzatti assembled genome (Table S6 in File S1). Interestingly, this analysis showed an equal number of euchromatic arrays present on chromosomes 2 and 3 (11 arrays each), followed by chromosomes 4 and 5 (six arrays each). The fewer euchromatic arrays found in the D. buzzatii genome may result from the computational challenge of repetitive element assembly (Treangen and Salzberg 2012), reinforcing the need for hybridization experiments of satDNA families spread throughout euchromatin. In line with this, it is relevant to suggest that some CDSTR198 arrays identified by FISH may be absent on assembled genomes. FISH on polytene chromosomes of D. seriema showed CDSTR198 located only in a few euchromatic sites (Figure 3B).
In contrast to TEs, satDNAs do not have the ability to transpose by themselves. However, there are some reported examples showing that TEs may act as a substrate for satDNA emergence and mobility (Dias et al. 2015; Meštrović et al. 2015; Satović et al. 2016). We created a database containing the 500-bp sequences immediately before and after each CDSTR198 array (37 in total; Table S6 in File S1) found in the assembled scaffolds of D. buzzatii. Comparative analysis of all flanking sequences did not show association to a specific TE or TE family or to any other specific sequence common to all arrays. These results raise the question about the dispersion mechanism of CDSTR198 in the D. buzzati genome.
Tandemly repeated sequences may undergo small recombination events involving copies of the same array in the same orientation. These events may result in the formation of extrachromosomal circular DNAs (eccDNAs) (Cohen and Segal 2009). The occasional presence of a replication-initiating region may provide further amplification and new eccDNA copies. Apparently, these eccDNAs can be inserted again into the genome by recombination. This mechanism was proposed to explain the dispersion of copies of the satDNA TCAST2 in Tribolium castaneum (Brajković et al. 2012), as well as of the D. melanogaster 1.688 satDNA (Cohen and Segal 2009), which also show an euchromatic dispersed distribution (Kuhn et al. 2012). In order to test this hypothesis, it would be interesting to look for the presence of eccDNA-containing CDSTR198 repeats in D. buzzatii.
CDSTR198 satDNA may contribute to telomeric function in D. buzzatii
Unlike most eukaryotes, Drosophila telomeric regions are maintained by a sequence complex organized in three subdomains: (i) arrays of TEs (Het-A/TART) responsible for maintaining telomeric sequences; (ii) telomere-associated sequences (TAS), formed by complex repetitive sequences, usually satDNAs, and (iii) a protein complex HOAP required for telomere stability (Silva-Sousa and Casacuberta 2012). Although the structure of telomeres is conserved among all Drosophila species, the TEs and TAS sequences are highly variable even among phylogenetically close species (Villasante et al. 2007). Based on the widespread presence of TAS in Drosophila and other species (including humans), Biessmann et al. (2000) proposed that homologous recombination between terminal satDNA repeats could have been an “ancient” mechanism for telomere extension. Today, TAS regions probably function as a buffer zone between the telomeres and internal chromosome domains (Sharma and Raina 2005).
We could not identify conserved domains for telomeric Het-A and TART TEs in the sequenced genome of D. buzzatii, even though these TEs were described in D. mojavensis and D. virilis (Villasante et al. 2007). Similarly, a recent screening of the D. buzzatii sequenced genome for the whole TE content did not identify Het-A or TART elements (Rius et al. 2016). The apparent absence of Het-A and TART in D. buzzatii may be related to the high evolutionary rate of these sequences (Villasante et al. 2007). Alternatively, there may be a different mechanism for telomere elongation operating in this species.
The CDSTR198 satDNA is located in the telomeric and subtelomeric regions of five (out of six) chromosomes of D. buzzatii (Figure 3A and Figure 4B). The presence of CDSTR198 in the telomeres associated with the apparent absence of Het-A and TART sequences open the possibility that CDSTR198 plays a role in telomere elongation through a recombination-based mechanism (e.g., unequal crossing over). Although not described in Drosophila, tandem repeat sequences are responsible for maintaining telomeres in the dipterous genus Chironomus (Löpez et al. 1996).
It is important to mention that a similar scenario described herein for the CDSTR198 of D. buzzatii was previously reported for D. virilis, which belongs to the virilis group. In this noncactophilic species, the terminal location of the pvB370 satDNA associated with the absence of telomere transposons led Biessmann et al. (2000) to propose the involvement of this satDNA in telomere elongation. However, TART-like and HeT-like elements were later described in the terminal regions of D. virilis, opening the possibility that these elements also participate in telomeric elongation in this species (Casacuberta and Pardue 2003; Pardue et al. 2005).
pBuM and CDSTR130 show regions of interspersed distribution in the microchromosomes
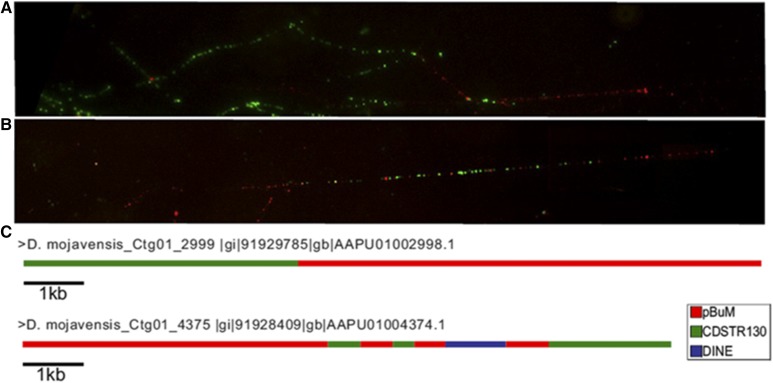
FISH with CDSTR130 and pBuM probes on D. mojavensis mitotic chromosomes revealed that these two satDNA colocalize on the microchromosome. In order to further investigate how these two satDNAs are organized, we performed double-FISH experiments on extended DNA fibers. We observed strong hybridization signals in fibers showing CDSTR130 long arrays followed by pBuM long arrays (Figure 7A). However, in some DNA fibers hybridization signals indicated an interspersed organization of both satDNAs (Figure 7B). These results were also confirmed in the analysis of D. mojavensis assembled contigs (Figure 7C). For example, the contig 2999 (AAPU01002998.1) is composed of 4435 bp of CDSTR130 copies adjacent to a pBuM array of 7716 bp. In the contig 4375 (AAPU01004374.1), we observed different arrays of pBuM and CDSTR130 interspersed with each other (Figure 7C).
Figure 7 (A and B).
FISH with CDSTR130 (green) and pBuM (red) probes onto extended DNA fibers of D. mojavensis. (C) Schematic representation of CDSTR130 and pBuM organization found on contigs Ctg01_2999(AAPU01002998.1) and Ctg01_4375(AAPU01004374.1) retrieved from the D. mojavensis assembled genome.
Nonhomologous satDNAs located in the same chromosome region are usually organized in separate arrays (e.g., Shiels et al. 1997; Lohe et al. 1993; Sun et al. 2003). However, there are some reports showing interspersion of repeats from different satellites (e.g., Žinić et al. 2000; Alkhimova et al. 2004; Wei et al. 2014). It has been suggested that interspersion between repeats may give rise to new higher order repeat structures (Mravinac and Plohl 2007; Wei et al. 2014). In a previous study conducted in cactophilic Drosophila species, Kuhn et al. (2009) showed high levels of interspersion between pBuM and DBC-150 in at least two species of the buzzatii cluster (D. gouveai and D. antonietae). Interestingly, such pattern was also observed in the microchromosomes. According to Kuhn et al. (2009), interspersion of repeats from nonhomologous satellites in the microchromosomes could be related to the peculiar characteristics of these chromosomes, such as highly heterochromatic nature and low content of genes, which could allow a more flexible interplay between repetitive elements without deleterious effects.
Differential transcription of cactophilic Drosophila satDNAs
SatDNAs do not code for proteins and have been traditionally viewed as “junk DNAs.” However, there is a growing number of studies showing satDNA transcription activity from yeast to mammals, and the biological function of these transcripts has now started to be appreciated. For example, satDNA transcripts were shown to be involved in heterochromatin assembly, kinetochore formation, and gene regulation (reviewed by Biscotti et al. 2015; Ferreira et al. 2015). Moreover, transcription of satDNAs is usually gender or stage specific and is often associated with differentiation and development (Usakin et al. 2007; Pecinka et al. 2010).
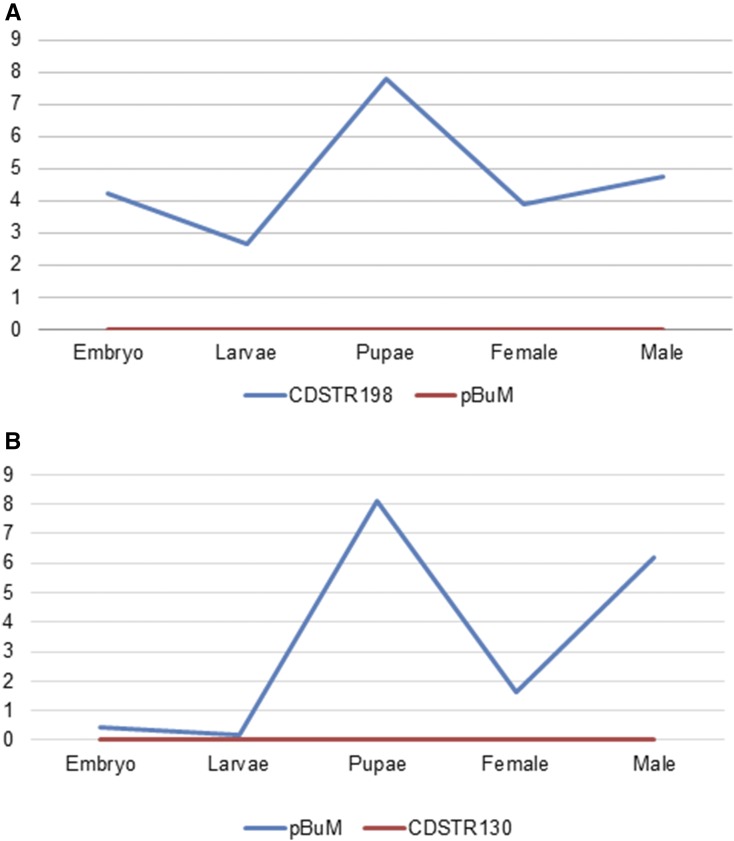
Herein, we investigated whether the satDNAs that we analyzed are transcribed by mapping the satDNA consensus sequences on the available RNA-seq data from D. buzzatii and D. mojavensis (Guillén et al. 2015; Rius et al. 2016). Read counts were calculated for embryos, third-staged larvae, pupae, and for male and female adult carcasses (Figure 8) (see Materials and Methods).
Figure 8.
Transcription profile of satDNA families in D. buzzatii (A) and D. mojavensis (B) at five different developmental stages. Counts were normalized to one million reads.
Our analysis did not identify transcripts from the most abundant satDNAs in the genome of D. buzzatii and D. mojavensis, pBuM and CDSTR130, respectively. As discussed previously, both are the main candidates for centromeric function in these species. This result was unexpected because previous studies in D. melanogaster showed that centromeric satellite RNAs in the form of long polyadenylated products play an important role in the formation of the kinetochore (Topp et al. 2004; Chan et al. 2012; Rošić et al. 2014). However, our results do not exclude the possibility that pBuM and CDSTR130 are transcribed. In this case, the absence of satDNA transcripts may be related to the methodology used for RNA extraction that preferentially captures poly(A) sequences. For example, satDNA transcripts of D. melanogaster involve non-coding RNAs that do not have poly(A) tails (Usakin et al. 2007).
Conversely, in all five analyzed tissues we detected transcripts derived from the CDSTR198 satDNA of D. buzzatii and from the pBuM satDNA of D. mojavensis. In both cases, the transcripts were particularly abundant in tissues from pupae and males. Interestingly, these two satDNAs are located in different genomic environments: while CDSTR198 arrays are located at several euchromatic loci (including some close to genes; Table S7 in File S1) in several D. buzzatii chromosomes, pBuM is exclusively located in the heterochromatic microchromosome of D. mojavensis (Figure 9). Future studies will be needed to address whether these transcripts participate in chromatin modulation and/or if they affect the transcription of neighboring genes, as observed for satDNA transcripts of Drosophila and other organisms (Menon et al. 2014; Feliciello et al. 2015).
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.042093/-/DC1.
Acknowledgments
We are grateful to Alfredo Ruiz (Universitat Autònoma de Barcelona) for several insightful discussions during different stages of this work and also for sharing the RNA-seq data we used. We thank Guilherme Borges Dias (Universidade Federal de Minas Gerais) for sequencing D. seriema. We also thank A. Bernardo Carvalho (Universidade Federal do Rio de Janeiro) for kindly sharing the D. mojavensis sequencing data with us. Genomic DNA quality control, library preparation, and sequencing were conducted at the Laboratório de Biotecnologia e Marcadores Moleculares of the Universidade Federal de Minas Gerais, with the aid of Anderson Oliveira do Carmo, Ana Paula Vimieiro Martins, and Evanguedes Kalapothakis. This work was supported by a grant from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (grant number APQ-01563-14) to G.C.S.K. L.G.d.L. was supported with a doctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Funding for sequencing was provided by the CAPES – Programa de Excelência Acadêmica (PROEX) – to Programa de Pós Graduação em Genética da Universidade Federal de Minas Gerais (process CAPES/PROEX 0529/2014).
Footnotes
Communicating editor: B. Oliver
Literature Cited
- Afgan E., Baker D., Van den Beek M., Blankenberg D., Bouvier D., et al. , 2016. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrup-MacDonald M. E., Kuo M. E., Sullivan L. L., Chew K., Sullivan B. A., 2016. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26(10): 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhimova O. G., Mazurok N. A., Potapova T. A., Zakian S. M., Heslop-Harrison J. S., et al. , 2004. Diverse patterns of the tandem repeats organization in rye chromosomes. Chromosoma 113(1): 42–52. [DOI] [PubMed] [Google Scholar]
- Baimal V., Sene F. M., Pereira M. A. O. R., 1983. Heterochromatin and karyotypic differentiation of some neotropical cactus-breeding species of the Drosophila repleta species group. Genetica 60(2): 81–92. [Google Scholar]
- Barghini E., Natali L., Cossu R. M., Giordani T., Pindo M., et al. , 2014. The peculiar landscape of repetitive sequences in the olive (Olea europaea L.) genome. Genome Biol. Evol. 6(4): 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2): 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beridze T., 2013. Satellite DNA. Springer Science and Business Media, Berlin. [Google Scholar]
- Biessmann H., Zurovcova M., Yao J. G., Lozovskaya E., Walter M. F., 2000. A telomeric satellite in Drosophila virilis and its sibling species. Chromosoma 109(6): 372–380. [DOI] [PubMed] [Google Scholar]
- Biscotti M. A., Canapa A., Forconi M., Olmo E., Barucca M., 2015. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res. 23(3): 463–477. [DOI] [PubMed] [Google Scholar]
- Blattes R., Monod C., Susbielle G., Cuvier O., Wu J., et al. , 2006. Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J. 25(11): 2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Campbell P., Leiva-Neto J. T., Markow T. A., 2007. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177: 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brajković J., Feliciello I., Bruvo-Mađarić B., Ugarković Đ., 2012. Satellite DNA-like elements associated with genes within euchromatin of the beetle Tribolium castaneum. G3 2: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M., Ranz J. M., Barbadilla A., Long M., Ruiz A., 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285(5426): 415–418. [DOI] [PubMed] [Google Scholar]
- Casacuberta E., Pardue M. L., 2003. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc. Natl. Acad. Sci. USA 100(6): 3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., et al. , 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA 109: 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan L. W., 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371(6494): 215–220. [DOI] [PubMed] [Google Scholar]
- Cohen S., Segal D., 2009. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet. Genome Res. 124(3–4): 327–338. [DOI] [PubMed] [Google Scholar]
- Craddock E. M., Gall J. G., Jonas M., 2016. Hawaiian Drosophila genomes: size variation and evolutionary expansions. Genetica 144(1): 107–124. [DOI] [PubMed] [Google Scholar]
- Dias G. B., Heringer P., Svartman M., Kuhn G. C. S., 2015. Helitrons shaping the genomic architecture of Drosophila: enrichment of DINE-TR1 in α- and β-heterochromatin, satellite DNA emergence, and piRNA expression. Chromosome Res. 23(3): 597–613. [DOI] [PubMed] [Google Scholar]
- Dover G., 1982. Molecular drive: a cohesive mode of species evolution. Nature 229(5879): 111–117. [DOI] [PubMed] [Google Scholar]
- Dover G. A., Tautz D., 1986. Conservation and divergence in multigene families: alternatives to selection and drift. Philos. Trans. R. Soc. Lond. B Biol. Sci. 312(1154): 275–289. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450(7167): 203–218. [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciello I., Akrap I., Ugarković Đ., 2015. Satellite DNA modulates gene expression in the beetle Tribolium castaneum after heat stress. PLoS Genet. 11(8): e1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D., Meles S., Escudeiro A., Mendes-da-Silva A., Adega F., et al. , 2015. Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer. Chromosome Res. 23(3): 479–493. [DOI] [PubMed] [Google Scholar]
- Fitzgerald D. J., Dryden G. L., Bronson E. C., Williams J. S., Anderson J. N., 1994. Conserved patterns of bending in satellite and nucleosome positioning DNA. J. Biol. Chem. 269(33): 21303–21314. [PubMed] [Google Scholar]
- Franco F. F., Sene F. M., Manfrin M. H., 2008. Molecular characterization of SSS139, a new satellite DNA family in sibling species of the Drosophila buzzatii cluster. Genet. Mol. Biol. 31(1): 155–159. [Google Scholar]
- Franco F. F., Silva‐Bernardi E. C. C., Sene F. M., Hasson E. R., Manfrin M. H., 2010. Intra‐and interspecific divergence in the nuclear sequences of the clock gene period in species of the Drosophila buzzatii cluster. J. Zoological Syst. Evol. Res. 48(4): 322–331. [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L., 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33(3): 319–344. [DOI] [PubMed] [Google Scholar]
- Gent J. I., Dawe R. K., 2012. RNA as a structural and regulatory component of the centromere. Annu. Rev. Genet. 46: 443–453. [DOI] [PubMed] [Google Scholar]
- Gregory T. R., Johnston J. S., 2008. Genome size diversity in the family Drosophilidae. Heredity 101(3): 228–238. [DOI] [PubMed] [Google Scholar]
- Guillén Y., Rius N., Delprat A., Williford A., Muyas F., et al. , 2015. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol. Evol. 7(1): 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532): 1098–1102. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. S., Brandes A., Schwarzacher T., 2003. Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res. 11(3): 241–253. [DOI] [PubMed] [Google Scholar]
- Jagannathan M., Warsinger-Pepe N., Watase G. J., Yamashita Y. M., 2017. Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 7: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T., Pagni M., 2000. Dotlet: diagonal plots in a web browser. Bioinformatics 16(2): 178–179. [DOI] [PubMed] [Google Scholar]
- Jurka J., 2012. LTR retrotransposons from fruit fly. Repbase Rep. 12(7): 1257. [Google Scholar]
- Khost D. E., Eickbush D. G., Larracuente A. M., 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 27: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn G. C. S., Heslop-Harrison J. S., 2011. Characterization and genomic organization of PERI, a repetitive DNA in the Drosophila buzzatii cluster related to DINE-1 transposable elements and highly abundant in the sex chromosomes. Cytogenet. Genome Res. 132: 79–88. [DOI] [PubMed] [Google Scholar]
- Kuhn G. C. S., Sene F. M., 2005. Evolutionary turnover of two pBuM satellite DNA subfamilies in the Drosophila buzzatii species cluster (repleta group): from alpha to alpha/beta arrays. Gene 349: 77–85. [DOI] [PubMed] [Google Scholar]
- Kuhn G. C. S., Ruiz A., Alves M. A., Sene F. M., 1996. The metaphase and polytene chromosomes of Drosophila seriema (repleta group; mulleri subgroup). Braz. J. Genet. 19: 209–216. [Google Scholar]
- Kuhn G. C. S., Bollgönn S., Sperlich D., Bachmann L., 1999. Characterization of a species‐specific satellite DNA of Drosophila buzzatii. J. Zoological Syst. Evol. Res. 37(2): 109–112. [Google Scholar]
- Kuhn G. C. S., Franco F. F., Manfrin M. H., Moreira-Filho O., Sene F. M., 2007. Low rates of homogenization of the DBC-150 satellite DNA family restricted to a single pair of microchromosomes in species from the Drosophila buzzatii cluster. Chromosome Res. 15(4): 457–470. [DOI] [PubMed] [Google Scholar]
- Kuhn G. C. S., Sene F. M., Moreira-Filho O., Schwarzacher T., Heslop-Harrison J. S., 2008. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res. 16(2): 307–324. [DOI] [PubMed] [Google Scholar]
- Kuhn G. C. S., Teo C. H., Schwarzacher T., Heslop-Harrison J. S., 2009. Evolutionary dynamics and sites of illegitimate recombination revealed in the interspersion and sequence junctions of two nonhomologous satellite DNAs in cactophilic Drosophila species. Heredity 102(5): 453–464. [DOI] [PubMed] [Google Scholar]
- Kuhn G. C. S., Küttler H., Moreira-Filho O., Heslop-Harrison J. S., 2012. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol. Biol. Evol. 29: 7–11. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9(4): 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., 2014. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol. Biol. 14(1): 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Hilliker A. J., Roberts P. A., 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134: 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löpez C. C., Nielsen L., Edström J. E., 1996. Terminal long tandem repeats in chromosomes from Chironomus pallidivittatus. Mol. Cell. Biol. 16(7): 3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrin M. H., Sene F. M., 2006. Cactophilic Drosophila in South America: a model for evolutionary studies. Genetica 126(1–2): 57–75. [DOI] [PubMed] [Google Scholar]
- Marques A., Ribeiro T., Neumann P., Macas J., Novák P., et al. , 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. USA 112(44): 13633–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters D. P., Bradnam K. R., Young H. A., Telis N., May M. R., et al. , 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14(1): R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon D. U., Coarfa C., Xiao W., Gunaratne P. H., Meller V. H., 2014. siRNAs from an X-linked satellite repeat promote X-chromosome recognition in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 111(46): 16460–16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meštrović N., Mravinac B., Pavlek M., Vojvoda-Zeljko T., Šatović E., et al. , 2015. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 23(3): 583–596. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5(7): 621–628. [DOI] [PubMed] [Google Scholar]
- Mravinac B., Plohl M., 2007. Satellite DNA junctions identify the potential origin of new repetitive elements in the beetle Tribolium madens. Gene 394(1): 45–52. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R., Schwarzacher T., de la Herrán R., Rejón C. R., Rejón M. R., et al. , 2006. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61–71. [DOI] [PubMed] [Google Scholar]
- Negre B., Casillas S., Suzanne M., Sánchez-Herrero E., Akam M., et al. , 2005. Conservation of regulatory sequences and gene expression patterns in the disintegrating Drosophila Hox gene complex. Genome Res. 15(5): 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [Google Scholar]
- Novák P., Neumann P., Pech J., Steinhaisl J., Macas J., 2013. RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29(6): 792–793. [DOI] [PubMed] [Google Scholar]
- Ohzeki J. I., Nakano M., Okada T., Masumoto H., 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159(5): 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. C., Almeida F. C., O’Grady P. M., Armella M. A., DeSalle R., et al. , 2012. Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of the Drosophila repleta species group. Mol. Phylogenet. Evol. 64(3): 533–544. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Rashkova S., Casacuberta E., DeBaryshe P. G., George J. A., et al. , 2005. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 13(5): 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlek M., Gelfand Y., Plohl M., Meštrović N., 2015. Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res. 22(6): 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Dinh H. Q., Baubec T., Rosa M., Lettner N., et al. , 2010. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22(9): 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M., Meštrović N., Mravinac B., 2014. Centromere identity from the DNA point of view. Chromosoma 123(4): 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., 1997. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press, New York. [Google Scholar]
- Rius N., Guillén Y., Delprat A., Kapusta A., Feschotte C., et al. , 2016. Exploration of the Drosophila buzzatii transposable element content suggests underestimation of repeats in Drosophila genomes. BMC Genomics 17(1): 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Soriano V., Burlet N., Vela D., Fontdevila A., Vieira C., et al. , 2016. Drosophila females undergo genome expansion after interspecific hybridization. Genome Biol. Evol. 8(3): 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rošić S., Köhler F., Erhardt S., 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 207(3): 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Heed W. B., Wasserman M., 1990. Evolution of the mojavensis cluster of cactophilic Drosophila with descriptions of two new species. J. Hered. 81(1): 30–42. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ruano F. J., López-León M. D., Cabrero J., Camacho J. P. M., 2016. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 6: 28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4(4): 406–425. [DOI] [PubMed] [Google Scholar]
- Satović E., Zeljko T. V., Luchetti A., Mantovani B., Plohl M., 2016. Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genomics 17(1): 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison P., 2000. Practical in situ Hybridization. BIOS Scientific Publishers Ltd., Oxford. [Google Scholar]
- Sharma S., Raina S. N., 2005. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 109(1–3): 15–26. [DOI] [PubMed] [Google Scholar]
- Shiels C., Coutelle C., Huxley C., 1997. Contiguous arrays of satellites 1, 3, and β form a 1.5-Mb domain on chromosome 22p. Genomics 44(1): 35–44. [DOI] [PubMed] [Google Scholar]
- Silva-Sousa R., Casacuberta E., 2012. Drosophila telomeres: an example of co-evolution with transposable elements, pp. 46–67 in Repetitive DNA, Vol. 7 Karger Publishers, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 2013 RepeatMasker Open-4.0. Available at: http://www. repeatmasker.org. Accessed: February 11, 2016.
- Stephan W., Cho S., 1994. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics 136: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T., Webb D., Dover G. A., 1985. Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J. 4(7): 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wahlstrom J., Karpen G., 1997. Molecular structure of a functional Drosophila centromere. Cell 91(7): 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Le H. D., Wahlstrom J. M., Karpen G. H., 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13(2): 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Nei M., 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10(3): 512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28(10): 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., 1993. Notes on the definition and nomenclature of tandemly repetitive DNA sequences, pp. 21–28 in DNA Fingerprinting: State of the Science. Birkhäuser, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- Topp C. N., Zhong C. X., Dawe R. K., 2004. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101(45): 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Salzberg S. L., 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13(1): 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrego R., Bernal-Ulloa S. M., Chavarría N. A., Herrera-Puerta E., Lucas-Hahn A., et al. , 2017. Satellite DNA methylation status and expression of selected genes in Bos indicus blastocysts produced in vivo and in vitro. Zygote 25: 131–140. [DOI] [PubMed] [Google Scholar]
- Usakin L., Abad J., Vagin V.V., De Pablos B., Villasante A., et al. , 2007. Transcription of the 1.688 satellite DNA family is under the control of RNA interference machinery in Drosophila melanogaster ovaries. Genetics 176: 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A., Abad J. P., Planelló R., Méndez-Lago M., Celniker S. E., et al. , 2007. Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase. Genome Res. 17(12): 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. H. C., Grenier J. K., Barbash D. A., Clark A. G., 2014. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 111(52): 18793–18798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. P., Barbash D. A., 2008. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 9(2): R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žinić S. D., Ugarković D., Cornudella L., Plohl M., 2000. A novel interspersed type of organization of satellite DNAs in Tribolium madens heterochromatin. Chromosome Res. 8(3): 201–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.