Abstract
One of the most powerful ways to develop hypotheses regarding the biological functions of conserved genes in a given species, such as humans, is to first look at what is known about their function in another species. Model organism databases and other resources are rich with functional information but difficult to mine. Gene2Function addresses a broad need by integrating information about conserved genes in a single online resource.
Keywords: functional genomics, orthologs, human genetic disease, model organism databases, data mining
The availability of full-genome sequences has uncovered a striking level of conservation among genes from single-celled organisms such as yeast, invertebrates such as flies or nematode worms, and vertebrates such as fish, mice, and humans. This conservation is not limited to amino acid identity or structure, or RNA sequence. Indeed, gene conservation often extends to conservation of biochemical function (e.g., common enzymatic functions); cellular function (e.g., specific role in intracellular signal transduction); and function at the organ, tissue, and whole-organism levels (e.g., control of organ formation, tissue homeostasis, or behavior).
Researchers applying small- or large-scale approaches in any common model organism often come across genes that are poorly characterized in their species of interest. A common and powerful way to develop an hypothesis regarding the function of a gene poorly characterized in one species—or newly implicated in some processes in that species—is to ask whether the gene is conserved and, if so, find out what is known about the functions of its orthologs in other species. This commonly applied approach gains importance when the poorly characterized gene is implicated in a human disease; in many cases, what we know about human gene function is largely based on what was first uncovered for orthologs in other species.
Despite the importance and broad application of this approach among biologists and biomedical researchers, there are barriers to applying the approach to its fullest. First, ortholog mapping is not straightforward. Over the years, many approaches and algorithms have been applied to mapping of orthologs. The results do not always agree and, at a practical level, the use of different genome annotation versions, as well as different gene or protein identifiers, can make it difficult to identify or have confidence in an ortholog relationship. Second, even after one or more orthologs in common model species have been identified, it is not easy to quickly assess in which species the orthologs have been studied and determine what functional information was gained. Model organism databases (MODs) and human gene databases provide relevant, expertly curated information. Although InterMine (Smith et al. 2012) provides a mechanism for batch search of standardized information, and NCBI Gene provides information about individual genes in a standardized format, it remains a challenge to navigate, access, and integrate information about all of the orthologs of a given gene in well-studied organisms. As a result, useful information can be missed, contributing to inefficiency and needless delay in reaching the goal of functional annotation of genes, including genes relevant to human disease.
Clearly, there is a need for an integrated resource that facilitates the identification of orthologs and mining of information regarding ortholog function, in particular, in common genetic model organisms supported by MODs. Previously, we developed approaches for integration of various types of gene- or protein-related information, including ortholog predictions [DRSC Integrative Ortholog Prediction Tool (DIOPT); Hu et al. 2011], disease–gene mapping based on various sources [DIOPT–Diseases and Traits (DIOPT–DIST); Hu et al. 2011], and transcriptomics data [Drosophila Gene Expression Tool (DGET); Hu et al. 2017]. Importantly, these can serve as individual components of a more comprehensive, integrated resource. Indeed, our DIOPT approach to identification of high-confidence ortholog predictions is now used in other contexts, including at FlyBase (Gramates et al. 2017) and at MARRVEL for mining information starting with human gene variant information (Wang et al. 2017; www.marrvel.org).
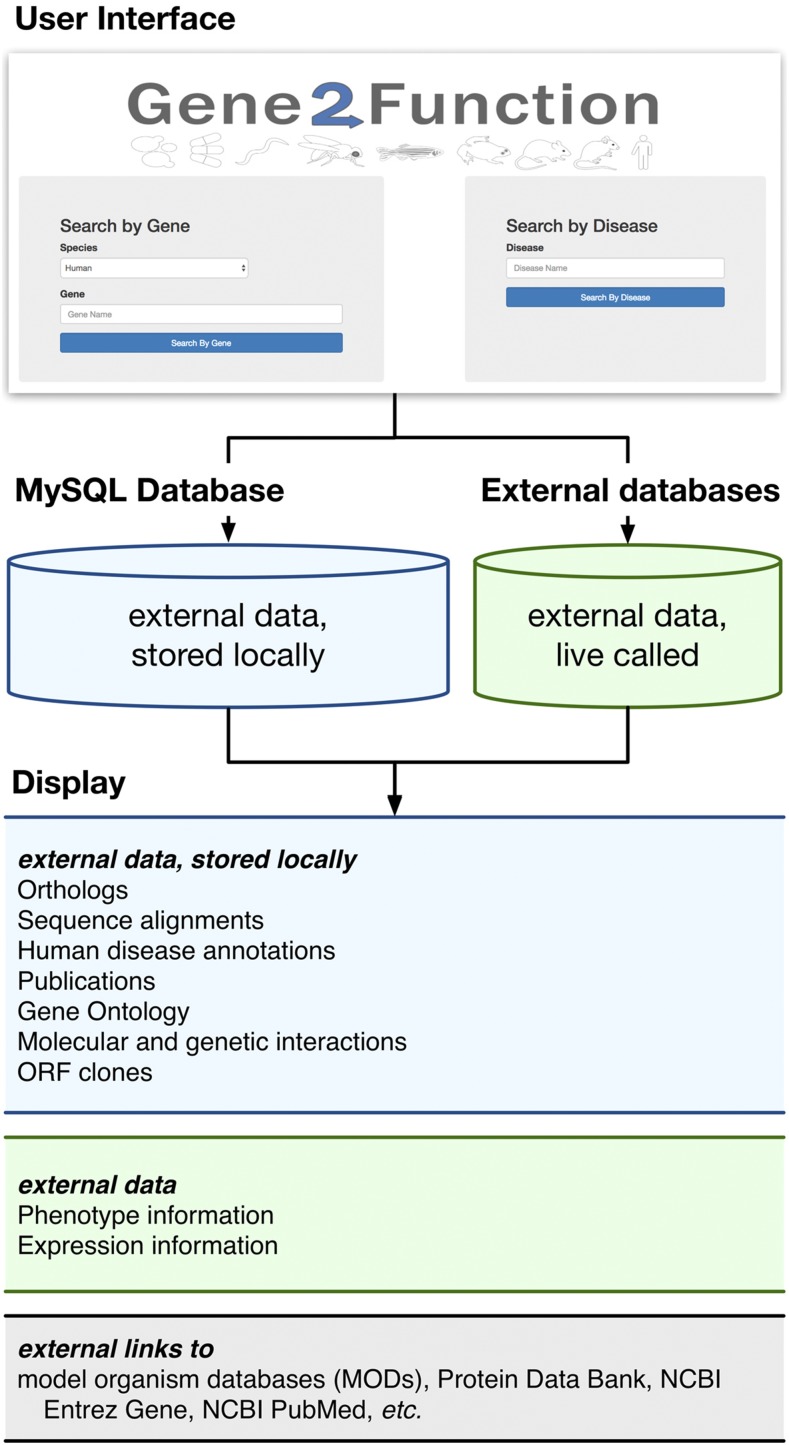
To address the broad need for an integrated resource, we developed Gene2Function (G2F; www.gene2function.org), an online resource that maps orthologs among human genes and common genetic model species supported by MODs, and displays summary information for each ortholog. G2F makes it easy to survey the wealth of information available for orthologs and navigate from one species to another, and connects users to detailed reports and information at individual MODs and other sources. The integration approach and set of information sources are outlined in Figure 1 and Table 1, and described in the Supplemental Material, File S1 (Supplemental Methods).
Figure 1.
Overview of the Gene2Function (G2F) online resource. For detailed information about the database, logic flow, and information sources, see File S1.
Table 1. Summary of disease and gene reports displayed in Gene2Function (G2F).
| Column Header | Content | Source of Content |
|---|---|---|
| Disease report | ||
| Gene symbol human | Official gene symbol | NCBI gene |
| Gene ID human | NCBI gene ID | NCBI gene |
| Count disease terms | Number of disease terms | OMIM, EBI GWAS |
| Disease terms | Disease terms | OMIM, EBI GWAS |
| Ortholog overview | Link to G2F gene report | Internal |
| Gene report | ||
| NCBI gene ID | NCBI gene ID | NCBI gene |
| Symbol | Official gene symbol | NCBI gene |
| Human disease counts | Number of disease terms; link to MARRVELa | OMIM, EBI GWAS |
| Species name | Species name | |
| Species-specific gene ID | Species-specific gene ID | Links to HGNC or MOD gene reportb |
| Species-specific database | Relevant database name | Links to HGNC or MOD home page |
| DIOPT score | DIOPT scorec | DIOPT |
| Best score | Yes or no, this pair has best score at DIOPT | DIOPT |
| Best score reverse | Yes or no, this pair has best score if opposite search | DIOPT |
| Confidence | DIOPT confidenced | DIOPT |
| Publication count | Number of publications on the ortholog | NCBI gene2pubmed |
| GO component counts | Number of cellular component GO terms assigned to the ortholog | NCBI gene2go |
| GO function counts | Number of molecular function GO terms assigned to the ortholog | NCBI gene2go |
| GO process counts | Number of biological processes GO terms assigned to the ortholog | NCBI gene2go |
| Protein interaction counts | Number of protein interactions assigned to the ortholog | BioGrid |
| Genetic interaction counts | Number of genetic interactions assigned to the ortholog | BioGrid |
| Mine phenotype data | Number of phenotype entries from Minese | HumanMine, MouseMine, XenMine, ZebrafishMine, FlyMine, WormBase, SGD |
| Mine expression data | Number of expression entries from Minese | HumanMine, MouseMine, XenMine, ZebrafishMine, FlyMine, WormBase, SGD |
| Mine disruption phenotype | Number of disruption phenotype entries | UniProt |
| 3D structure | Number of 3D structures available for the ortholog | Protein data bank |
| ORF clones | Number of ORF clones | PlasmID clone repositoryf |
| Protein alignment | Multiple or pairwise alignment of orthologs | DIOPT |
OMIM, Online Mendelian Inheritance in Man; EBI, European Bioinformatics Institute; GWAS, genome-wide association study; HGNC, HUGO Gene Nomenclature Committee; MOD, model organism database; DIOPT, DRSC Integrative Ortholog Prediction Tool; GO, gene ontology; SGD, Saccharomyces Genome Database; ORF, open reading frame.
MARRVEL, Model organism Aggregated Resources for Rare Variant ExpLoration (Wang et al. 2017).
The databases included at G2F are MGI (Blake et al. 2017), RGD (Shimoyama et al. 2015), Xenbase (Karpinka et al. 2015), ZFIN (Howe et al. 2017), FlyBase (Gramates et al. 2017), WormBase (Howe et al. 2016), SGD (Cherry et al. 2012), and PomBase (McDowall et al. 2015).
DIOPT score, number of ortholog prediction tools included at DIOPT (Hu et al. 2011) that cover both species and predict the displayed ortholog match.
In this column, “High” indicates that the ortholog pair has the best score among all pairs with both a forward and a reverse direction score and a DIOPT ≥ 2; “Moderate” indicates that the ortholog pair has the best score with the forward or the reverse search and a DIOPT ≥ 2, or has a DIOPT score ≥ 4 but is not the best score with either a forward or reverse search; and “Low” includes all other predicted ortholog pairs.
Mines (or MODs serving that function): HumanMine, MouseMine, XenMine, ZebrafishMine, FlyMine, WormBase, and SGD (Cherry et al. 2012; Smith et al. 2012; Howe et al. 2016).
Links provided for one of several repositories in the United States and overseas that have ORF clones, many of which are from the ORFeome Collaboration (2016).
To demonstrate the utility of G2F, we focus on two use cases: (1) a search initiated with a single human or common model organism gene of interest, and (2) a search initiated with a single human disease term of interest.
A gene search at G2F connects users to ortholog information and an overview of functional information for orthologs (Table 1). Specifically, starting with a search of a human, mouse, frog, fish, fly, worm, or yeast gene, users reach a summary table of orthologs and information. Information displayed includes the number of gene ontology (GO) terms assigned based on experimental evidence; the number of publications; and the number of molecular and genetic interactions reported. When available, the table also includes links to expression pattern annotations, phenotype annotations, three-dimensional structure information (Rose et al. 2017), and open reading frame (ORF) clones from the ORFeome collaboration consortium (Lamesch et al. 2004; Hu et al. 2007; ORFeome Collaboration 2016) which are available in a public repository (Zuo et al. 2007). The summary allows a user to quickly (1) evaluate conservation across major model organisms based on DIOPT score, pairwise alignment of the query protein to another species, and multiple-sequence alignment; (2) assess in what species the query gene has been well studied based on original publications, annotation, and data; and (3) identify reagents for follow-up studies. The summary table also allows a user to view detailed reports and is hyperlinked to more detailed information at original sources, such as data on specific gene pages at MODs.
A disease search at G2F first connects from disease terms to associated human genes, then uses the gene search results table format to display orthologs of the human gene and summary information (Table 1). After a search with a human disease term, users are first shown a page that helps to disambiguate terms, expanding or focusing the search, and also allows users to limit the results to disease–gene relationships curated in the Online Mendelian Inheritance in Man database and/or based on genome-wide association studies (GWAS) from the National Human Genome Research Institute–European Bioinformatics Institute GWAS Catalog (MacArthur et al. 2017). Next, users access a table of human genes that match the subset of terms, along with summary information regarding the genes and associated disease terms. On the far right-hand side of the table, users can connect to the same single gene-level report that is described above for a gene search.
Over the past two decades, GWAS have begun to reveal genetic risk factors for many common disorders (Wangler et al. 2017). As of February 2017, the GWAS Catalog (MacArthur et al. 2017) included 2385 publications, with 10,499 reported genes associated with 1682 diseases or traits. For some of the human genes, there are no publications or GO annotations. We used G2F to survey information in model organisms for this subset of genes and found many cases where one or more orthologous genes have been studied (File S1). The results of the ortholog studies appear in some cases to support the disease association, and the corresponding model systems could provide a foundation for follow-up studies (Table S1). The human gene SAMD10, for example, has been shown (using the iCOGS custom genotyping array) to be one of 23 new prostate cancer susceptibility loci (Eeles et al. 2013), but there is no information about this human gene available, aside from sequence and genome location. The results of a G2F search show that the gene is conserved in the mouse, rat, fish, fly, and worm. The mutant phenotypes of the fly ortholog suggest that the gene is involved in compound eye photoreceptor cell differentiation, EGFR signaling, positive regulation of Ras signaling, and ERK signaling, providing starting points for the development of new hypotheses regarding the function of SAMD10. Several uncharacterized human genes associated by GWAS with schizophrenia, namely IGSF9B, NT5DC2, C2orf69, and ASPHD1 (Ripke et al. 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014), are expressed at higher levels in the nervous system than in other tissues in one or more model organisms, suggesting a potential role in the nervous system in these models and supporting the idea that the models might be appropriate for follow-up studies aimed at understanding human gene function. These examples are extreme in that they represent human genes for which there are no publications describing functional information. For a large number of human genes, limited information is available. Functional annotations in model systems, as accessed through G2F, can help in the development of new hypotheses regarding the functions of these genes, as well as help researchers to choose an appropriate model organism or organisms for further study of the conserved gene.
Altogether, G2F provides a highly integrated resource that facilitates efficient use of existing gene function information by providing a big-picture view of the information landscape and building bridges between different islands of information, including MODs. This approach complements approaches designed for searches starting with long gene lists (e.g., InterMine; Smith et al. 2012) or those based on a phenotype-centered model (e.g., the Monarch Initiative; Mungall et al. 2017). The modular nature of the G2F resource makes it possible to easily update the information sources (e.g., replace a module) and add new types of information (e.g., an expanded summary of reagents or new types of experimental data).
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.043885/-/DC1.
Acknowledgments
The authors would like to thank Joseph Loscalzo, Richard Maas, and Calum MacRae of Brigham and Women’s Hospital for critical guidance at an early stage. We also thank Shinya Yamamoto of Baylor College of Medicine, Verena Chung, and members of the Perrimon laboratory for helpful feedback on content, programming, and functionality of the resource; and Cathryn King for help with the graphic design of the Gene2Function (G2F) home page. The Drosophila RNAi Screening Center is supported by National Institutes of Health (NIH) National Institute of General Medical Science grant R01 GM067761, with additional relevant funding from NIH grants R24 RR032668 and R24 OD021997. FlyBase is also supported by NIH National Human Genome Research Institute grant U41 HG000739. S.E.M. is supported in part by the Dana–Farber/Harvard Cancer Center, which is supported in part by NIH National Cancer Institute Cancer Center Support grant NIH 5 P30 CA06516. N.P. is an investigator at the Howard Hughes Medical Institute.
Footnotes
The FlyBase Consortium members at the time of writing include the following: Julie Agapite,† Kris Broll,† Madeline Crosby,† Gilberto Dos Santos,† David Emmert,† Kathleen Falls,† Susan Russo Gelbart,† L. Sian Gramates,† Beverley Matthews,† Norbert Perrimon,† Carol Sutherland,† Chris Tabone,† Pinglei Zhou,† Mark Zytkovicz,† Giulia Antonazzo,‡ Helen Attrill,‡ Nicholas Brown,‡ Silvie Fexova,‡ Phani Garapati,‡ Tamsin Jones,‡ Aoife Larkin,‡ Steven Marygold,‡ Gillian Millburn,‡ Alix Rey,‡ Vitor Trovisco,‡ Jose-Maria Urbano,‡ Brian Czoch,§ Josh Goodman,§ Gary Grumbling,§ Thomas Kaufman,§ Victor Strelets,§ James Thurmond,§ Phillip Baker,** Richard Cripps,** and Margaret Werner-Washburne**.
Communicating editor: B. J. Andrews
Literature Cited
- Blake J. A., Eppig J. T., Kadin J. A., Richardson J. E., Smith C. L., et al. , 2017. Mouse genome database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 45: D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G., et al. , 2012. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles R. A., Olama A. A., Benlloch S., Saunders E. J., Leongamornlert D. A., et al. , 2013. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 45: 385–391, 391e381–391e382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J. M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D. G., Bradford Y. M., Eagle A., Fashena D., Frazer K., et al. , 2017. The Zebrafish model organism database: new support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acids Res. 45: D758–D768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K. L., Bolt B. J., Cain S., Chan J., Chen W. J., et al. , 2016. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44: D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T. V., Zhu C., et al. , 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Comjean A., Perrimon N., Mohr S. E., 2017. The Drosophila Gene Expression Tool (DGET) for expression analyses. BMC Bioinformatics 18: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinka J. B., Fortriede J. D., Burns K. A., James-Zorn C., Ponferrada V. G., et al. , 2015. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 43: D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Milstein S., Hao T., Rosenberg J., Li N., et al. , 2004. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 14: 2064–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., et al. , 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45: D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall M. D., Harris M. A., Lock A., Rutherford K., Staines D. M., et al. , 2015. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 43: D656–D661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C. J., McMurry J. A., Kohler S., Balhoff J. P., Borromeo C., et al. , 2017. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 45: D712–D722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORFeome Collaboration , 2016. The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat. Methods 13: 191–192. [DOI] [PubMed] [Google Scholar]
- Ripke S., O’Dushlaine C., Chambert K., Moran J. L., Kahler A. K., et al. , 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P. W., Prlic A., Altunkaya A., Bi C., Bradley A. R., et al. , 2017. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 45: D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium , 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M., De Pons J., Hayman G. T., Laulederkind S. J., Liu W., et al. , 2015. The rat genome database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 43: D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. N., Aleksic J., Butano D., Carr A., Contrino S., et al. , 2012. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 28: 3163–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Al-Ouran R., Hu Y., Kim S. Y., Wan Y. W., et al. , 2017. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 100: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M. F., Hu Y., Shulman J. M., 2017. Drosophila and genome-wide association studies: a review and resource for the functional dissection of human complex traits. Dis. Model. Mech. 10: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo D., Mohr S. E., Hu Y., Taycher E., Rolfs A., et al. , 2007. PlasmID: a centralized repository for plasmid clone information and distribution. Nucleic Acids Res. 35: D680–D684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.