Abstract
Background:
To date, only a few studies have investigated associations between ERCC2, NBN, and RAD51 variants and risk of developing osteosarcoma. In this systematic review and meta-analysis, we focused on clarifying links.
Materials and Methods:
We systematically searched PubMed, Google Scholar, and ISI web of knowledge databases to identify relevant studies. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to calculate the strength of associations with fixed effect models.
Results:
No statistical evidence of association was found between ERCC2 rs13181 (G vs. T: OR= 1.224, 95% CI: 0.970-1.545, p= 0.088; GT vs. TT OR= 1.135, 95% CI: 0.830-1.552, p= 0.428; GG vs. TT: OR= 1.247, 95% CI: 0.738-2.108, p= 0.409; GG+GT vs. TT: OR= 1.174, 95% CI: 0.929-1.484, p= 0.179; GG vs. GT+ TT: OR= 1.476, 95% CI: 0.886-2.460, p= 0.135), ERCC2 rs1799793 (GA+AA vs. GG: OR= 1.279, 95% CI: 0.912-1.793, p= 0.154), NBN rs709816 (OR= 1.047, 95% CI: 0.763-1.437, p= 0.775), NBN rs1805794 (OR= 1.126, 95% CI: 0.789-1.608, p= 0.513), RAD51 rs1801320 (OR= 0.977, 95% CI: 0.675-1.416, p= 0.904), RAD51 rs1801321 (TT+GT vs. GG OR= 1.167, 95% CI: 0.848-1.604, p= 0.343), RAD51 rs12593359 (GG+GT vs. TT OR= 0.761, 95% CI: 0.759-1.470, p= 0.744) polymorphisms and osteosarcomas. The lack of the original data limited our further evaluation of the adjusted ORs concerning age and gender; however, the previous individual studies results indicated the age-and gender-specific effects of two ERCC2 rs1799793 and NBN rs1805794 variants on osteosarcoma risk.
Conclusion:
The results suggested a lack of association between the ERCC2 (rs13181 and rs1799793), NBN (rs709816 and rs1805794), and RAD51 (rs1801320, rs1801321, and rs12593359) variants with osteosarcoma risk. Further comprehensive and well-designed studies are required to assess the role for ERCC2, NBN, RAD51 variants in osteosarcoma development more adequately.
Keywords: Osteosarcoma, ERCC2, NBN, RAD51, variant, meta-analysis
Introduction
Osteosarcoma is the most common primary bone tumor in children and adolescents (Arndt et al., 2012). According to the statistics, osteosarcoma representing approximately 3.4% of all childhood cancers and 56% of malignant bone tumors in children (Geller et al., 2010). It was reported that osteosarcoma is the second most frequent cause of cancer-related death in adolescents (Liu et al., 2016; Sobhan et al., 2017). In humans, the peak incidence of osteosarcoma occurs in the second decade of life, during periods of rapid skeletal growth (Gorlick et al., 2003; Geller et al., 2010). Based on epidemiological studies, osteosarcoma predominantly occurred in the femur (42%, with 75% of tumors in the distal femur), followed by the tibia (19%, with 80% of tumors in the proximal tibia), and the humerus (10%, with 90% of tumors in the proximal humerus) (Ottaviani et al., 2009).
Osteosarcoma is a complex, multistep, and multifactorial disease (Sobhan et al., 2017). The molecular mechanism involved in the tumorigenesis of osteosarcoma is not well understood yet (Li et al., 2016). Many environmental factors (such as radiation and chemicals exposure) and genetic (family history and several putative single genes) are known to contribute to the development of osteosarcoma (Ottaviani et al., 2009; Tang et al., 2016; Sobhan et al., 2017).
To date, several studies identified different putative genetic susceptibility polymorphisms for risk of osteosarcoma development, which participates in several critical cellular functions such as repair of damaged DNA; however, the statistical power of these studies was limited by small sample sizes. Recently, a few studies have investigated the genetic link between the important DNA repair genes variants, including ERCC2, NBN, and RAD51 and increased risk of developing osteosarcoma. However, the results have been inconsistent. In addition, the data regarding the role of those genetic polymorphisms in osteosarcoma risk are sparse. It is clear that meta-analysis can be used to increase power and answer questions not posed by the individual studies. Therefore, the objective of this systematic review and meta-analysis was to detect the potential association between osteosarcoma susceptibility and ERCC2 rs13181 and rs1799793, NBN rs709816 and rs1805794, RAD51 rs1801320, rs1801321, and rs12593359 polymorphisms.
Materials and Methods
Search Strategy
We searched PubMed, Google Scholar, and ISI web of knowledge electronic databases to identify eligible studies that were published before February 2017. The used search terms were: “DNA repair gene” and “ERCC2”, “NBN, “RAD51” and “bone tumors”, “bone malignancy”, “osteosarcomas”, and “polymorphism”, “polymorphisms”, “variant”, or “mutation”. In addition, we checked references of all those retrieved articles to identify more relevant papers.
Selection Criteria
We used the following inclusion criteria to identify articles for our meta-analysis: (1) the studies assessed the association between the ERCC2, NBN, RAD51 polymorphisms, and osteosarcoma risk; (2) studies with a case-control or cohort design; and (3) studies provided sufficient published data to estimate an odds ratio (OR) with a 95% confidence interval (95% CI). In case of more than one eligible article with overlapping data by the same authors, we included the most recent or the largest one.
Data Extraction
In the current meta-analysis, two authors independently searched and identified the eligible articles based on the inclusion criteria. The following information were extracted from each study: first author, year of publication, country (ethnicity), number of cases and controls, and the genotype frequencies of the cases and controls.
Statistical Analysis
The statistical analysis for the current meta-analysis study was performed using the comprehensive meta-analysis (CMA) V2 software (Biostat, USA). In the current meta-analysis, all P values were considered two-sided, and P = 0.05 was set as the threshold value for statistical significance. To evaluate the associations between ERCC2 rs13181 polymorphism and osteosarcoma risk, the pooled odds ratio (OR) and associated 95% confidence interval (CI) were calculated under five genetic contrasts, including allelic (G vs. T), heterozygote (GT vs. TT), homozygote (GG vs. TT), dominant (GG+GT vs. TT), and recessive (GG vs. GT+TT). In addition, due to insufficient data, we used only the dominant genetic contrast to calculate ORs for ERCC2 rs1799793 (GA+AA vs. GG), NBN rs709816 (TT+TC vs. CC) and rs1805794 (CC+CG vs. GG), RAD51 rs1801320 (CC+CG vs. GG), rs1801321 (TT+GT vs. GG), and rs12593359 (GG+GT vs. TT) polymorphisms. The I2 statistics was also employed to assess the risks of heterogeneity. Therefore, in the current meta-analysis the I2 values of 25, 50, and 75% meant a low, moderate, and high heterogeneity, respectively, (Higgins et al., 2003; Khoram-Abadi et al., 2016). Moreover, a random effects model using the DerSimonian was utilized to calculate the OR and 95% CI for comparisons with moderate to high heterogeneity (P-value > 0.1 and I2 > 25%) (DerSimonian et al., 1986). If the Q test and the I2 statistics (P-value < 0.1 and I2 < 25%) both indicated no existence of heterogeneity, a fixed model with the Mantel-Haenszel method would be employed to pool data. We removed each study in turn for sensitivity analyses, and the remaining studies were reanalyzed to assess the stability of the meta-analysis results. Funnel plots and Egger’s test were used to examine publication bias (P<0.05) (Egger et al., 1997). Chi-Square test was used to determine the frequency distribution of genotypes, which were met the Hardy–Weinberg equilibrium (HWE).
Results
Characteristics of the Pooled Case-Control Ctudies
Through electronic search and considering the inclusion criteria, a total of seven relevant studies concerning ERCC2, NBN, and RAD51 gene polymorphisms and osteosarcoma were initially identified. However, three studies were excluded because they were about DNA repair genes association with osteosarcoma treatment. The final pool of eligible studies included four articles (Goričar et al., 2015; Jin et al., 2015; Ma et al., 2015; Wang et al., 2016) with 540 cases, and 1228 controls for ERCC2, NBN, and RAD51 gene polymorphisms in a sample of Slovenian population (Goricar et al. 2015) and three in Chinese population (Jin et al. 2015; Ma et al. 2015; Wang et al. 2016). All studies were case–control in design. In searching reference lists, relevant meta-analyses and reviews, we did not find additional papers. General characteristics of the included studies are summarized in Table 1.
Table 1.
Characteristics of Studies Included in the Meta-Analysis of ERCC2, NBN, and RAD51 Gene Polymorphisms and Osteosarcoma.
| First author | Country (Ethnicity) | Case/Control | Cases | Controls | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | ||||||||||
| ERCC2 rs13181 | TT | TG | GG | T | G | TT | TG | GG | T | G | |||
| Jin et al. 2015 | China (Asian) | 148/298 | 86 | 62 | 181 | 117 | |||||||
| Ma et al. 2015 | China (Asian) | 141/282 | 96 | 32 | 13 | 224 | 58 | 206 | 58 | 18 | 470 | 94 | <0.001 |
| Wang et al. 2016 | China (Asian) | 172/275 | 90 | 66 | 16 | 246 | 98 | 155 | 103 | 18 | 413 | 139 | 0.873 |
| ERCC2 rs1799793 | AA | AG | GG | AA | AG | GG | A | G | |||||
| Jin et al. 2015 | China (Asian) | 148/298 | 84 | 64 | 201 | 97 | |||||||
| Ma et al. 2015 | China (Asian) | 141/282 | 60 | 62 | 19 | 182 | 100 | 134 | 117 | 31 | 385 | 179 | 0.475 |
| NBN rs709816 | CC | CT | TT | CC | CT | TT | |||||||
| Goricar et al. 2015 | Slovenia (Caucasian) | 79/373 | 29 | 50 | 139 | 234 | |||||||
| Jin et al. 2015 | China (Asian) | 148/298 | 55 | 93 | 115 | 183 | |||||||
| NBN rs1805794 | GG | GC | CC | GG | GC | CC | |||||||
| Goricar et al. 2015 | Slovenia (Caucasian) | 79/373 | 11 | 68 | 33 | 340 | |||||||
| Jin et al. 2015 | China (Asian) | 148/298 | 52 | 96 | 127 | 171 | |||||||
| RAD51 rs1801320 | GG | GC | CC | GG | GC | CC | |||||||
| Goricar et al. 2015 | Slovenia (Caucasian) | 78/373 | 69 | 9 | 304 | 69 | |||||||
| Jin et al. 2015 | China (Asian) | 148/298 | 101 | 47 | 213 | 85 | |||||||
| RAD51 rs1801321 | GG | GT | TT | GG | GT | TT | |||||||
| Goricar et al. 2015 | Slovenia (Caucasian) | 79/373 | 25 | 54 | 133 | 240 | |||||||
| Jin et al. 2015 | China (Asian) | 148/298 | 88 | 60 | 187 | 111 | |||||||
| RAD51 rs12593359 | TT | TG | GG | TT | TG | GG | |||||||
| Goricar et al. 2015 | Slovenia (Caucasian) | 79/373 | 22 | 57 | 103 | 270 | |||||||
| Jin et al. 2015 | China (Asian) | 148/298 | 49 | 99 | 105 | 193 |
Meta-Analysis Results
Table 2 lists the main results of the meta-analysis for ERCC2, NBN, and RAD51 polymorphisms. For ERCC2 rs13181 polymorphism, no significant association was found between this polymorphism and osteosarcoma when all eligible studies were pooled into meta-analysis in any genetic contrast (G vs. T: OR= 1.224, 95% CI: 0.970-1.545, p= 0.088; GT vs. TT: OR= 1.135, 95% CI: 0.830-1.552, p= 0.428; GG vs. TT: OR= 1.247, 95% CI: 0.738-2.108, p= 0.409; GG+GT vs. TT: OR= 1.174, 95% CI: 0.929-1.484, p= 0.179; GG vs. GT+TT: OR= 1.476, 95% CI: 0.886-2.460, p= 0.135).
Table 2.
Results of Meta-Analysis for ERCC2, NBN, and RAD51 Gene Polymorphisms and Osteosarcoma
| Polymorphism | Genetic Model | Type of Model | Heterogeneity | Odds ratio | |||
|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | POR | |||
| ERCC2 rs13181 | |||||||
| G vs. T | Fixed | 0 | 0.696 | 1.224 | 0.970-1.545 | 0.088 | |
| GT vs. TT | Fixed | 0 | 0.829 | 1.135 | 0.830-1.552 | 0.428 | |
| GG vs. TT | Fixed | 0 | 0.432 | 1.247 | 0.738-2.108 | 0.409 | |
| GG+GT vs. TT | Fixed | 0 | 0.909 | 1.174 | 0.929-1.484 | 0.179 | |
| GG vs. GT+TT | Fixed | 0 | 0.974 | 1.476 | 0.886-2.460 | 0.135 | |
| ERCC2 rs1799793 | Dominant | Fixed | 70.49 | 0.066 | 1.279 | 0.912-1.793 | 0.154 |
| NBN rs709816 | Dominant | Fixed | 0 | 0.911 | 1.047 | 0.763-1.437 | 0.775 |
| NBN rs1805794 | Dominant | Fixed | 73.32 | 0.053 | 1.126 | 0.789-1.608 | 0.513 |
| RAD51 rs1801320 | Dominant | Fixed | 61.85 | 0.105 | 0.977 | 0.675-1.416 | 0.904 |
| RAD51 rs1801321 | Dominant | Fixed | 0 | 0.902 | 1.167 | 0.848-1.604 | 0.343 |
| RAD51 rs12593359 | Dominant | Fixed | 0 | 0.761 | 1.057 | 0.759-1.470 | 0.744 |
In the current meta-analysis only the dominant genetic contrast was determined for ERCC2 rs1799793, NBN rs709816 and rs1805794, RAD51 rs1801320, rs1801321, and rs12593359 polymorphisms, due to insufficient data. However, we did not find a significant association between the ERCC2 rs1799793 (GA+AA vs. GG: OR= 1.279, 95% CI: 0.912-1.793, p= 0.154), NBN rs709816 (OR= 1.047, 95% CI: 0.763-1.437, p= 0.775), NBN rs1805794 (OR= 1.126, 95% CI: 0.789-1.608, p= 0.513), RAD51 rs1801320 (OR= 0.977, 95% CI: 0.675-1.416, p= 0.904), RAD51 rs1801321 (TT+GT vs. GG: OR= 1.167, 95% CI: 0.848-1.604, p= 0.343), RAD51 rs12593359 (GG+GT vs. TT: OR= 0.761, 95% CI: 0.759-1.470, p= 0.744) polymorphisms and osteosarcomas.
Heterogeneity Test
We used Q-test and I2 statistics to test the heterogeneity among the studies. No heterogeneity was observed in either polymorphisms (A vs. G) as well as the dominant genotype contrast for overall cancer, which was included for the analysis (Overall allele, A vs. G: Q=31.08, Pheterogeneity = 0.186, I2= 19.58; Overall dominant contrast, AA+GG vs. GG: Q= 25.76, Pheterogeneity = 0.11, I2 = 25.76).
Publication Bias
Funnel plot and Egger’s linear regression were performed to assess the publication bias of the included studies. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry (Figure 2). The results of Egger’s test also showed no strong statistical evidence of publication bias (PBeggs = 1.000, PEggers= 0.417, Figure 2).
Figure 1.
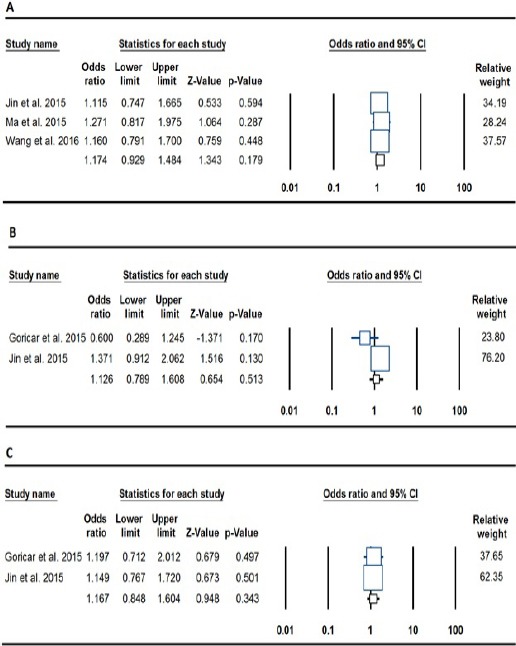
Forest Plot for the Association of the ERCC2, NBN, and RAD51 Gene Polymorphisms and Osteosarcoma. A: ERCC2 rs13181 (dominant: GG+GT vs. TT), B: NBN rs1805794 (dominant: TT+TC vs. CC) and C: RAD51 rs1801321 (GG+GT vs. TT).
Figure 2.
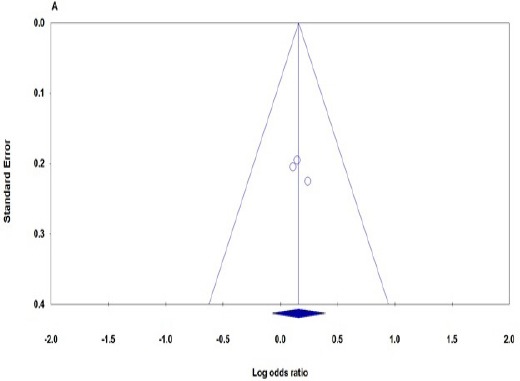
Begg’s Funnel Plots of the ERCC2 rs13181 Polymorphism and Osteosarcoma Risk for Publication Bias Test in Dominant Genetic Contrast (GG+GT vs. TT). Each point represents a separate study for the indicated association.
Discussion
ERCC2, NBN, and RAD51 genes play an important role in DNA repair pathways (Damaraju et al., 2006). Several important single nucleotide polymorphisms (SNPs) have been identified in the loci. Recently, several case control epidemiological studies have evaluated the association between RAD51, ERCC2 and NBN polymorphisms and risk of a large range of cancers (Ji et al., 2015).
The Human RAD51 gene, one of the key proteins for homologous recombination (HR), plays a crucial role in maintaining the genetic stability of the cell through HR repair of DNA double-strand breaks (DSBs) (Richardson, 2005). It is well known that RAD51 is the major component of the HR pathway in combination of BRCA1 and BRCA2 (Roy et al., 2011). However, the nature of interaction between BRCA1 and RAD51 is still unknown. It is postulated that the association is likely to be indirect and possibly mediated by BRCA2 (O’Donovan et al., 2010). RAD51 is involved in the search for homology and strand pairing stages of the process. RAD51 overcomes internal DNA bonding forces to unwind and separate DNA strands and also to compete with other DNA binding proteins (Thacker, 2005). The human RAD51 gene is found to be located on chromosome 15q15.1, span >39 kb, contains 10 exons and encodes a 339 amino acid protein (Chen et al., 2016). RAD51 gene belongs to a region which exhibits a high loss of heterozygosity (LOH) in a large range of malignancies such lung, the colorectum, and the breast (Lose et al., 2006).
The excision repair cross-complementing rodent repair deficiency, group 2 (ERCC2) or XPD (Xeroderma Pigmentosum Complementary group D) plays an important role in transcription-coupled nucleotide excision repair (NER) pathway (Vashisht et al., 2015). ERCC2 is a 5’-3’ ATP-dependent helicase as a subunit of the basal transcription factor TFIIH (Fukuda et al., 2001; Compe et al., 2012). Human XRCC1 gene maps to chromosome 19q13.32 and is composed of 23 exons. It spans approximately 54336 bp and encodes a protein of 760 amino acids with a molecular weight of 86,900 (Li et al., 2013; Forat-Yazdi et al., 2015). Several validated single nucleotide polymorphisms (SNPs) in ERCC2 are listed in the dbSNP database, which play a causative role in DNA repair-deficiency diseases, including Xeroderma Pigmentosum (XP), Trichothiodystrophy (TTD), and Cockayne Syndrome (CS) (Lehmann 2003). These diseases characterized by high ultraviolet-light hypersensitivity, a high mutation frequency, and cancer-proneness, as well as some mental and growth retardation and probably aging (Benhamou ET AL., 2005). Additionally, it is reported that different mutations in the ERCC2 (XPD) gene may lead to one of 6 different clinical disorders: XP, XP neurological disease, TTD, the XP/CS complex, XP/TTD complex or a severe form of CS known as COFS (cerebral, ocular, facial, skeletal syndrome) (Kraemer et al., 2007).
The NBN gene encoding a tumor suppressor protein belongs to the DNA DSBs repair genes and encodes a protein called nibrin (also known NBS1, p95 or NBN), which participates in several critical cellular functions, including the repair of damaged DNA (Czornak et al., 2008; Berardinelli et al., 2013). The gene is located on chromosome 8 at 8q21and consists of 16 exons, which spans a region of more than 50 kb. Its product nibrin is a 95-kDa protein, which forms a multiprotein complex with hMRE11 and hRAD50 (MRN) (OMIM; Uzunoglu et al., 2016). Mutations in the NBN gene lead to an autosomal recessive disorder named Nijemen breakage syndrome (NBS), a radiation-sensitivity disorder with multiple deficiencies, which may result in susceptibility to cancers such as breast cancer (Schröder-Heurich et al., 2014; Uzunoglu et al., 2016).
Considering the functional significance of ERCC2, NBN, and RAD51 genes in carcinogenesis, it is speculated that ERCC2, RAD51, and NBN polymorphisms may be a potential susceptibility factor for osteosarcoma. However, a few studies evaluated the association between ERCC2, NBN, and RAD51 polymorphisms and osteosarcoma risk (Goričar et al., 2015; Jin et al., 2015; Ma et al., 2015; Wang et al., 2016). To the best of our knowledge, this meta-analysis represents the first one investigating the association between ERCC2 (rs13181 and rs1799793), NBN (rs709816 and rs1805794), RAD51 (rs1801320, rs1801321, and rs12593359) polymorphisms and osteosarcoma risk. Notwithstanding, our results suggested that ERCC2 (rs13181 and rs1799793), NBN (rs709816 and rs1805794), RAD51 (rs1801320, rs1801321, and rs12593359) polymorphisms might not be associated with osteosarcoma risk. The finding was consistent with the individual study by Jin et al., reported the association of ERCC2 rs1799793 and NBN rs1805794 polymorphisms with risk of osteosarcoma (2015). Additionally, they held that these two polymorphisms are associated with an increased risk of osteosarcoma in females (Jin et al., 2015). However, the other included study, with 79 osteosarcoma cases and 373 controls from Slovenia, by Goricar et al. found no association between NBN rs709816 and rs1805794, RAD51 rs1801320, rs1801321, and rs12593359 polymorphisms and risk of osteosarcoma (2015). However, they demonstrated that the NBN rs1805794 polymorphism is significantly associated with risk of osteosarcoma in younger patients (<30 years) (Goričar et al., 2015; Jin et al., 2015). Therefore, these two individual studies indicated the age-and gender-specific effects of ERCC2 rs1799793 polymorphism and in particular NBN rs1805794 polymorphism on risk of the osteosarcoma development.
Although we conducted the first meta-analysis investigating the association between ERCC2, NBN and RAD51 genes polymorphism and risk of osteosarcoma, there were some limitations. First, the present meta-analysis involved only four studies, the meta-analysis was relatively small in comparison with other meta-analyses; therefore, we cannot generalize our findings. To validate the present meta-analysis results, more comprehensive studies with larger sample size must be included in future investigations. Second, we only included published articles and also searched literature written in English; hence, the existence of high risk of potential bias must be considered. Finally, the current meta-analysis was based on unadjusted ORs estimates, the eligible studies did not describe the age and gender distribution between cases and controls. Therefore, further evaluation of adjustments of risk factors such as age, gender and the potential interactions among gene–gene and gene–environment was not possible for us.
In summary, the current meta-analysis findings indicated no significant association between ERCC2 (rs13181 and rs1799793), NBN (rs709816 and rs1805794), RAD51 (rs1801320, rs1801321, and rs12593359) polymorphisms and osteosarcoma risk. Due to small sample size, this conclusion should be interpreted with caution. Further well-designed and large-scale studies are recommended considering gene–gene and gene–environment interactions for investigation of such associations.
References
- Arndt C, Rose P, Folpe A, et al. Common musculoskeletal tumors of childhood and adolescence. Mayo Clinic Proceedings. 2012;87:475–87. doi: 10.1016/j.mayocp.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou S, Sarasin A. ERCC2 /XPD gene polymorphisms and lung cancer:a huge review. Am J Epidemiol. 2005;161:1–14. doi: 10.1093/aje/kwi018. [DOI] [PubMed] [Google Scholar]
- Berardinelli F, Masi A, Antoccia A. NBN gene polymorphisms and cancer susceptibility:A systemic review. Current Genomics. 2013;14:425–40. doi: 10.2174/13892029113146660012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang H, Pu F. Association between a functional variant in RAD51 gene’s 3’untranslated region and its mRNA expression in lymphoblastoid cell lines. Springerplus. 2016;5:1688. doi: 10.1186/s40064-016-3339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E, Egly JM. TFIIH:when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–54. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- Czornak K, Chughtai S, Chrzanowska KH. Mystery of DNA repair:the role of the MRN complex and ATM kinase in DNA damage repair. J Appl Genet. 2008;49:383–96. doi: 10.1007/BF03195638. [DOI] [PubMed] [Google Scholar]
- Damaraju S, Murray D, Dufour J, et al. Association of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancer. Clin Cancer Res. 2006;12:2545–54. doi: 10.1158/1078-0432.CCR-05-2703. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177, 88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, et al. Bias in metaanalysis detected by a simple, graphical test. BMJ. 1997;315:629, 34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forat-Yazdi M, Gholi-Nataj M, Neamatzadeh H, et al. Association of XRCC1 Arg399Gln polymorphism with colorectal cancer risk:A HuGE meta-analysis of 35 studies. Asian Pac J Cancer Prev. 2015;16:3285–91. doi: 10.7314/apjcp.2015.16.8.3285. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Yamauchi J, Wu SY, et al. Reconstitution of recombinant TFIIH that can mediate activator-dependent transcription. Genes Cells. 2001;6:707–19. doi: 10.1046/j.1365-2443.2001.00456.x. [DOI] [PubMed] [Google Scholar]
- Geller DS, Gorlick R. Osteosarcoma:a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–18. [PubMed] [Google Scholar]
- Gorlick RG, Toretsky JA, Marina N, et al. Osteosarcoma. In: Kufe DW, Pollock RE, Weichselbaum RR, et al., editors. Holland-Frei cancer medicine. 6th edition. Vol. 2003. Hamilton (ON): BC Decker; 2003. Available from: https://www.ncbi.nlm.nih.gov/books/NBK13901/ [Google Scholar]
- Gray K, Kumar S, Figg N, et al. Effects of DNA damage in smooth muscle cells in atherosclerosis. Circ Res. 2015;116:816–26. doi: 10.1161/CIRCRESAHA.116.304921. [DOI] [PubMed] [Google Scholar]
- Goričar K, Kovač V, Jazbec J, et al. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer Epidemiol. 2015;39:182–8. doi: 10.1016/j.canep.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Ji WP, He NB. Investigation on the DNA repaired gene polymorphisms and response to chemotherapy and overall survival of osteosarcoma. Int J Clin Exp Pathol. 2015;8:894, 9. [PMC free article] [PubMed] [Google Scholar]
- Jin G, Wang M, Chen W, et al. Single nucleotide polymorphisms of nucleotide excision repair and homologous recombination repair pathways and their role in the risk of osteosarcoma. Pak J Med Sci. 2015;31:269–73. doi: 10.12669/pjms.312.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoram-Abadi KM, Forat-Yazdi M, Kheirandish S, et al. DNMT3B -149 C>T and -579 G>T polymorphisms and risk of gastric and colorectal cancer:a meta-analysis. Asian Pac J Cancer Prev. 2016;17:3015–20. [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome:a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–11. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Li L, Li Y, Zhao J, et al. CX-5461 induces autophagy and inhibits tumor growth via mammalian target of rapamycin-related signaling pathways in osteosarcoma. Onco Targets Ther. 2016;9:5985–97. doi: 10.2147/OTT.S104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Peng Q, Chen Y, et al. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk:a meta-analysis. PLoS One. 2013;8:e73448. doi: 10.1371/journal.pone.0073448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MH, Cui YH, Guo QN, et al. Elevated ASCL2 expression is associated with metastasis of osteosarcoma and predicts poor prognosis of the patients. Am J Cancer Res. 2016;6:1431–40. [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Y, Sun TS, et al. Role of ERCC2 and ERCC3 gene polymorphisms in the development of osteosarcoma. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15017302. gmr.15017302. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan PJ, Livingston DM. BRCA1 and BRCA2:breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 2010;31:961–7. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2:different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder-Heurich B, Bogdanova N, Wieland B, et al. Functional deficiency of NBN, the Nijmegen breakage syndrome protein, in a p.R215W mutant breast cancer cell line. BMC Cancer. 2014;14:434. doi: 10.1186/1471-2407-14-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhan MR, Forat-Yazdi M, Mazaheri M, et al. Association between the DNA Repair Gene XRCC3 rs↧39 polymorphism and risk of osteosarcoma:A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:549–55. doi: 10.22034/APJCP.2017.18.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhan MR, Mostafavi S, Mazaheri M, et al. Genetics of pediatric bone tumors:a systematic review. IJPHO. 2017;7:117–29. [Google Scholar]
- Tang Y, Wang J, Xie K, et al. Association of interleukin 16 gene polymorphisms and plasma IL16 level with osteosarcoma risk. Sci Rep. 2016;6:34607. doi: 10.1038/srep34607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunoglu H, Korak T, Ergul E, et al. Association of the nibrin gene (NBN) variants with breast cancer. Biomed Rep. 2016;4:369, 73. doi: 10.3892/br.2016.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Li S, Boyer T, et al. Lessons learned from BRCA1 and BRCA2. Oncogene. 2000;19:6159–75. doi: 10.1038/sj.onc.1203968. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Yu CC, Sharma T, et al. The association of the xeroderma pigmentosum group D DNA helicase (XPD) with transcription factor IIH is regulated by the cytosolic iron-sulfur cluster assembly pathway. J Biol Chem. 2015;290:14218–25. doi: 10.1074/jbc.M115.650762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu N. Association between XRCC1 and ERCC2 gene polymorphisms and development of osteosarcoma. Int J Clin Exp Pathol. 2016;9:223–9. [Google Scholar]


