Abstract
Objective:
To identify natural bioactive molecules with potential to inhibit DNA methyltransferase 1 (DNMT1) and cause reactivation of genes silenced due to promoter hypermethylation.
Methods and Results:
-(-) Menthol and epigallocatechin-3-gallate (EGCG) (reference molecule) were investigated using an in vitro methylation assay, which indicated potential of -(-) menthol as an epigenetic modulator with the ability to directly inhibit M.SssI (an analogue of DNMT1) activity at 100µM. Methylation specific PCR and bisulphite sequencing revealed complete hypomethylation of 15 CpG sites in the Fanconi anemia, complementation group F (FANCF) gene between +280 and + 432 nucleotides relative to the transcription start site, which resulted in significant (P<0.001) up-regulation of FANCF gene expression by 2.1 and 2.5 fold respectively after treatment with menthol (80µM) and EGCG (80µM) for 4 days in the SiHa cell line as analyzed by qRT PCR.
Conclusion:
The present work highlighted the potential of -(-) menthol, a naturally occurring cyclic monoterpene, as an epigenetic modulator causing promoter hypomethylation induced reactivation of the FANCF gene mediated by possible inhibition of DNMT1 activity in the SiHa cell line.
Keywords: (-) Menthol, EGCG, promoter hypomethylation, FANCF, cervical cancer, re expression
Introduction
Epigenetic modifications have been implicated in induction and progression of cancer (Teng et al., 2006). DNA methylation being the most studied and understood phenomenon among epigenetic modifications is a significant therapeutic target due to its reversible format (Robert et al. 2003; Siedlecki et al., 2006). In cancer, there occurs global DNA hypomethylation and regional promoter hypermethylation in tumor suppressor genes (Soengas et al., 2001). Promoter hypermethylation associated loss of gene expression has been reported in cervical carcinoma (Wentzensen et al., 2009). Fanconi anemia (FA) is an autosomal recessive chromosomal instability syndrome characterized by hypersensitivity to DNA cross-linking agents and predisposes to cancer (Siddique et al., 2001). Altogether eight FA proteins are required for the formation of FA core complex with FANCF functioning as a linker within this core complex required for monoubiquitylation of FANCD2 and FANCI, the downstream effectors in the maintenance of genome integrity (Tumini et al., 2011).
Ganzinelli et al (2011) have associated low expression of FANCF in III stage patients of ovarian carcinomas with promoter hypermethylation. Development of ovarian tumors in mice has also been associated with loss of FANCF gene function (Bakker et al., 2012). FANCF promoter hypermethylation is frequently reported in cervical cancer, squamous cell carcinomas of lung and oral cavity (Narayan et al., 2003; Marsit et al., 2004; Narayan et al., 2004) and has been linked to cisplatin resistance in ovarian cancer due to disruption of FA BRCA pathway (Taniguchi et al., 2003).
-(-)Menthol is a naturally occurring cyclic monoterpene (Faridi et al., 2011) which can inhibit the growth of human cancer cells, including neuroendocrine tumor cells (Mergler et al.,2007), human melanoma cells (Yamamura et al., 2008), human promyelocytic leukemia HL-60 cells (Lu et al., 2006), human colon cancer cells (Bernhardt et al.,2008), human liver tumor cells (MacDougall et al., 2003), and also acts on human glioblastoma cells (Wondergem et al., 2008). Role of menthol in regulating DNA methylation is still unexplored despite multiple activities shown by this compound. EGCG has been shown to cause promoter hypomethylation of p16, RARß2, MGMT, hMLH1, WIF-1, GSTP1 and RECK genes across different cancer types and has been reviewed extensively (Singh et al., 2013). The present study determines the potential of menthol to cause reversal of hypermethylated FANCF gene promoter, its effect on FANCF re expression in SiHa cell line and the possible mechanism involved. EGCG was used as a reference molecule for comparative analysis due to its well established potential as a DNMT1 inhibitor and epigenetic modulator. 5-aza-2’-deoxycytidine was used as a positive control.
Materials and Methods
Compounds
5-aza-2’-deoxycytidine (HPLC >97%) and epigallocatechin-3-gallate (EGCG) (HPLC >97%) were obtained from Sigma Aldrich, USA. – (-) menthol (GLC >99.7%) was procured from MP Bio, USA.
M.SssI activity inhibition assay
Menthol and EGCG (used as a reference molecule) were investigated for their potential to inhibit M.SssI (an analogue of DNMT1) activity at 50 and 100 µM, as described previously (Parashar et al., 2012).
Cell lines and treatment
HeLa, SiHa and Caski cell lines were procured and maintained as reported previously (Parashar et al., 2012). 1x106 SiHa cells were treated with 5-aza-2’-deoxycytidine (20µM) (positive control), EGCG (reference molecule) (80µM) and menthol (80µM) for 4 days. Medium and respective compound were replaced after every 48 h. Untreated cells were taken as control.
Cytotoxicity analysis using MTT assay
Cytotoxic concentrations (IC50) of EGCG and menthol were evaluated in SiHa cervical cancer cell line (Parashar and Capalash, 2016).
DNA methylation analysis
DNA methylation analysis of FANCF gene was carried out as described in (Parashar et al., 2012). Briefly, DNA was isolated from untreated, 5-aza-2’-deoxycytidine (20µM), EGCG (80µM) and menthol (80µM) treated SiHa cells and subjected to bisulphite modification and methylation specific PCR (MSP). MSP was also carried out to determine FANCF promoter methylation status in untreated HeLa and Caski cells. Methylation profile of CpG sites was determined and analyzed using bisulphite sequencing and BiQ analyzer software respectively (Bock et al., 2005).
Expression analysis
Expression analysis of FANCF and DNMT1 gene was performed using qRT PCR as described previously (Parashar and Capalash, 2016) with forward and reverse gene specific primers for FANCF, β actin and DNMT1 (Forward-5’-ACCGCTTCTACTTCCTCGAGGCCTA-3’; Reverse-5’-GTTGCAGTCCTCTGTGAACACTGTGG-3’) (Parashar et al., 2012).
Statistical Analysis
Data were assessed by Student’s t test. Each value represents the mean ± SE of three experiments. The results were considered as statistically significant at P<0.05.
Results
Interaction between compounds and M.SssI
Epigallocatechin-3-gallate (EGCG) and menthol were evaluated for their potential to inhibit M.SssI activity (DNMT1 analogue). Active BstU1 (methylation sensitive) leads to restriction digestion of 360 bp template into 319 bp band suggesting M.SssI inhibition. EGCG (Fig 1A) was found to inhibit M.SssI activity at 50 and 100 µM concentration while menthol (Fig 1B), was active at 100µM. Inhibition of M.SssI activity was reflected with the presence of 319 bp band (-M.SssI, positive control) after digestion with methylation sensitive Bst U1 (CG/CG) and 360 bp band showed no effect on M.SssI (+M.SssI, negative control) (Figure 1C).
Figure 1.
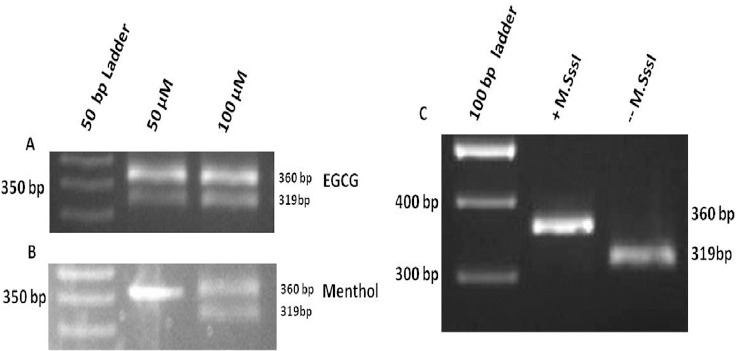
In Vitro Methylation assay with M.SssI in the Presence of EGCG (A) and menthol (B). Inhibition of M.SssI activity is reflected with the presence of 319 bp band (-M.SssI, positive control) after digestion with methylation sensitive Bst U1 and 360 bp band shows no effect on M.SssI (+M.SssI, negative control) (C).
Cytotoxicity Assay
IC50 value was found to be 140 and 176 µM for EGCG and menthol respectively (Figure 2). SiHa cell line was treated with EGCG and menthol at 80µM respectively, below their IC50 values.
Figure 2.
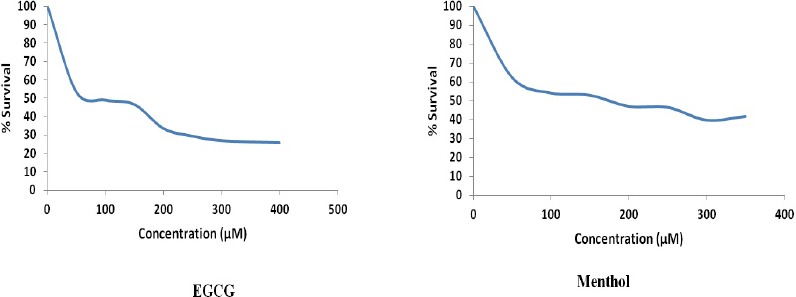
MTT assay Showing IC50 Values for EGCG (140µM) and Menthol (176µM) in SiHa Cell Line
Reversal of DNA hypermethylation
Methylation Specific PCR
FANCF promoter is methylated in SiHa cell line only Figure 3A (Parashar et al., 2012). Hence, SiHa cell line was used to study the effect of EGCG and menthol to cause promoter hypomethylation. Methylation specific PCR showed complete reversal of promoter hypermethylation in case of EGCG treated sample with disappearance of methylation specific band whereas treatment with menthol resulted in increase of unmethylated template as evident with the increased intensity of unmethylated band Figure 3B.
Figure 3.
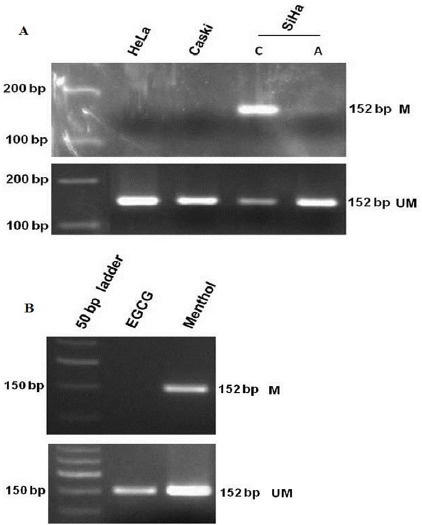
(A) Promoter Methylation Analysis of FANCF Gene in HeLa, SiHa and Caski Cell Lines by MSP. (B) Reversal of Promoter Hypermethylation after Treatment with EGCG (80µM) and Menthol (80µM) for 4 Days in SiHa Cell Line. M-Methylation Specific Band; U, Unmethylation Specific Band; C, Untreated Control and A, 5-Aza-2’-Deoxycytidine Treated Cells.
Bisulphite Sequencing
Reversal of promoter hypermethylation of FANCF gene after treatment with EGCG and menthol was comparatively evaluated with untreated cells using bisulphite sequencing. 15 CpG sites were screened between +280 to +432 region spanned by the amplicon obtained after MSP. The obtained sequences were analyzed using BiQ Analyzer (Bock et al., 2005). Untreated cells had a methylated signature with all the 15 CpG sites methylated whereas treatment with EGCG and menthol led to hypomethylation of all 15 CpG sites which explained the complete loss of methylation band after treatment with EGCG and increased intensity of unmethylated band after treatment with menthol, together confirmed reversal of CpG methylation by EGCG and menthol (Figure 3).
Quantification of FANCF and DNMT1 expression
The transcription of FANCF gene was found to be significantly upregulated by 9 folds in 5-aza-2’-deoxycytidine (P<0.05), 2.1 folds in menthol (P<0.001) and 2.5 folds in EGCG (P<0.001) treated samples by qRT PCR (Figure 5A). 5-aza-2’-deoxycytidine, EGCG and menthol did not have any significant effect on the expression of DNMT1 in treated SiHa cells as shown by qRT PCR (Figure 5B).
Figure 4.
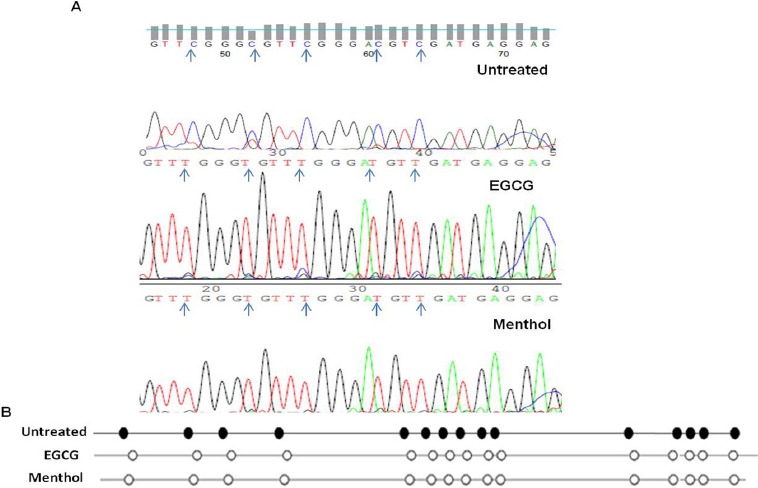
(A) Reversal of Promoter Hypermethylation in EGCG and Menthol Treated Cells as Compared to Untreated Cells of SiHa Cell Line. Blue arrow Represent “C” in a CpG Dinucleotide (B) Bisulphite Sequencing Analysis of 15 CpG Sites of FANCF Gene Promoter Spanning +280 to +432 Region in Untreated, EGCG and Menthol Treated SiHa Cell Line. Black Dot, Methylated CpG Site; White Dot, Unmethylated CpG Site.
Figure 5.
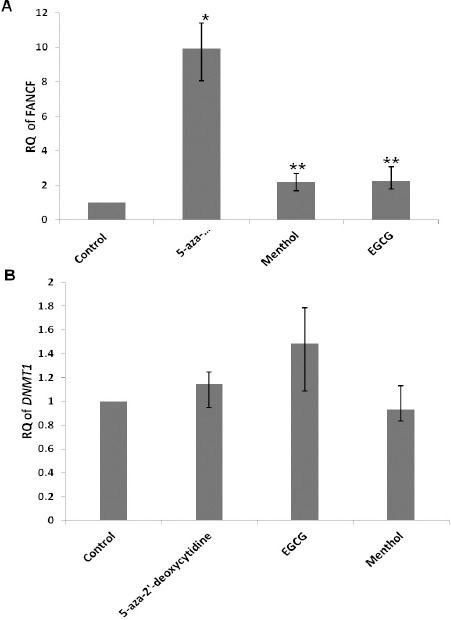
Relative Quantification (RQ) Of (A) FANCF and (B) DNMT1 Expression in SiHa Cells after Treatment with Natural Compounds for 4 Days. EGCG (80µm), Menthol (80µm) and 5-Aza-2’-Deoxycytidine (20µm) Significantly Upregulated FANCF Expression. β Actin was Used as the Internal Control. Bars Represent ± S.E With N=3. *P<0.05; **P<0.01.
Discussion
Dependence of cancer cells on normal DNMT1 activity so as to maintain their abnormal methylation profile makes it an attractive therapeutic target, due to reversible format of epigenetic processes. The limitations and ’endangered universal status’ of 5-aza-2’-deoxycytidine has changed the direction towards the search of potent, non-toxic and mechanism independent natural compounds having the feasibility to inhibit DNMT1. Promoters of the eight genes (FANCA, B, C, E, F, G, L and M) that form the Fanconi anemia core complex have been characterized and the region around transcription start site was found to be regulating its expression (Meier et al., 2011). However, CpG methylation on promoter region located upstream transcription start site may not exclusively regulate the gene expression (Parashar and Capalash, 2012); but previous reports have shown that +280 to +432 region is involved in promoter methylation dependent regulation of FANCF (Narayan et al., 2003; Narayan et al., 2004). Hence, this region of FANCF promoter was selected to study the reversal of promoter hypermethylation.
Promoter hypermethylation and down regulation of FANCF expression was observed only in SiHa cells and not in HeLa and Caski cells. Treatment of the SiHa cell line with 5’-aza-2’-deoxycytidine, a known hypomethylating agent (Brueckner et al., 2005) showed reversal of hypermethylation indicated by the amplification of unmethylation specific band, further supported its promoter hypermethylation. Hence reversal of promoter hypermethylation and reactivation of FANCF with natural compounds was studied in SiHa cells only. Previous studies have also reported methylation of FANCF promoter in SiHa cells and not in HeLa cell line (Narayan et al., 2003; Parashar et al., 2012; Li and Zhang, 2013).
EGCG and menthol reverted promoter hypermethylation of FANCF and induced its expression in SiHa cell line. Menthol and EGCG were found to hypomethylate all the 15 CpG sites under consideration. Maintenance of methyl group on cytosine in hemi-methylated DNA is primarily done by DNA methyltransferase 1(DNMT1). Loss of methylation from cytosine in CpG site can be related to loss of DNMT1 activity after treatment with natural compounds.
The potential of EGCG and menthol to inhibit DNMT1 directly by binding to its catalytic domain was assessed by M.SssI based in vitro assay. M.SssI is a universal CpG methyltransferase which methylates the cytosine residue within a CpG dinucleotide and does not require hemimethylated DNA template after replication for CpG methylation (Brueckner et al., 2005). The catalytic domain of DNA methyltransferases is evolutionary conserved across prokaryotes and eukaryotes (Santi et al., 1983).
EGCG showed inhibition of M.SssI indicating its potential activity against DNMT1. EGCG has also been shown to interact with DNMT1 at Cys1226 residue through in silico approach (Yoo et al., 2011). Anti-DNMT1 activity of EGCG (50 µM) has also been reported using nuclear extract from KYSE 510 cells based on DNA methyltransferase assay (Fang et al., 2003). No change in DNMT1 expression was observed after treatment with EGCG at 80 µM in SiHa cells. A similar observation has been reported with EGCG (5-50 µM) in KYSE 510 esophageal cells (Fang et al., 2003). However, downregulation of DNMT1 expression by higher concentration (152µM) of EGCG has been shown in Jurkat cell line (Achour et al., 2013).
Menthol leads to apoptosis in cancer cells mediated by Ca2+ influx (Mergler et al., 2007) but its role in hypomethylation is not known. In the present study it was observed that menthol did not affect DNMT1 expression but caused M.SssI inhibition at 100µM suggesting its potential to inhibit DNMT1 activity which may explain the loss of promoter hypermethylation and increased expression of FANCF in SiHa cells treated with menthol. Menthol being a stable compound with high solubility and bioavailability (Yong et al., 2004) can be a promising compound for future studies.
The present work highlights the potential of menthol in causing promoter hypomethylation induced reactivation of FANCF gene mediated by possible inhibition of DNMT1 activity in SiHa cell line.
Conflict of interest
The author(s) declare that they have no competing interests.
Funding Statement
This study was supported by Council of Scientific and Industrial Research (CSIR), New Delhi, India (No. 37(1363)/09/EMR-II) and CSIR-SRF to Gaurav Parashar (CSIR Award No.: 9/135/(0659)/2012/EMR-I).
Acknowledgements
Gaurav Parashar and Neena Capalash duly acknowledge the support and fellowship provided by Council of Scientific and Industrial Research (CSIR), India. Nidarshana Chaturvedi Parashar kindly acknowledges the Research Associateship provided by Department of Biotechnology, India.
References
- Achour M, Mousli M, Alhosin M, et al. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem Biophys Res Commun. 2013;430:208–12. doi: 10.1016/j.bbrc.2012.11.087. [DOI] [PubMed] [Google Scholar]
- Bakker ST, van de Vrugt HJ, Visser JA, et al. Fancf-deficient mice are prone to develop ovarian tumours. J Pathol. 2012;226:28–39. doi: 10.1002/path.2992. [DOI] [PubMed] [Google Scholar]
- Bernhardt G, Biersack B, Bollwein S, Schobert R, Zoldakova M. Terpene conjugates of diaminedichloridoplatinum(II) complexes:antiproliferative effects in HL-60 leukemia,518A2 melanoma, and HT-29 colon cancer cells. Chem Biodivers. 2008;5:1645, 59. doi: 10.1002/cbdv.200890152. [DOI] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, et al. BiQ Analyzer:visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Boy RG, Siedlecki P, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–11. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563, 70. [PubMed] [Google Scholar]
- Faridi U, Sisodia BS, Shukla AK, et al. Proteomics indicates modulation of tubulin polymerization by L-menthol inhibiting human epithelial colorectal adenocarcinoma cell proliferation. Proteomics. 2011;11:2115, 9. doi: 10.1002/pmic.201000691. [DOI] [PubMed] [Google Scholar]
- Ganzinelli M, Mariani P, Cattaneo D, et al. Expression of DNA repair genes in ovarian cancer samples:Biological and clinical considerations. Eur J Cancer. 2011;47:1086–94. doi: 10.1016/j.ejca.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang C. Demethylation of FANCF gene may be a potential treatment through inhibiting the proliferation of cervical cancer. Chinese-German J Clin Oncol. 2013;12:339–42. [Google Scholar]
- Lu HF, Hsueh SC, Yu FS, et al. The role of Ca 2+in (–)-menthol-induced human promyelocytic leukemia HL-60 cell death. In Vivo. 2006;20:69, 75. [PubMed] [Google Scholar]
- MacDougall JM, Fandrick K, Zhang X, Serafin SV, Cashman JR. Inhibition of human liver microsomal (S)-nicotine oxidation by (–)-menthol and analogues. Chem Res Toxicol. 2003;16:988, 93. doi: 10.1021/tx0340551. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Liu M, Nelson HH, et al. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers:implications for treatment and survival. Oncogene. 2004;23:1000–4. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- Meier D, Schindler D. Fanconi anemia core complex gene promoters harbor conserved transcription regulatory elements. PLoS One. 2011;6:e22911. doi: 10.1371/journal.pone.0022911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergler S, Strowski MZ, Kaiser S, et al. Transient receptor potential channel TRPM8 agonists stimulate calcium influx and neurotensin secretion in neuroendocrine tumor cells. Neuroendocrinology. 2007;85:81, 92. doi: 10.1159/000101693. [DOI] [PubMed] [Google Scholar]
- Narayan G, Arias-Pulido H, Koul S, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri:its relationship to clinical outcome. Mol Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan G, Arias-Pulido H, Nandula SV, et al. Promoter hypermethylation of FANCF:disruption of Fanconi anemia-BRCA pathway in cervical cancer. Cancer Res. 2004;64:2994–7. doi: 10.1158/0008-5472.can-04-0245. [DOI] [PubMed] [Google Scholar]
- Parashar G, Parashar NC, Capalash N. Curcumin causes promoter hypomethylation and increased expression of FANCF gene in SiHa cell line. Mol Cell Biochem. 2012;365:29, 35. doi: 10.1007/s11010-012-1240-z. [DOI] [PubMed] [Google Scholar]
- Parashar G, Capalash N. Expression of the TIMP2 gene is not regulated by promoter hypermethylation in the Caski cell line. Oncol Lett. 2012;3:1079–82. doi: 10.3892/ol.2012.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar G, Capalash N. Promoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clin Exp Med. 2016;16:471–8. doi: 10.1007/s10238-015-0366-1. [DOI] [PubMed] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61, 5. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9, 10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Siddique MA, Nakanishi K, Taniguchi T, Grompe M, D’Andrea AD. Function of the Fanconi anemia pathway in Fanconi anemia complementation group F and D1 cells. Exp Hematol. 2001;29:1448–55. doi: 10.1016/s0301-472x(01)00754-8. [DOI] [PubMed] [Google Scholar]
- Siedlecki P, Garcia BR, Musch T, et al. Discovery of two novel, small-molecule inhibitors of DNA methylation. J Med Chem. 2006;49:678, 83. doi: 10.1021/jm050844z. [DOI] [PubMed] [Google Scholar]
- Singh V, Sharma P, Capalash N. DNA Methyltransferase-1 inhibitors as epigenetic therapy for cancer. Curr Cancer Drug Targets. 2013;21:379–99. doi: 10.2174/15680096113139990077. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207, 11. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nature Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- Teng IW, Hou PC, Lee KD, et al. Targeted methylation of two tumor suppressor genes is sufficient to transform mesenchymal stem cells into cancer stem/initiating cells. Cancer Res. 2006;71:4653, 63. doi: 10.1158/0008-5472.CAN-10-3418. [DOI] [PubMed] [Google Scholar]
- Tumini E, Plevani P, Muzi-Falconi M, Marini F. Physical and functional crosstalk between Fanconi anemia core components and the GINS replication complex. DNA Repair. 2011;10:149, 58. doi: 10.1016/j.dnarep.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection:Appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293, 9. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondergem R, Ecay TW, Mahieu F, Owsianik G, Nilius B. HGF/SF and menthol increase human glioblastoma cell calcium and migration. Biochem Biophys Res Commun. 2008;372:210, 5. doi: 10.1016/j.bbrc.2008.05.032. [DOI] [PubMed] [Google Scholar]
- Yamamura H, Ugawa S, Ueda T, Morita A, Shimada S. TRPM8 activation suppresses cellular viability in human melanoma. Am J Physiol Cell Physiol. 2008;295:296, 301. doi: 10.1152/ajpcell.00499.2007. [DOI] [PubMed] [Google Scholar]
- Yong CS, Yang CH, Rhee JD, et al. Enhanced rectal bioavailability of ibuprofen in rats by poloxamer 188 and menthol. Int J Pharm. 2004;269:169–76. doi: 10.1016/j.ijpharm.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Yoo J, Medina-Franco JL. Homology modeling, docking and structure-based pharmacophore of inhibitors of DNA methyltransferase. J Comput Aided Mol Des. 2011;25:555–67. doi: 10.1007/s10822-011-9441-1. [DOI] [PubMed] [Google Scholar]


