Abstract
Growth differentiation factor 15 (GDF15) is a strong predictor of cardiovascular events and mortality in individuals with or without cardiovascular diseases. Single nucleotide polymorphisms (SNPs) in microRNA (miRNA) target sites, also known as miRSNPs, are known to enhance or weaken miRNA-mRNA interactions and have been linked to diseases such as cardiovascular disease and cancer. In this study, we aimed to elucidate the functional significance of the miRSNP rs1054564 in regulating GDF15 levels. Two rs1054564-containing binding sites for hsa-miR-873-5p and hsa-miR-1233-3p were identified in the 3′ untranslated region (UTR) of the GDF15 transcript using bioinformatics tools. Their activities were further characterized by in vitro reporter assays. Bioinformatics prediction suggested that miRNA binding sites harboring the rs1054564-G allele had lower free energies than those with the C allele and therefore were better targets with higher affinities for both hsa-miR-873-5p and hsa-miR-1233-3p. Reporter assays showed that luciferase activity was significantly decreased by rs1054564-G-containing 3′ UTRs for both miRNAs (P < 0.05) and was restored by miRNA inhibitors. Comparing the fold suppression of the two miRNAs, only that of hsa-miR-1233-3p showed significant changes between the rs1054564-G- and C-containing 3′ UTRs (P = 0.034). In addition, western blots showed that transfection of both miRNA mimics significantly decreased endogenous GDF15 expression in a melanoma cell line (P < 0.05). Taken together, our findings demonstrate that GDF15 is a target of hsa-miR-873-5p and hsa-miR-1233-3p and that the rs1054564-C allele partially abolishes hsa-miR-1233-3p-mediated translational suppression of GDF15. These results suggest that rs1054564 confers allele-specific translational repression of GDF15 via hsa-miR-1233-3p. Our work thus provides biological insight into the previously reported clinical association between rs1054564 and plasma GDF15 levels.
Introduction
Growth differentiation factor 15 (GDF15) is a member of the transforming growth factor-β cytokine superfamily and its expression is low in all organs under normal conditions but increases in response to stress signals in adults [1]. GDF15 is secreted by cells in response to ischemia, proinflammatory cytokine stimulation, and oxidative or mechanical stress [1], and it diffuses rapidly via circulation [2]. Circulating GDF15 levels have been used to predict disease progression in cancer, cardiovascular disease, chronic renal and heart failure, and pulmonary embolism [3]. GDF15 is also a strong predictor of cardiovascular, non-cardiovascular, and all-cause mortality in community-dwelling and disease populations [4]. Although GDF15 appears to have anti-inflammatory and antiapoptotic effects in the heart [5], the co-localization of GDF15 with apoptotic markers in active macrophages suggests it may have proinflammatory effects. Thus, it remains unknown whether GDF15 is a simple biomarker or whether it is an active protective or detrimental mediator of cardiovascular events. Previous studies have shown associations between GDF15 levels and genetic polymorphisms, clinical parameters, and levels of circulating metabolic and inflammatory markers, albeit with controversial results [6–11].
MicroRNAs (miRNAs) are a class of single-stranded, endogenous, non-coding RNAs of approximately 22 nt that play vital regulatory roles in animals and plants by targeting mRNAs for degradation or translational repression [12]. It is estimated that an average miRNA has approximately 100–200 target sites, and a large fraction (~30%) of protein-coding genes appear to be regulated by miRNAs. Recent studies have shown crucial correlations between single nucleotide polymorphisms (SNPs) in miRNA-related pathways and many pathological conditions [13–16]. SNPs in microRNA (miRNA) target sites, also known as miRSNPs, in the 3′ untranslated regions (UTRs) of target genes, in particular, represent a specific mode of control of genetic information amplification, whose dysregulation may lead to substantial differences in posttranscriptional gene expression. By definition, miRSNPs in the seed sequence (i.e., the region of base-pairing between nucleotides 2–8 of the miRNA and the complementary sequence in the target mRNA) can create, destroy, or modify miRNA–mRNA binding [17–19], and as a result, these function as gain- or loss-of-function mutations. Whereas gain-of-function mutations in 3′ UTRs create new miRNA target sites and attenuate protein translation, loss-of-function mutations in 3′ UTRs reduce or abolish miRNA–mRNA interactions and augment protein expression. For example, a mismatch in the seed sequence pairing of miR-22 and its target site in the TNFAIP8 3′ UTR has been shown to abolish translational repression of TNFAIP8 [20].
Owing to their potential to alter protein translational efficiency, miRSNPs are likely to contribution to phenotypic variation and disease susceptibility. Several studies have used computational approaches to predict miRSNPs in the genome, and significant associations between these miRSNPs and respective protein levels or related disease traits have been found [20–24]. However, it is difficult to prove that these associations are not instead due to linkage disequilibrium with other SNPs or some other mechanisms. Although increasingly sophisticated computational tools to predict miRSNPs are becoming available, target prediction still remains a major challenge and requires in vitro experiments for functional validation.
Among the SNPs near the 3′ UTR of the GDF15 locus, rs1054564 showed the most significant association with circulating GDF15 levels [25, 26]. The Framingham study also revealed that rs1054564 was associated with cis-gene expression of PGPEP1 and LRRC25 in blood cell lines and lower circulating HDL cholesterol levels [9]. Ek et al. [27] further indicated that genetically increased GDF15 levels, such as via GDF15 SNP rs1054564, directly influence methylation levels at several CpG sites. The aim of this study was to determine if specific miRNAs are capable of regulating GDF15 expression via translational repression.
Materials and methods
In silico analyses
MirSNP (http://cmbi.bjmu.edu.cn/mirsnp), a publicly available online database, is a collection of human SNPs in predicted miRNA–mRNA binding sites. Analyses of the miRNA binding sites in the GDF15 3′ UTR were performed using microRNA.org (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/). We used the NCBI database of SNPs (dbSNP; http://www.ncbi.nlm.nih/gov/SNP) to obtain information about genetic variations. Using these bioinformatics tools, we identified two miRNAs, hsa-miR-873-5p and hsa-miR-1233-3p, that potentially bind to a stretch of sequence harboring rs1054564 in the 3′ UTR of GDF15. The miRNA–target binding structures and energies were predicted using RNAhybird (http://bibiserv.techfak.uni-bielefeld.de/rnahybird).
Construct
A genomic DNA fragment of the GDF15 3′ UTR was amplified by PCR from one individual who was heterozygous for rs1054564 in our previous association study [26]. The upstream and downstream primers used were 5′-ACTAGCTGCATATGAGCAGTCCTGGTCC-3′ and 5′-AAGCTTCACCACAGGGAACAGTTCAG-3′, which were tagged with the SpeI and HindIII restriction enzyme sites (underlined), respectively. PCR products were subcloned into the pCR®2.1 vector (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Plasmid DNA was subsequently isolated from recombinant colonies and sequenced to ensure accuracy. The GDF15 3′ UTR inserts were then extracted by SpeI/HindIII digestion, gel-purified, and subcloned into the SpeI/HindIII site of pMIR-REPORT™ (Ambion) downstream of the firefly luciferase reporter gene. The validity and orientation of the inserts relative to the luciferase gene were confirmed by sequencing.
Cell culture
Human embryonic kidney HEK293T (from Dr. Tzung-Chieh Tsai) and melanoma A2058 cell lines (Food Industry Research and Development Institute, Taiwan) were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT, USA), 80 units/ml penicillin, 80 μg/ml streptomycin, and 0.0175 mg/ml l-proline (Sigma).
Transfection and luciferase assay
Mimics and inhibitors for hsa-miR-873-5p and hsa-miR-1233-3p as well as miRNA negative control #1 were obtained from Ambion (Carlsbad, CA, USA). Cells were plated in a 24-well plate and grown to 80–90% confluence. Firefly luciferase constructs (500 ng) were cotransfected with 20 nM mimic miRNAs with or without Ambion® Anti-miR™ miRNA inhibitors into HEK293T cells using Lipofectamine 2000 (Invitrogen). To monitor transfection efficiency, cells were additionally cotransfected with 50 ng pRL-TK (Promega, Madison, WI, USA), which encoded the Renilla luciferase. Luminescence was measured 48 h after transfection using a dual-luciferase reporter assay system (Promega). All transfections were performed in triplicate, and data were analyzed by normalizing firefly luciferase activity with that of the Renilla luciferase for each sample. Each construct was tested in three independent transfections.
Western blot
Equal amounts of total cell lysate proteins were loaded, separated by 10% SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Membranes were incubated with primary antibodies against GDF15 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h and subsequently with horseradish peroxidase-conjugated secondary antibodies for 1 h. Band densities were detected by ECL chemiluminescence (Amersham Biosciences, Buchs, Switzerland) as described by the manufacturer. Tubulin (Cell Signaling Technology, Beverly, MA, USA) was used as an internal control. Images were scanned with a master imager (Microtek ScanMaker 9800XL, Shanghai, China) and semi-quantified with Photoshop 7.0 (Adobe, San Jose, CA, USA).
Statistical analyses
All statistical analyses were performed using the SPSS 12.0 statistical software package (SPSS, Chicago, IL, USA). Relative luciferase activity data are presented as means ± SD and were analyzed with Student’s t-test. All P-values reported are two-sided. P < 0.05 was considered statistically significant.
Results
Rs1054564 is located in the predicted binding sites of hsa-miR-873-5p and hsa-miR-1233-3p in the GDF15 3′ UTR
Using a publicly available online database, MirSNP (http://cmbi.bjmu.edu.cn/mirsnp), hsa-miR-873-5p was identified as the only conserved miRNA predicted to bind to the GDF15 3′ UTR in the region containing rs1054564 with a good mirSVR score (-0.1265) (Table 1). The prediction indicated an enhanced binding activity based on the rs1054564-G allele adjacent to the 3′ end of the conserved non-seed region of hsa-miR-873-5p (Fig 1). In order to search for other possible miRNAs that could involve rs1054564, we further used two miRNA target prediction tools, MiRanda and TargetScan. A non-conserved miRNA, hsa-miR-1233-3p, was predicted to bind to the GDF15 3′ UTR in the region that contains rs1054564 with a low mirSVR score (-0.0096) (Table 1), and rs1054564 was located inside the hsa-miR-1233-3p seed sequence (Fig 1).
Table 1. Prediction results of miRNAs targeting the GDF15 3′ UTR SNP rs1054564.
| Gene | SNP | Allele | miRNA | Conserveda | mirSVRb | Energyc | Scored | Seed region |
|---|---|---|---|---|---|---|---|---|
| GDF15 | rs1054564 | G | hsa-miR-873-5p | yes | -0.1265 | -28.4 | 97 | no |
| C | -26.0 | |||||||
| G | hsa-miR-1233-3p | no | -0.0096 | -27.1 | 89 | yes | ||
| C | -21.4 |
aConservation information among vertebrates from microRNA.org
bmirSVR score of binding site from microRNA.org
cFree energy of miRNA–mRNA duplex from RNAhybird
dPredicted score of miRNA–mRNA binding by TargetScan. The higher the score the more stable the binding.
Fig 1. Schematic of GDF15 mRNA harboring putative miR-873-5p/miR-1233-3p binding sites and SNP rs1054564 G>C in the 3′ UTR.
Alignment shows the GDF15 3′ UTR variant rs1054564 G>C region with the miRNA hsa-miR-873-5p and hsa-miR-1233-3p motifs. The location of the rs1054564 G>C polymorphism is designated with an arrow in the sequence alignment. Allele G forms a Watson-Crick base-pair with C in the miRNAs (solid line), whereas allele C does not (no line). The seed regions of both miRNAs are designated with an open box. The dots between the base pairs G:U represent GU wobble pairs.
Predicted structures and binding energies of miRNA–target duplexes
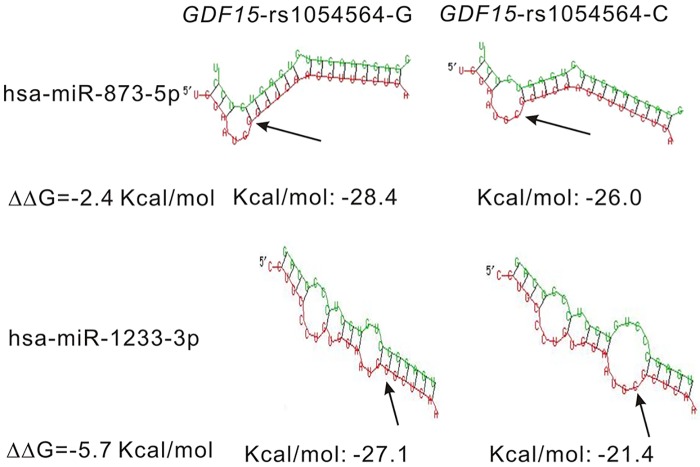
To determine the potential of rs1054564 alleles to alter predicted miRNA–mRNA interactions, we conducted in silico hybridization between the predicted miRNAs and GDF15 3′ UTRs containing the major or minor allele of rs1054564. The affinity between each miRNA and its target sequence can be assessed by computing the minimal free energy of the double-stranded RNA hybrid [28], and this allowed us to compare the stabilities of the miRNA-target interactions among hybrids with different rs1054564 alleles. The program RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) was used to evaluate the Gibbs free energy (ΔG, expressed as kcal/mole) for each rs1054564-containing 21-mer target sequence hybridized to the miRNA of interest. RNAhybrid determined the most favorable hybridization site for each miRNA and subsequently computed the hybridization energy and a potential base-pairing pattern.
The predicted minimal folding energies of the hsa-miR-873-5p- and hsa-miR-1233-3p-target duplexes differed for the rs1054564-G and -C alleles (−28.4 vs. −26.0 kcal/mole for the hsa-miR-873-5p-rs1054564-G and -rs1054564-C duplexes respectively, and −27.1 vs. −21.4 kcal/mole for the hsa-miR-1233-3p-rs1054564-G and -rs1054564-C duplexes, respectively) (Fig 2). The lower minimal folding energies of the rs1054564-G-containing duplexes indicated more stable binding of hsa-miR-873-5p and hsa-miR-1233-3p to the target mRNA and hence more efficient translational repression of the rs1054564-G-containing GDF15 transcript. The difference in Gibbs free energy (ΔΔG) between rs1054564-G- and -C-containing hsa-miR-1233-3p–mRNA hybrids (−5.7 kcal/mole) was lower than that between rs1054564-G- and -C-containing hsa-miR-873-5p–mRNA hybrids (−2.4 kcal/mole), suggesting a more pronounced effect of rs1054564 variation on the mRNA-hsa-miR-1233-3p interaction than on the mRNA-hsa-miR-873-5p interaction.
Fig 2. In silico analysis of the pairing of miR-873-5p and miR-1233-3p to the binding site in the 3′ UTR of GDF15 showing the effect of the minor allele of rs1054564 on the hybrid structures formed between hsa-miR-873-5p/hsa-miR-1233-3p and the GDF15 3′ UTR.
Allele C disrupts the stem part of the typical stem-loop RNA folding structure. The arrow indicates the SNP site in the GDF15 3′ UTR in each folding structure.
Functional analyses of rs1054564
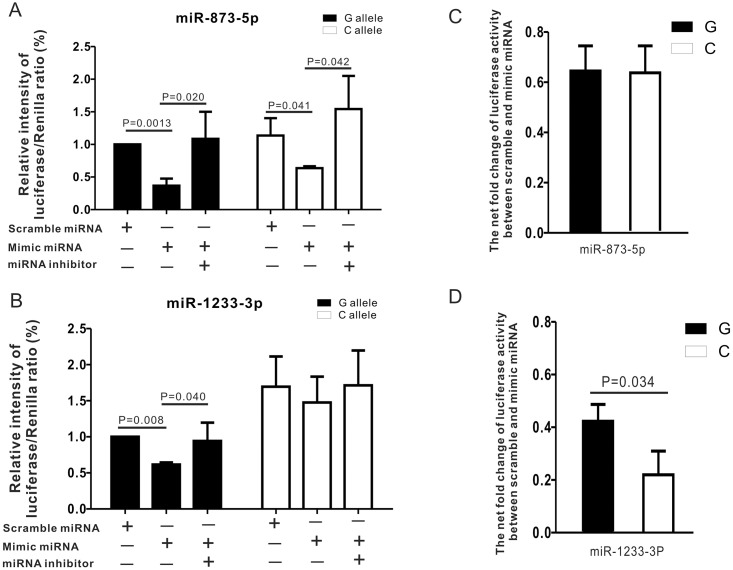
To examine whether the rs1054564 variants affected the translational regulation of the GDF15 protein, we generated luciferase reporter constructs containing the GDF15 3′ UTR with different rs1054564 alleles. These constructs were designated as pMIR-G and pMIR-C, containing the rs1054564-G and -C alleles, respectively. These constructs were cotransfected with either a scrambled or a mimic miRNA of interest into HEK293T cells. Compared with the results of the scrambled miRNA control, luciferase expression was significantly reduced following transfection with either pMIR-G or pMIR-C in the presence of hsa-miR-873-5p (63.2% decrease for pMIR-C, P = 0.041; 64.5% decrease for pMIR-G, P = 0.0013) (Fig 3A). However, in the presence of hsa-miR-1233-3p, expression was only reduced following transfection with pMIR-G (42.1% decrease, P = 0.008) (Fig 3B). In other words, while hsa-miR-873-5p suppressed luciferase expression of both constructs, resulting in a 1.7% difference in the fold-changes between the constructs (P > 0.05) (Fig 3C), hsa-miR-1233-3p only significantly suppressed luciferase expression from pMIR-G, resulting in a 20.1% difference in fold-changes between the two constructs (P = 0.034) (Fig 3D). We further used miRNA inhibitors to assess whether the translational suppression was indeed caused by the respective miRNAs, and both miRNA inhibitors fully reversed this suppression. Our data thus indicate that hsa-miR-1233-3p may preferentially target the GDF15 3′ UTR carrying the major rs1054564-G allele for translational suppression.
Fig 3. Mimic miRNAs regulate human GDF15 3′ UTR expression in human HEK293T cells.
Results of luciferase reporter activity were analyzed by Student’s t-test and are expressed as mean ± SEMs. (A) Effect of mimic miR-873-5p. (B) Effect of mimic miR-1233-3p. (C) Difference in net suppressive effect of mimic miR-873-5p between rs1054564 alleles G and C. (D) Difference in net suppressive effect of mimic miR-1233-3p between rs1054564 alleles G and C.
GDF15 protein levels are regulated by both hsa-miR-873-5p and hsa-miR-1233-3p
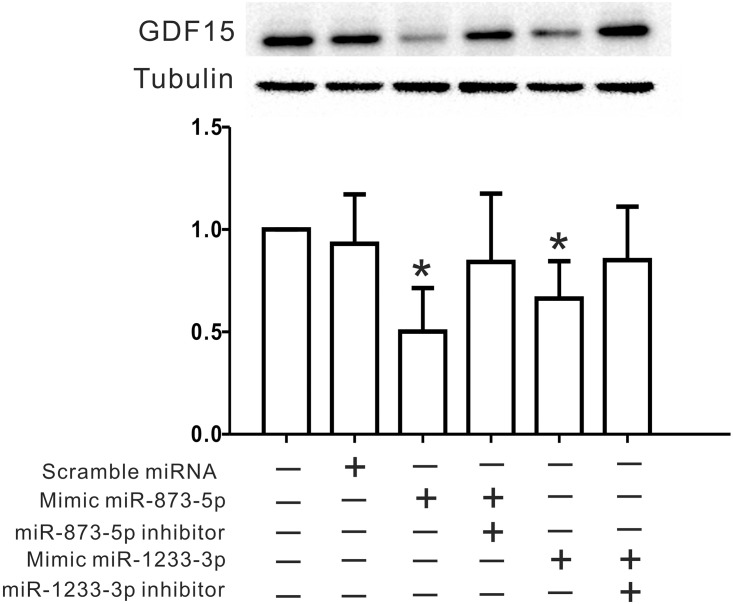
To determine the effect of hsa-miR-873-5p and hsa-miR-1233-3p on endogenous GDF15 protein expression, we transfected the two mimic miRNAs into the A2058 melanoma cell line. Western blot showed that both hsa-miR-873-5p and hsa-miR-1233-3p significantly downregulated GDF15 protein levels (42.7% decrease by hsa-miR-873-5p, P = 0.018; 26.7% decrease by hsa-miR-1233-3p, P = 0.030). This effect was significantly reversed by treatment with corresponding miRNA inhibitors (Fig 4). However, downregulation of GDF15 expression by hsa-miR-873-5p was more effective than that by hsa-miR-1233-3p.
Fig 4. Effect of hsa-miR-873-5p and hsa-miR-1233-3p mimic miRNAs on GDF15 protein expression in human melanoma A2058 cells.
Endogenous GDF15 was suppressed by both mimic miRNAs and restored by miRNA inhibitors. Data represent expression levels relative to those of control samples and indicate the mean of three independent experiments; error bars indicate SE. p-values were determined by a Student’s t-test, and * denotes a p-value < 0.05.
Our findings demonstrate that GDF15 is a target of hsa-miR-873-5p and hsa-miR-1233-3p and that the rs1054564-C allele partially abolishes hsa-miR-1233-3p-mediated translational suppression of GDF15.
Discussion
In this study, we discovered that both hsa-miR-873-5p and hsa-miR-1233-3p repressed endogenous GDF15 translation in a melanoma cell line, indicating that the GDF15 transcript is indeed a target of these miRNAs. Moreover, we are the first to report that variants of the SNP rs1054564 differentially regulate hsa-miR-1233-3p-mediated translational repression. Higher luciferase expression and weaker binding to hsa-miR-1233-3p tentatively suggest that individuals carrying the minor rs1054564-C allele may exhibit elevated GDF15 protein expression. Under pathological conditions like chronic systemic inflammation where GDF15 levels are already high [29–31], the presence of this C-allele may predispose carriers to an even greater risk of undesirable disease outcomes and poor prognoses.
Polymorphisms in miRNA target sites can affect the binding efficacy of miRNA. As a result, they can alter target gene expression and may even contribute to the development of certain diseases. In fact, a number of studies have already reported associations between miRSNPs and human diseases [32–38]. Most of these studies focused on miRSNPs within the target sequence complementary to the seed region of the conserved miRNAs, as a mismatch between the two sequences in this region can alter miRNA-target interactions. For instance, Yuan et al. identified an MMP-9 SNP, rs1056628, that was located in the MMP-9 3′ UTR region complementary to the miR-491 seed sequence and that contributed to an increased risk of atherosclerotic cerebral infarction by increasing MMP-9 expression through destruction of a miR-491 target site [34]. Moreover, Saba et al. showed that SNP rs9291296 is located in the 3' UTR of gamma-aminobutyric acid receptor subunit alpha-4 in a region complementary to the seed sequence of miR-26a-5p. This SNP strengthens miR-26a-5p binding by creating a target site and was found to be associated specifically with degenerating neurons, such as prion disease and other neurodegenerative disorders [35].
In the current study, we identified a non-conserved miRNA, hsa-miR-1233-3p, the binding affinity of which was significantly affected by miRSNP rs1054564. Functional data showed that luciferase expression was only significantly reduced by the rs1054564-G allele, implying that the rs1054564-C allele destroys a target site in the GDF15 3′ UTR complementary to the seed region of hsa-miR-1233-3p. It is worth noting that hsa-miR-1233-3p may play a significant role in the regulation of GDF15, even though its lower ranking score from the prediction tools reflected a lack of evolutionary conservation. Indeed, two recent studies have identified and validated significant roles of non-conserved miRNAs [37, 39]. Cui et al. [39] showed that the C allele of SNP rs2266788 destroys the miRNA hsa-miR-3201 binding site at in the APOA5 3′ UTR, thereby increasing translation of APOA5 and subsequently increasing plasma APOA5 levels. In addition, Ryan et al. [37] identified a SNP that disrupted a novel binding site for miR-516a-3p, leading to moderate increases in CXCR2 mRNA and protein expression and increased MAPK signaling that was associated with lung cancer risk. These results indicate that non-conserved miRNAs and genetic polymorphisms can play potentially significant roles in protein regulation.
As found in this study, several previous studies have identified two miRNAs regulating gene expression via overlapping target sites that contain the same SNP [28, 39, 40]. Consistent with our study, Minguzzi et al. found that MTHFD1L SNP rs7646 creates a target site complementary to the seed region of miR-197 that contributes to hybrid stability; in contrast, SNP rs7646 is located in a region complementary to the non-seed region of miR-9. Their results demonstrated that rs7646 significantly affects miR-197 binding affinity, causing greater suppression when miR-197 is bound to MTHFD1L mRNA containing the G allele rather than the A allele. However, rs7646 did not cause any significant changes in miR-9 binding affinity.
Using computational modeling, we predicted and computed the minimal free energies of the secondary structures of has-miR-873-5p and has-miR-1233-3p when bound to the GDF15 3′ UTR containing either a G or C allele at SNP rs1054564. Because a SNP in this sequence could influence the interactions of several miRNAs, the sum of the ΔΔG value was used to assess the impact of this SNP. Landi et al. [41] proposed that the sum of the ΔΔG reflects the influence of a SNP in the interactions between miRNAs and the target sequence. Namely, the larger the sum of the ΔΔG of a SNP, the more likely it is to be a functional SNP. Our results showed that rs1054564-C might significantly change the has-miR-1233-3p binding affinity, causing a loss of the suppressive effect, whereas it results in only a mild change in the has-miR-873-5p binding affinity. Thus, our data provide evidence that GDF15 is a direct miR-1233-3p target and that the GDF15-associated SNP rs1054564 affects miR-1233-3p binding efficacy.
MiR-1233 has been found to play a role in a plethora of diseases. For example, it is a potential biomarker for cancer and cardiovascular disease [42–45], and its overexpression in placenta significantly decreases the proliferation and invasive ability of trophoblasts in patients with hypertensive disorder complicating pregnancy [46]. Interestingly, while its expression changes in opposite directions in patients with gastric cancer and renal cell carcinoma [42, 43], it is consistently upregulated in patients with heart failure and acute pulmonary embolism [44, 45]. As higher GDF15 levels are also found in these patients [47], these results thus imply the existence of a feedback mechanism in which miR-1233 transcription is increased in order to compensate for elevated GDF15 levels under disease conditions.
Our results clearly indicate that hsa-miR-873-5p represses GDF15 translation regardless of rs1054564 genotype. Although hsa-miR-873-5p and GDF15 levels appeared to be negatively correlated in our in vitro assays, elevated hsa-miR-873-5p or GDF15 levels have been previously associated with different stages of cancer-like cell proliferation [48–50], metastasis [49, 51, 52], and chemoresistance [47, 53–55]. One reason for these findings is that hsa-miR-873-5p represses the translation of several proteins in addition to GDF15, and this may in turn increase the transcription or prolong the mRNA stability of GDF15. Furthermore, it is possible that hsa-miR-873-5p and GDF15 are often not co-expressed in the same type of cancer. Simply overexpressing miR-873-5p or GDF15 may therefore generate completely different results depending on the cell type-specific machinery and underlying gene expression profiles of the cancer cells in question [49,56]. Nevertheless, our combined results of hsa-miR-873-5p-mediated repression of luciferase and endogenous GDF15 expression in two different cell types suggest that miR-873-5p may represent a novel therapeutic target for the treatment of cancers that exhibit increased activity of the GDF15 gene.
In addition to cancer, it has been reported that the chromosome 9p21 locus, where miR-873-5p resides, is strongly associated with coronary artery disease (CAD) [57–63] and peripheral arterial disease (PAD) [64, 65]. Intriguingly, we have previously showed that GDF15 serves as a prognostic factor for all-cause mortality in diverse human disorders including CAD and PAD [26]. Based on the findings of this study, it is tantalizing to speculate that miR-873-5p participates in CAD and PAD through cis-regulation of GDF15 expression. However, other open reading frames in the vicinity of 9p21, including cyclin-dependent kinase inhibitor 2A/B, may also contribute to these diseases [66]. Future studies are needed to determine whether miR-873-5p plays a role in the cardiovascular system and whether it loses its ability to repress GDF15 expression under pathological conditions.
There are several limitations to our study. First, we could not find a cell line heterozygous for rs1054564 to examine the allelic differences in GDF15 translation. Second, we could not detect endogenous hsa-miR-873-5p and hsa-miR-1233-3p expression in the cell lines used in this study and therefore could not address their modes of interaction in greater detail. Third, a suitable animal model was not available, hampering further investigation. Finally, the mechanism by which hsa-miR-873-5p and hsa-miR-1233-3p regulate GDF15 translation is not yet fully understood and requires further elucidation.
Conclusions
Our work delineates a comprehensive framework for miRNA-mediated translational repression of GDF15. We propose that in a certain percentage of the human population, the rs1054564-C mutation weakens the hsa-miR-1233-3p binding site in the GDF15 3′ UTR and thereby increases the chances for higher GDF15 protein expression. This upregulation might at least partially account for the strong associations between rs1054564 and elevated GDF15 levels reported in previous study [26]. Further investigations regarding the interactions between the studied miRNAs and GDF15 levels and their involvement in the progression of cardiovascular diseases and cancers should be interesting and may help to uncover novel therapies against related complications.
Acknowledgments
This study was supported by grants from the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation (TCRD-TPE-103-RT-2, TCRD-I103-01-01, TCRD-TPE-MOST-103-01, TCRD-TPE-MOST-104-09, TCRD-TPE-106-C1-1, TCRD-TPE-106-RT-3, TCRD-TPE-MOST-105-03); a grant from the Tzu Chi University, Hualien, Taiwan (TCMMP104-06-03); a grant from the National Science Council (MOST 104-2314-B-303-013-MY3, NSC 101-2314-B-303–023 -MY3) to Y. L. Ko; and grants from the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation (TCRD-TPE-103-22, TCRD-TPE-104-18, TCRD-TPE-106-46) to M. S. Teng. We greatly appreciate technical support from the Core Laboratory of the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation. We appreciate Dr. Tzung-Chieh Tsai (Department of Microbiology, Immunology and Biopharmaceuticals, National Chiayi University, Chiayi, Taiwan) supply us the Human embryonic kidney HEK293T cell line.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by grants from the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation (TCRD-TPE-103-RT-2, TCRD-I103-01-01, TCRD-TPE-MOST-103-01, TCRD-TPE-MOST-104-09, TCRD-TPE-106-C1-1, TCRD-TPE-106-RT-3), a grant from the Tzu Chi University, Hualien, Taiwan (TCMMP104-06-03), a grant from the National Science Council (MOST 104-2314-B-303-013-MY3) to Y. L. Ko, and grants from the Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation (TCRD-TPE-103-22, TCRD-TPE-104-18, TCRD-TPE-106-46) to M. S. Teng. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Xu X, Li Z, Gao W. Growth differentiation factor 15 in cardiovascular diseases: from bench to bedside. Biomarkers. 2011;16(6):466–75. doi: 10.3109/1354750X.2011.580006 . [DOI] [PubMed] [Google Scholar]
- 2.Bauskin AR, Brown DA, Junankar S, Rasiah KK, Eggleton S, Hunter M, et al. The propeptide mediates formation of stromal stores of PROMIC-1: role in determining prostate cancer outcome. Cancer Res. 2005;65(6):2330–6. doi: 10.1158/0008-5472.CAN-04-3827 . [DOI] [PubMed] [Google Scholar]
- 3.Wiklund FE, Bennet AM, Magnusson PK, Eriksson UK, Lindmark F, Wu L, et al. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell. 2010;9(6):1057–64. doi: 10.1111/j.1474-9726.2010.00629.x ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels LB, Clopton P, Laughlin GA, Maisel AS, Barrett-Connor E. Growth-differentiation factor-15 is a robust, independent predictor of 11-year mortality risk in community-dwelling older adults: the Rancho Bernardo Study. Circulation. 2011;123(19):2101–10. doi: 10.1161/CIRCULATIONAHA.110.979740 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ago T, Sadoshima J. GDF15, a cardioprotective TGF-beta superfamily protein. Circ Res. 2006;98(3):294–7. doi: 10.1161/01.RES.0000207919.83894.9d [DOI] [PubMed] [Google Scholar]
- 6.Breit SN, Johnen H, Cook AD, Tsai VW, Mohammad MG, Kuffner T, et al. The TGF-beta superfamily cytokine, MIC-1/GDF15: a pleotrophic cytokine with roles in inflammation, cancer and metabolism. Growth Factors. 2011;29(5):187–95. doi: 10.3109/08977194.2011.607137 . [DOI] [PubMed] [Google Scholar]
- 7.Dostalova I, Roubicek T, Bartlova M, Mraz M, Lacinova Z, Haluzikova D, et al. Increased serum concentrations of macrophage inhibitory cytokine-1 in patients with obesity and type 2 diabetes mellitus: the influence of very low calorie diet. Eur J Endocrinol. 2009;161(3):397–404. doi: 10.1530/EJE-09-0417 . [DOI] [PubMed] [Google Scholar]
- 8.Eggers KM, Kempf T, Lagerqvist B, Lindahl B, Olofsson S, Jantzen F, et al. Growth-differentiation factor-15 for long-term risk prediction in patients stabilized after an episode of non-ST-segment-elevation acute coronary syndrome. Circ Cardiovasc Genet. 2010;3(1):88–96. doi: 10.1161/CIRCGENETICS.109.877456 . [DOI] [PubMed] [Google Scholar]
- 9.Ho JE, Mahajan A, Chen MH, Larson MG, McCabe EL, Ghorbani A, et al. Clinical and genetic correlates of growth differentiation factor 15 in the community. Clin Chem. 2012;58(11):1582–91. doi: 10.1373/clinchem.2012.190322 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempf T, von Haehling S, Peter T, Allhoff T, Cicoira M, Doehner W, et al. Prognostic utility of growth differentiation factor-15 in patients with chronic heart failure. J Am Coll Cardiol. 2007;50(11):1054–60. doi: 10.1016/j.jacc.2007.04.091 . [DOI] [PubMed] [Google Scholar]
- 11.Vila G, Riedl M, Anderwald C, Resl M, Handisurya A, Clodi M, et al. The relationship between insulin resistance and the cardiovascular biomarker growth differentiation factor-15 in obese patients. Clin Chem. 2011;57(2):309–16. doi: 10.1373/clinchem.2010.153726 . [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. . [DOI] [PubMed] [Google Scholar]
- 13.Glinsky GV. Disease phenocode analysis identifies SNP-guided microRNA maps (MirMaps) associated with human "master" disease genes. Cell Cycle. 2008; 7(23):3680–94. doi: 10.4161/cc.7.23.7153 . [DOI] [PubMed] [Google Scholar]
- 14.Glinsky GV. SNP-guided microRNA maps (MirMaps) of 16 common human disorders identify a clinically accessible therapy reversing transcriptional aberrations of nuclear import and inflammasome pathways. Cell Cycle. 2008;7(22):3564–76. doi: 10.4161/cc.7.22.7073 . [DOI] [PubMed] [Google Scholar]
- 15.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14(23):7956–62. doi: 10.1158/1078-0432.CCR-08-1199 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10(3):399–416. doi: 10.2217/14622416.10.3.399 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35(Database issue):D51–4. doi: 10.1093/nar/gkl797 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hariharan M, Scaria V, Brahmachari SK. dbSMR: a novel resource of genome-wide SNPs affecting microRNA mediated regulation. BMC Bioinformatics. 2009;10:108 doi: 10.1186/1471-2105-10-108 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010;38(Database issue):D640–51. doi: 10.1093/nar/gkp926 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi TY, Cheng X, Yu KD, Sun MH, Shao ZM, Wang MY, et al. Functional variants in TNFAIP8 associated with cervical cancer susceptibility and clinical outcomes. Carcinogenesis. 2013;34(4):770–8. doi: 10.1093/carcin/bgt001 [DOI] [PubMed] [Google Scholar]
- 21.Qiao Y, Ma N, Wang X, Hui Y, Li F, Xiang Y, et al. MiR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett. 2011. October 3;585(19):3095–100. doi: 10.1016/j.febslet.2011.08.039 [DOI] [PubMed] [Google Scholar]
- 22.Lill CM, Schilling M, Ansaloni S, Schröder J, Jaedicke M, Luessi F, et al. Assessment of microRNA-related SNP effects in the 3' untranslated region of the IL22RA2 risk locus in multiple sclerosis. Neurogenetics. 2014. May;15(2):129–34. doi: 10.1007/s10048-014-0396-y [DOI] [PubMed] [Google Scholar]
- 23.Goda N, Murase H, Kasezawa N, Goda T, Yamakawa-Kobayashi K. Polymorphism in microRNA-binding site in HNF1B influences the susceptibility of type 2 diabetes mellitus: a population based case-control study. BMC Med Genet. 2015. September 2;16:75 doi: 10.1186/s12881-015-0219-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Q, Chen Y, Dai J, Wang B, Liu M, Wang Y, et al. Methylenetetrahydrofolate reductase polymorphisms at 3'-untranslated region are associated with susceptibility to preterm birth. Transl Pediatr. 2015. January;4(1):57–62. doi: 10.3978/j.issn.2224-4336.2015.01.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown DA, Lindmark F, Stattin P, Balter K, Adami HO, Zheng SL, et al. Macrophage inhibitory cytokine 1: A new prognostic marker in prostate cancer. Clin Cancer Res. 2009;15:6658–6664. doi: 10.1158/1078-0432.CCR-08-3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu LA, Wu S, Jimmy Juang JM, Chiang FT, Teng MS, Huang HL, Ko YL. Growth differentiation factor 15 may predict mortality of peripheral and coronary artery diseases and correlate with their risk factors. Mediators of Inflammation. vol. 2017, Article ID 9398401, 13 pages,. doi: 10.1155/2017/9398401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ek WE, Hedman ÅK, Enroth S, Morris AP, Lindgren CM, Mahajan A, Gustafsson S, Gyllensten U, Lind L, Johansson Å. Genome-wide DNA methylation study identifies genes associated with the cardiovascular biomarker GDF-15. Hum Mol Genet. 2016. February 15;25(4):817–27. doi: 10.1093/hmg/ddv511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minguzzi S, Selcuklu SD, Spillane C, Parle-McDermott A. An NTD-associated polymorphism in the 3' UTR of MTHFD1L can affect disease risk by altering miRNA binding. Hum Mutat. 2014. January;35(1):96–104. doi: 10.1002/humu.22459 [DOI] [PubMed] [Google Scholar]
- 29.Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci USA 1997; 94:11514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlittenhardt D, Schober A, Strelau J, Bonaterra GA, Schmiedt W, Unsicker K, et al. Involvement of growth differentiation factor-15/macrophage inhibitory cytokine-1 (GDF-15/MIC-1) in oxLDL-induced apoptosis of human macrophages in vitro and in arteriosclerotic lesions. Cell Tissue Res 2004; 318:325–33. doi: 10.1007/s00441-004-0986-3 [DOI] [PubMed] [Google Scholar]
- 31.Zimmers TA, Jin X, Hsiao EC, McGrath SA, Esquela AF, Koniaris LG. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock 2005; 23:543–8. [PubMed] [Google Scholar]
- 32.Zhang L, Liu Y, Song F, Zheng H, Hu L, Lu H, et al. Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci U S A. 2011. August 16;108(33):13653–8. doi: 10.1073/pnas.1103360108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas U, Sczakiel G, Laufer SD. MicroRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3'-UTR via altered RNA structure. RNA Biol. 2012. June;9(6):924–37. doi: 10.4161/rna.20497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan M, Zhan Q, Duan X, Song B, Zeng S, Chen X, et al. A functional polymorphism at miR-491-5p binding site in the 3'-UTR of MMP-9 gene confers increased risk for atherosclerotic cerebral infarction in a Chinese population. Atherosclerosis. 2013. February;226(2):447–52. doi: 10.1016/j.atherosclerosis.2012.11.026 [DOI] [PubMed] [Google Scholar]
- 35.Saba R, Medina SJ, Booth SA. A functional SNP catalog of overlapping miRNA-binding sites in genes implicated in prion disease and other neurodegenerative disorders. Hum Mutat. 2014. October;35(10):1233–48. doi: 10.1002/humu.22627 [DOI] [PubMed] [Google Scholar]
- 36.Yao Y, Shao J, Wu J, Zhang Q, Wang J, Xiao D, et al. The Functional Variant in the 3'UTR of PTPRT with the Risk of Esophageal Squamous Cell Carcinoma in a Chinese Population. Cell Physiol Biochem. 2015;36(1):306–14. doi: 10.1159/000374073 [DOI] [PubMed] [Google Scholar]
- 37.Ryan BM, Robles AI, McClary AC, Haznadar M, Bowman ED, Pine SR, et al. Identification of a functional SNP in the 3'UTR of CXCR2 that is associated with reduced risk of lung cancer. Cancer Res. 2015. February 1;75(3):566–75. doi: 10.1158/0008-5472.CAN-14-2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Wu J, Zhao Y, Guo Z. miR-502 medaited histone methyltransferase SET8 expression is associated with outcome of esophageal squamous cell carcinoma. Sci Rep. 2016. September 8;6:32921 doi: 10.1038/srep32921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui G, Li Z, Li R, Huang J, Wang H, Zhang L, et al. A functional variant in APOA5/A4/C3/A1 gene cluster contributes to elevated triglycerides and severity of CAD by interfering with microRNA 3201 binding efficiency. J Am Coll Cardiol. 2014. July 22;64(3):267–77. doi: 10.1016/j.jacc.2014.03.050 [DOI] [PubMed] [Google Scholar]
- 40.Caussy C, Charrière S, Marçais C, Di Filippo M, Sassolas A, Delay M, et al. An APOA5 3' UTR variant associated with plasma triglycerides triggers APOA5 downregulation by creating a functional miR-485-5p binding site. Am J Hum Genet. 2014. January 2;94(1):129–34. doi: 10.1016/j.ajhg.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008;29:579–84. doi: 10.1093/carcin/bgm304 [DOI] [PubMed] [Google Scholar]
- 42.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6(9):e25787 doi: 10.1371/journal.pone.0025787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang ZZ, Wang CJ, Niu L, Xu J, Wang M, Cao H, et al. Analysis of plasma MicroRNAs to identifying early diagnostic molecule for gastric cancer. Int J Clin Exp Med. 2015. March 15;8(3):3700–6. [PMC free article] [PubMed] [Google Scholar]
- 44.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015. April;17(4):393–404. doi: 10.1002/ejhf.223 [DOI] [PubMed] [Google Scholar]
- 45.Kessler T, Erdmann J, Vilne B, Bruse P, Kurowski V, Diemert P, et al. Serum microRNA-1233 is a specific biomarker for diagnosing acute pulmonary embolism. J Transl Med. 2016. May 5;14(1):120 doi: 10.1186/s12967-016-0886-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong W, Peng H, Tian A, Wei Y, Li H, Tian J, et al. Expression of miRNA-1233 in placenta from patients with hypertensive disorder complicating pregnancy and its role in disease pathogenesis. Int J Clin Exp Med. 2015. June 15;8(6):9121–7. [PMC free article] [PubMed] [Google Scholar]
- 47.Chan MM, Santhanakrishnan R, Chong JP, Chen Z, Tai BC, Liew OW, et al. Growth differentiation factor 15 in heart failure with preserved vs. reduced ejection fraction. Eur J Heart Fail. 2016. January;18(1):81–8. doi: 10.1002/ejhf.431 [DOI] [PubMed] [Google Scholar]
- 48.Yang CZ, Ma J, Zhu DW, Liu Y, Montgomery B, Wang LZ, et al. GDF15 is a potential predictive biomarker for TPF induction chemotherapy and promotes tumorigenesis and progression in oral squamous cell carcinoma. Ann Oncol. 2014. June;25(6):1215–22. doi: 10.1093/annonc/mdu120 [DOI] [PubMed] [Google Scholar]
- 49.Gao Y, Xue Q, Wang D, Du M, Zhang Y, Gao S. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am J Transl Res. 2015. November 15;7(11):2519–26. [PMC free article] [PubMed] [Google Scholar]
- 50.Urakawa N, Utsunomiya S, Nishio M, Shigeoka M, Takase N, Arai N, et al. GDF15 derived from both tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Lab Invest. 2015. May;95(5):491–503. doi: 10.1038/labinvest.2015.36 [DOI] [PubMed] [Google Scholar]
- 51.Wang XB, Jiang XR, Yu XY, Wang L, He S, Feng FY, et al. Macrophage inhibitory factor 1 acts as a potential biomarker in patients with esophageal squamous cell carcinoma and is a target for antibody-based therapy. Cancer Sci. 2014. February;105(2):176–85. doi: 10.1111/cas.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Wang J, Kong J, Tang J, Wu Y, Xu E, et al. GDF15 promotes EMT and metastasis in colorectal cancer. Oncotarget. 2016. January 5;7(1):860–72. doi: 10.18632/oncotarget.6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji H, Lu HW, Li YM, Lu L, Wang JL, Zhang YF, et al. Twist promotes invasion and cisplatin resistance in pancreatic cancer cells through growth differentiation factor 15. Mol Med Rep. 2015. September;12(3):3841–8. doi: 10.3892/mmr.2015.3867 [DOI] [PubMed] [Google Scholar]
- 54.Wu DD, Li XS, Meng XN, Yan J, Zong ZH. MicroRNA-873 mediates multidrug resistance in ovarian cancer cells by targeting ABCB1. Tumour Biol. 2016. August;37(8):10499–506. doi: 10.1007/s13277-016-4944-y [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Hua W, Niu LC, Li SM, Wang YM, Shang L, et al. Elevated growth differentiation factor 15 expression predicts poor prognosis in epithelial ovarian cancer patients. Tumour Biol. 2016. July;37(7):9423–31. doi: 10.1007/s13277-015-4699-x [DOI] [PubMed] [Google Scholar]
- 56.Tsui KH, Hsu SY, Chung LC, Lin YH, Feng TH, Lee TY, et al. Growth differentiation factor-15: a p53- and demethylation-upregulating gene represses cell proliferation, invasion, and tumorigenesis in bladder carcinoma cells. Sci Rep. 2015. August 7;5:12870 doi: 10.1038/srep12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, Asano H, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet. 2012. March;20(3):333–40. doi: 10.1038/ejhg.2011.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan K, Patel RS, Newcombe P, Nelson CP, Qasim A, Epstein SE, et al. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol. 2013. March 5;61(9):957–70. doi: 10.1016/j.jacc.2012.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Setten J, Isgum I, Smolonska J, Ripke S, de Jong PA, Oudkerk M, et al. Genome-wide association study of coronary and aortic calcification implicates risk loci for coronary artery disease and myocardial infarction. Atherosclerosis. 2013. June;228(2):400–5. doi: 10.1016/j.atherosclerosis.2013.02.039 [DOI] [PubMed] [Google Scholar]
- 60.Dichgans M, Malik R, König IR, Rosand J, Clarke R, Gretarsdottir S, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014. January;45(1):24–36. doi: 10.1161/STROKEAHA.113.002707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munir MS, Wang Z, Alahdab F, Steffen MW, Erwin PJ, Kullo IJ, et al. The association of 9p21-3 locus with coronary atherosclerosis: a systematic review and meta-analysis. BMC Med Genet. 2014. June 6;15:66 doi: 10.1186/1471-2350-15-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee IT, Liang KW, Wang JS, Lee WJ, Chen YD, Lin SY, et al. Value of Chromosome 9p21 Polymorphism for Prediction of Cardiovascular Mortality in Han Chinese Without Coronary Lesions: An Observational Study. Medicine (Baltimore). 2015. September;94(39):e1538 doi: 10.1097/MD.0000000000001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zanetti D, Via M, Carreras-Torres R, Esteban E, Chaabani H, Anaibar F, et al. Analysis of Genomic Regions Associated With Coronary Artery Disease Reveals Continent-Specific Single Nucleotide Polymorphisms in North African Populations. J Epidemiol. 2016. May 5;26(5):264–71. doi: 10.2188/jea.JE20150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murabito JM, White CC, Kavousi M, Sun YV, Feitosa MF, Nambi V, et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet. 2012. February 1;5(1):100–12. doi: 10.1161/CIRCGENETICS.111.961292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian LB, Fang H, Gao L, Tan Z, Zhen YF, Tian JL, et al. 9p21 polymorphisms increase the risk of peripheral artery disease in the Han Chinese population. J Int Med Res. 2013. February;41(1):106–14. doi: 10.1177/0300060512474569 [DOI] [PubMed] [Google Scholar]
- 66.Hannou SA, Wouters K, Paumelle R, Staels B. Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab. 2015. April;26(4):176–84. doi: 10.1016/j.tem.2015.01.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.