Abstract
RNA interference (RNAi)-based gene regulation has recently emerged as a promising strategy to silence genes that drive disease progression. RNAi is typically mediated by small interfering ribonucleic acids (siRNAs), which, upon delivery into the cell cytoplasm, trigger degradation of complementary messenger RNA molecules to halt production of their encoded proteins. While RNAi has enormous clinical potential, its in vivo utility has been hindered because siRNAs are rapidly degraded by nucleases, cannot passively enter cells, and are quickly cleared from the bloodstream. To overcome these delivery barriers, siRNAs can be conjugated to nanoparticles (NPs), which increase their stability and circulation time to enable in vivo gene regulation. Here, we present methods to conjugate siRNA duplexes to NPs with gold surfaces. Further, we describe how to quantify the resultant amount of siRNA sense and antisense strands loaded onto the NPs using a fluorescence-based assay. This method focuses on the attachment of siRNAs to 13 nm gold NPs, but it is adaptable to other types of nucleic acids and nanoparticles as discussed throughout the protocol.
Keywords: Nanoparticles, siRNA, Nucleic acids, Gene regulation, Conjugation, Loading
1 Introduction
RNA interference (RNAi) is a potent method to regulate gene expression that is under intense investigation as a therapy for a variety of diseases including cancer, hepatitis C, Alzheimer’s, and Parkinson’s [1]. In RNAi, exogenous small interfering RNAs (siRNAs) delivered into cells initiate the degradation of complementary messenger RNA (mRNA) molecules by the cells’ internal machinery; this halts production of the proteins encoded by the mRNAs, resulting in reduced gene expression [1]. While RNAi has potential to transform our ability to treat disease, there are several challenges associated with delivering siRNAs to diseased sites for gene therapy. For example, siRNAs rapidly degrade in the presence of nucleases, have a poor biodistribution profile, and cannot passively enter cells due to their negative charge [1, 2]. To facilitate passage across negatively charged cell membranes, siRNAs are typically complexed with cationic transfection agents. These cationic agents are useful for in vitro studies, but their high toxicity precludes in vivo use [3, 4]. Accordingly, researchers are developing new strategies to enable in vivo siRNA delivery, and the most common approach is to use nanoparticles as siRNA carriers [3–5]. Nanoparticles are advantageous as siRNA delivery vehicles because they can overcome several of the aforementioned challenges related to siRNA delivery [3–5]. The two main methods of siRNA delivery using nanoparticles are encapsulation, wherein siRNAs are entrapped inside porous nanoparticles or within layers of positively charged materials surrounding nanoparticles [4, 6–10], and conjugation, wherein siRNAs are bound to nanoparticle surfaces and exposed as the outer layer [5, 11–14]. Both of these methods have been shown to protect siRNAs from degradation, promote their cellular uptake, and improve gene regulation both in vitro and in vivo [6–15]. Therefore, there is substantial evidence to support continued development of nanoparticles for siRNA delivery. In this chapter, we describe the synthesis and characterization of siRNA nanocarriers prepared by the conjugation method.
Thorough and consistent characterization of siRNA nanocarriers is critical for their successful implementation as mediators of RNAi. Researchers must accurately quantify the amount of siRNA loaded within or on nanoparticle carriers to precisely dose therapies. Further, loading density is known to influence the cell uptake of nanocarriers coated with siRNA (or other nucleic acids), with high density favoring increased cell uptake [16–18], so quantitative characterization is essential to understand and enhance the interactions between siRNA-coated nanoparticles and cells. Several methods exist that could be used to qualitatively or quantitatively measure siRNA bound to or entrapped within nanoparticles. For example, zeta potential and dynamic light scattering measurements can confirm siRNA loading, as the addition of siRNA changes nanoparticles’ surface charge and hydrodynamic diameter [12]. While qualitatively confirming the presence of siRNA is useful, quantification of siRNA loading provides more valuable information. One common method to quantify siRNA loading involves determining the siRNA remaining in solution after nanoparticle conjugation and purification by recording the absorbance at 260 nm (the peak absorbance of siRNA) with a spectrophotometer and calculating the amount of siRNA present using the Beer-Lambert law [19]. The strength of this method is its simplicity, but it is not ideal because it does not directly measure siRNAs on nanoparticles, assumes no losses during processing, has low sensitivity, and cannot distinguish sense- and antisense strand loading. Some alternative approaches have been developed to directly and quantitatively measure siRNAs and other nucleic acids bound to nanoparticles, including methods based on real-time polymerase chain reaction (PCR) [20] and methods that use fluorophore-labeled nucleic acids to quantify loading via fluorescence measurements [21, 22]. The main limitation of the PCR approach is its complexity, and the fluorescence-based approaches are limited by the high cost of fluorophore-labeled siRNA and potential errors in quantification that may result from fluorophores being quenched due to their close proximity to each other and the nanoparticles’ surfaces. Additionally, fluorophore modifications may alter siRNA loading onto nanoparticles and, unless both strands are fluorophore labeled, fluorescence readout cannot quantify both antisense and sense RNA strands. Given that the number of sense and anti-sense oligonucleotides on nanoparticles may not be equal [23], it is imperative to measure both strands individually when quantifying loading.
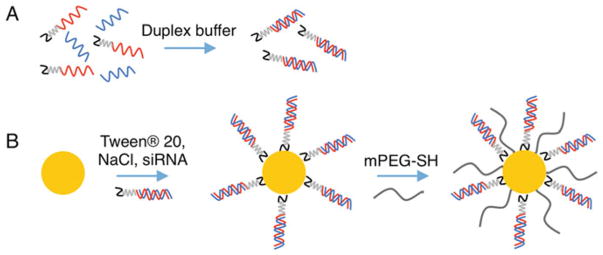
Here, we describe methods to coat nanoparticles containing gold surfaces with thiolated siRNAs (Fig. 1) and to quantify the amount of conjugated siRNAs using a simple, fluorescence-based approach that does not require the siRNAs to be modified with fluorophores (Fig. 2). We focus on gold-based nanoparticles since these are the most extensively studied nanocarriers that utilize a surface conjugation strategy [5]. In this protocol, antisense RNA strands are first hybridized to sense RNA strands containing a 3′ thiol (see Note 1), which facilitates siRNA duplex loading on the nanoparticles’ surfaces via gold–thiol bond formation (Fig. 1). Next, the conjugated siRNA is quantified as shown in Fig. 2. Briefly, the antisense strands are dehybridized from the nanoparticle-bound sense strands then the nanoparticles are pelleted by centrifugation. The antisense-containing supernatant is collected and incubated with the components of the Quant-IT OliGreen® kit to produce a fluorescent signal (see Note 2). The OliGreen® dye is weakly fluorescent in solution, but produces a strong fluorescent signal that is easily detected with a fluorescent plate reader upon binding to single-stranded nucleic acids. The concentration of antisense RNA in the sample is determined by comparing the sample fluorescence intensity to that of a standard curve of known antisense RNA concentration. Next, thiolated sense RNA strands are released from the nanoparticles by breaking the gold–thiol bonds in β-mercaptoethanol. The released sense RNA strands are collected and quantified using the Quant-iT™ OliGreen® kit. This method for quantifying siRNA duplexes loaded on nanoparticles is advantageous because it does not require siRNA that has been fluorophore-labeled, can be adapted to suit various nanoparticle core materials, and allows for individual measurement of both antisense and sense RNA strands. We have found that approximately twice as many sense strands bind to nanoparticles’ surfaces as antisense strands (Fig. 3), so it is useful to obtain measurements for each sequence.
Fig. 1.
Schematic representing siRNA conjugation to gold nanoparticle (AuNP) surfaces. (a) siRNA duplexes are prepared by mixing antisense (blue) and thiolated sense (red) oligonucleotides in duplex buffer at a 1:1 ratio, heating to 95 °C, and cooling to 37 °C slowly over 1 h. The black portion of the sense strand indicates a thiol group, and the gray portion indicates a PEG spacer. (b) Freshly prepared siRNA duplexes are conjugated to AuNP surfaces by adding excess siRNA to AuNPs (yellow) suspended in dilute Tween® 20 and NaCl. siRNA loading is maximized by slowly increasing the concentration of NaCl, which screens charges between siRNA duplexes. Last, any exposed surface area is passivated with methoxy-PEG-thiol (mPEG-SH, gray) to stabilize the siRNA-coated AuNPs
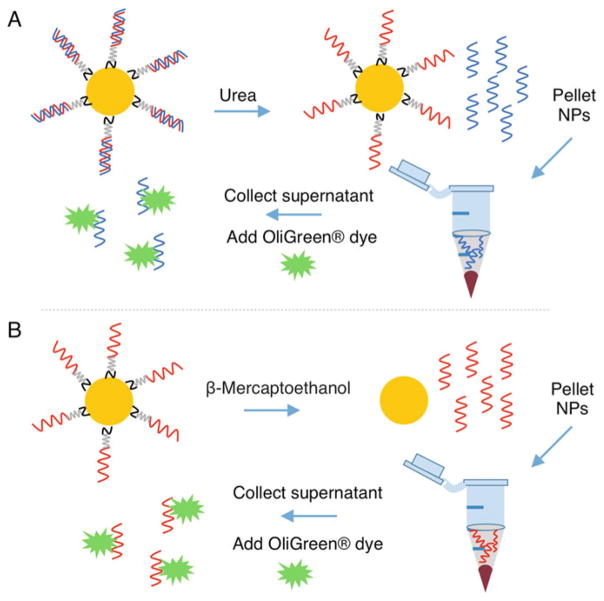
Fig. 2.
Schematic representing the procedure to quantify siRNA duplexes on nanoparticles (NPs). (a) Antisense RNA strands (blue) are dehybridized from sense RNA strands (red) on nanoparticles by incubating in 8 M urea at 45 °C. The sense-loaded nanoparticles are pelleted by centrifugation, and the supernatant containing the antisense strands is collected. Antisense RNA strands within the supernatant are measured using components of the Quant-iT™OliGreen® kit. (b) Sense RNA strands (red) are removed from the nanoparticles’ surfaces by breaking the gold–thiol bond with β-Mercaptoethanol. The nanoparticles are pelleted by centrifugation, and the supernatant containing the sense RNA strands is collected for analysis of RNA content with the Quant-iT™ OliGreen® kit
Fig. 3.
siRNA loading on nanoparticle surfaces increases with nanoparticle surface area. Representative antisense and sense RNA loading data is shown for 13 nm diameter AuNPs (a) and 150 nm diameter silica core/gold shell nanoshells (b). Data indicate mean ± standard deviation of three batches of siRNA-coated nanoparticles
We describe this protocol using 13 nm diameter spherical gold nanoparticles (AuNPs) as the siRNA nanocarrier, but it is translatable to other types of nanoparticles and oligonucleotides as well. For example, we and other researchers have previously described how this assay can be adapted to load and measure DNA (deoxyribonucleic acid) or microRNA on 13 nm AuNPs or larger AuNPs [24–26]. To demonstrate the versatility of this protocol for different nanocarriers, we have added notes to describe how it may be adapted to the synthesis and characterization of siRNA-coated nanoshells, which are nanoparticles that contain 120 nm silica cores and 15 nm-thick gold shells (Fig. 3). The 13 nm AuNPs and 150 nm nanoshells used in this protocol are meant to serve as model systems; readers should optimize the instructions provided as necessary for their specific nanoparticle formulation and application.
2 Materials
Prepare all solutions using ultrapure, RNase-free water (see Note 3) and store at appropriate temperatures as indicated below. In addition to the reagents listed in this section, readers should have access to the following: −80 °C freezer, 4 °C refrigerator, microcentrifuge tubes, pipettes, black-walled 96-well plates, thermomixer, sonicator bath, rocking platform, ice, ice bucket, vortex, microcentrifuge, spectrophotometer, fume hood, and a fluorescent plate reader.
2.1 siRNA Duplex Preparation
2.2 Oligonucleotide Conjugation to AuNPs
AuNPs, made RNase-free by treatment with DEPC (diethylpyrocarbonate) (see Note 6): Store at room temperature.
Tween® 20: Prepare a 10% solution of Tween® 20 by diluting 1 mL Tween® 20 in 9 mL water (see Note 7). Store at room temperature.
Sodium chloride (NaCl): Prepare 5 M NaCl solution by dissolving 58.44 g NaCl in 200 mL ultrapure water (this water does not have to be RNase-free since the final solution will be treated with DEPC). DEPC-treat the entire solution to deactivate RNases (see Note 3). Store at room temperature.
Methoxy PEG-thiol (mPEG-SH) at desired molecular weight (see Note 8): Store mPEG-SH as a lyophilized powder at −80 °C under argon. Fresh 1 mM solution should be made in ultrapure RNase-free water prior to use.
Phosphate buffered saline (PBS), 1×, RNase-free: Store at room temperature.
2.3 Quantifying Antisense Oligonucleotides Bound to AuNPs
siRNA-coated AuNPs: Prepare as described in the Methods (Subheading 3.2). Store at 4 °C.
8 M Urea, RNase-free: Prepare by dissolving 12.01 g urea with 25 mL RNase-free water. Store at room temperature.
Tween® 20: Prepare a 0.1% solution in Ultrapure RNase-free water by diluting the previously prepared 10% stock solution 1:100.
Quant-iT™ OliGreen® ssDNA Assay Kit: Store at 4 °C (see Note 2).
Antisense oligonucleotides: Store at −80 °C. These antisense RNA strands will be used to generate a standard curve.
2.4 Quantifying Sense Oligonucleotides Bound to AuNPs
Sense strand-coated AuNPs: Prepared as described in the Methods (Subheading 3.3) and stored at 4 °C.
PBS, 1×, RNase-free: Store at room temperature.
2 M β-Mercaptoethanol: Prepare a 2 M β-Mercaptoethanol solution by combining 0.7 μL of 14.3 M β-Mercaptoethanol with 4.3 mL TE buffer diluted 1:20 in water. (TE buffer is 10 mM Tris–HCl, 1 mM EDTA, pH 7.5 and is provided in the OliGreen® kit.)
Tween® 20: Prepare a 0.1% solution in ultrapure RNase-free water by diluting the previously prepared 10% stock solution 1:100. Store at room temperature.
Quant-iT™ OliGreen® ssDNA Assay Kit: Store at 4 °C (see Note 2).
Sense oligonucleotides: Store at −80 °C. These sense RNA strands will be used to generate a standard curve.
3 Methods
Subheadings 3.1 and 3.2 describe how to attach siRNA duplexes to AuNPs and are depicted in Fig. 1. Subheadings 3.3 and 3.4 describe how to quantify siRNA sense and antisense strands bound to AuNPs and are depicted in Fig. 2.
3.1 siRNA Duplex Preparation
Thaw sense and antisense oligonucleotides to room temperature and keep on ice throughout the procedure. Dilute oligonucleotides in RNase-free duplex buffer to the desired concentration (see Note 9).
Mix equal molar amounts of sense and antisense oligonucleotides in an RNase-free microcentrifuge tube (see Note 10).
Heat the combined oligonucleotides to 95 °C for 5 min, shaking at 400 rpm in a thermomixer.
Cool the oligonucleotides to 37 °C slowly over 1 h, shaking at 400 rpm (see Note 11).
Duplexed siRNA should be used immediately for conjugation or stored at −80 °C.
3.2 Oligonucleotide Conjugation to AuNPs
Add Tween® 20 to a final concentration of 0.2% Tween® 20 to a solution of 10 nM RNase-free 13 nm AuNPs.
Add NaCl to the Tween-stabilized AuNPs to a final concentration of 150 mM. Incubate 5 min at room temperature.
Add the previously prepared siRNA duplexes at a concentration of 1 nmole siRNA per mL of 10 nM AuNPs (see Note 12). Sonicate for 30 s using a sonicator bath, and incubate the particles at room temperature on a rocking platform at a moderate speed for 4 h.
After 4 h, increase the NaCl concentration of the solution to 350 mM. Sonicate again for 30 s in the sonicator bath, and incubate the particles overnight at room temperature on a rocking platform at a moderate speed (see Note 13).
After overnight incubation, backfill any remaining empty space on the AuNPs’ surfaces with mPEG-SH by adding mPEG-SH to a final concentration of 10 μM. Incubate at room temperature on a rocking platform at a moderate speed for 4 h (see Note 14).
Purify the siRNA-coated AuNPs by sequential centrifugation. For 13 nm AuNPs, centrifuge at 21,000 × g for 30 min to form a pellet. Remove the supernatant and dilute the AuNP pellet to half of the original volume with RNase-free 1× PBS. Repeat this procedure three times, resuspending to the desired concentration after the final wash (see Note 15).
To determine the concentration of the siRNA-coated AuNPs, dilute a small volume of the sample 1:100 in water and measure the extinction at 520 nm in a spectrophotometer. For 13 nm AuNPs, an extinction coefficient of 2.70 × 108 L/(mol·cm) can be used to calculate the concentration using Beer’s law (see Note 16).
Store siRNA-coated AuNPs at desired concentration at 4 °C. We typically store siRNA-coated AuNPs at or above 100 nM.
3.3 Quantification of Antisense Oligonucleotides Bound to AuNPs
Dilute siRNA-coated AuNPs (prepared as in Subheading 3.2 and stored at 100 nM) to 6.67 nM in 8 M urea to a final volume of 300 μL in an RNase-free microcentrifuge tube. Incubate at 45 °C for 20 min in a thermomixer shaking at 400 rpm to dehybridize the antisense strands from the sense strands, which will remain bound to the AuNPs (Fig. 2).
Add 300 μL of 0.1% Tween® 20 to the urea-treated AuNPs, then centrifuge the solution at 21,000 × g for 30 min.
Collect and transfer the supernatant, which contains the dehybridized antisense RNA strands, to an RNase-free tube. The AuNP pellet containing surface-bound sense strands can be stored at 4 °C until sense strand quantification.
Prepare a standard curve of known antisense concentration by diluting single-stranded antisense oligonucleotides in RNase-free water. The following antisense concentrations are recommended: 80, 40, 20, 10, 5, 0 nM antisense RNA (see Note 17).
In a black-walled 96-well plate, add 76.6 μL of each antisense RNA standard to individual wells. Add 23.4 μL of 8 M urea to each standard to match the concentration of urea in the samples prepared from siRNA-coated AuNPs. It is recommended to prepare the standard curve in duplicate or triplicate wells to minimize error.
In the 96-well plate, add 50 μL of each antisense sample obtained from steps 1–3 of Subheading 3.3 to three separate wells. Dilute the samples 1:2 by adding 50 μL of RNase-free water to each well.
Prepare the Quant-iT™ OliGreen® reagent. First, prepare 1× TE Buffer by adding 250 μL 20× TE Buffer to 4.75 mL RNase-free water. Dilute the OliGreen® reagent 1:200 by adding 25 μL OliGreen® reagent to the 1× TE Buffer.
Add 100 μL of the diluted OliGreen® reagent to each well containing antisense samples obtained from the AuNPs or containing antisense RNA standards.
On a microplate reader, measure the fluorescence intensity of the samples using an excitation wavelength of ~480 nm and an emission wavelength of ~520 nm (see Note 18).
Use the fluorescence intensities for each point in the standard curve to calculate the concentration of antisense RNA in the samples prepared from siRNA-coated AuNPs (see Note 19). The number of antisense strands loaded per AuNP can be calculated by dividing the sample antisense concentration by the AuNP concentration represented in the plate, taking into account the sample dilutions throughout the procedure. Typical antisense siRNA loading on 13 nm AuNPs and on 150 nm nanoshells is provided in Fig. 3.
3.4 Quantification of Sense Oligonucleotides Bound to AuNPs
Remove the AuNP pellet obtained from step 3 in Subheading 3.3 from the refrigerator. Gently pipette to remove all of the remaining supernatant, which contains antisense strands that have been dehybridized from the AuNPs. Be careful not to remove any of the AuNP pellet, which contains surface-bound sense RNA strands (Fig. 2).
Dilute the AuNP pellet in 500 μL RNase-free 1× PBS and vortex. The nanoparticles should disperse into the solution.
Dilute the AuNPs 1:2 by combining 250 μL sample and 250 μL RNase-free water and read the extinction at 520 nm. Calculate the AuNP concentration as previously described (Subheading 3.2, step 7).
In a fume hood, combine 100 μL of the AuNP solution from step 2 with 100 μL of 2 M β-Mercaptoethanol diluted in 1× TE buffer in an RNase-free microcentrifuge tube (Fig. 2) (see Note 20).
Wrap the microcentrifuge tube in aluminum foil and incubate on a rocking platform at a moderate speed at room temperature for 24 h (see Note 20).
In a fume hood, remove the aluminum foil wrapper from the sample. Add 200 μL 0.1% Tween® 20 to the AuNP sample, and then centrifuge at 21,000 × g for 30 min to form a pellet.
Working in a fume hood, collect the supernatant, which contains the sense strands that have been displaced from the AuNPs’ surfaces, and place it into a new RNase-free microcentrifuge tube.
Prepare a standard curve of known sense oligonucleotide concentrations by diluting sense oligonucleotides in RNase-free water. The following sense concentrations are recommended: 40, 20, 10, 5, 1, 0 nM sense RNA (see Note 21).
Prepare the Quant-iT™ OliGreen® reagent as previously described (Subheading 3.3, step 7).
In a black-walled 96-well plate, place 100 μL of each sense standard into individual wells in a 96-well plate. It is recommended to run each standard in duplicate or triplicate wells.
Place 50 μL of the sense-containing supernatant obtained in step 7 into three individual wells. Dilute the samples 1:2 by adding 50 μL RNase-free water to each well.
Add 100 μL of diluted OliGreen® reagent to each well containing sense RNA samples obtained from the AuNPs or containing sense RNA standards.
On a microplate reader, measure the fluorescence intensity of the samples using an excitation wavelength of ~480 nm and an emission wavelength of ~520 nm (see Note 18). Compare the fluorescence intensity of the samples obtained from the AuNPs to that of the standard curve to determine the amount of sense strands as described previously (step 10 of Subheading 3.3). Calculate the number of sense RNA strands per AuNP by dividing the sample sense concentration by the AuNP concentration determined in step 3 of Subheading 3.4, taking into account the sample dilutions throughout the procedure. Typical sense RNA loading on 13 nm AuNPs and on 150 nm nanoshells is provided in Fig. 3.
Acknowledgments
The authors acknowledge support from the University of Delaware Research Foundation, the W.M. Keck Foundation, Grant IRG14-251-07-IRG from the American Cancer Society, and an Institutional Development Award (IDeA) from the National Institutes of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under grant number U54-GM104941. J.R.M. received support from a National Defense Science and Engineering Graduate Fellowship from the Department of Defense.
Footnotes
We use siRNA that is thiolated at the 3′ end of the sense strand, but other positions for the thiol group would also be suitable.
We describe the use of the Quant-iT™ OliGreen® kit for this assay, but other dyes that are weakly fluorescent until they bind nucleic acids, which then amplifies their fluorescence, would be suitable as well. The OliGreen® dye is advantageous in that it is highly sensitive with the ability to accurately detect nucleic acids as dilute as 100 pg/mL.
It is imperative to use water that is nuclease-free. Ultrapure RNase/DNase-free water can be purchased from a variety of commercial vendors. Alternatively, purified water such as Milli-Q water can be made nuclease-free by adding diethylpyrocarbonate (DEPC) to the water at 0.1% (v/v), heating the solution to 37 °C for several hours, and autoclaving. We autoclave at 121 °C for 40 min for 500 mL of solution, but the time and temperature should be adjusted based on the volume of liquid to be autoclaved.
We use siRNAs with a sense sequence that contains, at the 3′ end, an overhang (such as dithymidine) to increase stability against intracellular nucleases, a short polyethylene glycol (PEG) spacer to enhance loading onto nanoparticles, and a thiol group to bind the gold surface. An example siRNA sense sequence against yellow fluorescence protein (YFP) mRNA is as follows: 5′–UGA CAG UCC AAC UAC AAC AGC TT–PEG36–SH–3′. We typically also use antisense sequences that contain a 3′ dithymidine overhang, but no additional modifications. This protocol may be adapted for use with other siRNA sense and antisense sequences, or for use with other nucleic acids (such as DNA and microRNA).
Upon thawing, RNA sense and antisense strands should be reconstituted in nuclease-free duplex buffer. Nuclease-free duplex buffer at pH 7.5 contains 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 100 mM potassium acetate. Excess RNA molecules may be stored in this duplex buffer.
This protocol uses 13 nm AuNPs produced by the Frens method [27] and stored at a concentration of 10 nM. The protocol can easily be adapted for particles of different sizes or concentrations by scaling the reagents up or down according to the available particle surface area. To render 13 nm AuNPs RNase-free, add 0.1% v/v DEPC, shake at 37 °C for several hours and autoclave. We have observed that some types of nanoparticles, including nanoshells, become unstable upon autoclaving. In this case, we have found that treating the particles with DEPC at 37 °C for 48 h allows sufficient time for DEPC to deactivate RNases and then completely hydrolyze to prevent future side reactions.
We use Tween® 20 in this protocol, but other surfactants such as sodium dodecyl sulfate would also be suitable.
The purpose of the mPEG-SH is to stabilize the nanoparticles. We have tested mPEG-SH molecular weights ranging from 400–5000 Da. In general, higher molecular weight PEG that extends beyond the length of the siRNA duplex will increase nuclease resistance, but may prevent the siRNA from interacting with cells in downstream applications, and lower molecular weight PEG that is much shorter than the siRNA duplex may not be sufficient to prevent the nanoparticles from aggregating. We typically use either 2000 or 5000 Da mPEG-SH for back-filling nanoparticles coating with siRNAs containing ~21 nucleotides per strand. The mPEG-SH utilized should be optimized for specific nucleic acid sequences and intended applications.
Typical resuspension concentration for individual RNA sense and antisense oligonucleotides is ~200 μM in RNase-free duplex buffer. Store lyophilized and resuspended oligonucleotides at −80 °C.
All methods should be completed using RNase-free materials including microcentrifuge tubes and pipette tips. These can be purchased RNase-free (recommended), or rendered RNase-free by DEPC treatment. We recommend spraying and wiping any materials, including pipettes and the lab bench, with an RNase-removal agent to remove any RNases.
To cool siRNA duplexes slowly, set the thermomixer to 37 °C and mix at 400 rpm for 1 h. The siRNA duplexes will anneal as they cool to 37 °C.
The amount of siRNA used during synthesis will require optimization for individual siRNA sequences. We have found that 1–2 nmole siRNA per mL of 13 nm AuNPs at a concentration of 10 nM works well for most sequences. For 150 nm diameter nanoshells, 0.1–0.2 nmole siRNA per mL of nanoshells at a concentration of 0.0045 nM works well.
Generally, increasing the concentration of NaCl added during synthesis will increase the amount of siRNA that loads onto the AuNPs since the NaCl screens charges between duplexes. Additionally, increasing the number of NaCl additions over several hours may improve siRNA loading. This should be optimized for individual sequences and types of nanoparticles.
The amount of mPEG-SH added and duration of incubation should be optimized for individual siRNA sequences and types of nanoparticles. mPEG-SH concentrations have been tested in the range of 5–30 μM, while recommended incubation times range from 1–4 h. Further, incubating mPEG-SH with nanoparticles at 4 °C may improve siRNA loading.
After washing the AuNPs three times, suspend the AuNP pellet in a small volume for storage at your desired concentration. For example, an original volume of 5 mL 10 nM AuNPs should be resuspended in 500 μL 1× PBS after the final wash for storage at ~100 nM. We have stored siRNA-conjugated AuNPs at 100 nM for several weeks without adversely impacting the functionality of the siRNA.
Beer’s Law states that A = εcl where A is the nanoparticle absorbance at its peak resonance wavelength as measured by the spectrophotometer, ε is the extinction coefficient, c is the concentration of the nanoparticles, and l is the path length of the sample. The extinction coefficient for 13 nm AuNPs is provided, but readers should confirm the extinction coefficient for other types of nanoparticles as they adapt this protocol to their needs.
The antisense standard curve will be further diluted in 8 M urea to mimic the urea concentration in the samples prepared from siRNA-coated AuNPs. It is recommended to prepare the standard curve such that the desired antisense concentrations (for example: 80, 40, 20, 10, 5, 0 nM antisense) are reached after adding 8 M urea to the standards. For example, to prepare the standard curve to reach the previously stated final concentrations, dilute antisense RNA to the following concentrations: 104.44, 52.22, 26.11, 13.05, 6.53, 0 nM antisense RNA. While these standard curve concentrations work well for the concentration and loading of siRNA on 13 nm AuNPs presented here, they may need to be optimized for different nanoparticles, nanoparticle concentrations, or loading densities. For example, the final antisense standard curve concentrations for 150 nm diameter nanoshells should be 20, 10, 5, 2, 0.5, 0 nM antisense. It is crucial for the sample fluorescence readings to be within the range of the standard curve.
It is recommended to use excitation ~480 nm and emission ~520 nm, but the exact wavelengths can be altered as long as the excitation and emission wavelengths do not overlap. We typically use excitation at 485 nm and emission at 515 nm.
If the standard curve is linear, the concentration of antisense RNA from the AuNPs can be calculated using y = mx + b where y is the measured fluorescence reading, m is the slope of the standard curve plot, x is the antisense concentration to be calculated, and b is the intercept of the standard curve plot.
We perform this step and all other steps involving β-Mercaptoethanol in a chemical fume hood because the β-Mercaptoethanol is odorous. To minimize odor when transferring the tube containing β-Mercaptoethanol outside the hood, we wrap it in aluminum foil. Alternatively, the rocking platform could be placed in the chemical fume hood.
For nanoshells, we recommend using the following sense standard curve concentrations: 10, 5, 2.5, 1, 0.5, and 0 nM sense RNA.
References
- 1.Deng Y, Wang CC, Choy K, et al. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene. 2014;538:217–227. doi: 10.1016/j.gene.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 2.Hill AB, Chen M, Chen C-K, et al. Overcoming gene-delivery hurdles: Physiological considerations for nonviral vectors. Trends Biotechnol. 2016;34(2):91–105. doi: 10.1016/j.tibtech.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Guo Z, Chen X. Production and clinical development of nanoparticles for gene delivery. Mol Ther Methods Clin Dev. 2016;3:16023. doi: 10.1038/mtm.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HJ, Kim A, Miyata K, et al. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev. 2016;104:61–77. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Ding Y, Jiang Z, Saha K, et al. Gold nanoparticles for nucleic acid delivery. Mol Ther. 2014;22(6):1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng ZJ, Morton SW, Ben-Akiva E, et al. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano. 2013;7(11):9571–9584. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbakry A, Zaky A, Liebl R, et al. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9(5):2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 8.Lee J-S, Green JJ, Love KT, et al. Gold, poly(β-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9(6):2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlman JE, Barnes C, Khan OF, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JB, Hong J, Bonner DK, et al. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giljohann DA, Seferos DS, Prigodich AE, et al. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen SA, Day ES, Ko CH, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5(209):209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randeria PS, Seeger MA, Wang X-Q, et al. siRNA-based spherical nucleic acids reverse impaired wound healing in diabetic mice by ganglioside GM3 synthase knockdown. Proc Natl Acad Sci U S A. 2015;112(18):5573–5578. doi: 10.1073/pnas.1505951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Giljohann DA, Chenc DL, et al. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci U S A. 2012;109(30):11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnaby SN, Lee A, Mirkin CA. Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proc Natl Acad Sci U S A. 2014;111(27):9739–9744. doi: 10.1073/pnas.1409431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi CHJ, Hao L, Narayan SP, et al. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci U S A. 2013;110(19):7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel PC, Giljohann DA, Daniel WL, et al. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug Chem. 2010;21(12):2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giljohann DA, Seferos DS, Patel PC, et al. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7(12):3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raja MAG, Katas H, Wen TH. Stability, intracellular delivery, and release of siRNA from chitosan nanoparticles using different cross-linkers. PLoS One. 2015;10(6):e1028963. doi: 10.1371/journal.pone.0128963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E-Y, Stanton J, Vega RA, et al. A real-time PCR-based method for determining the surface coverage of thiol-capped oligonucleotides bound onto gold nanoparticles. Nucleic Acids Res. 2006;34(7):e54. doi: 10.1093/nar/gkl147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demers LM, Mirkin CA, Mucic RC, et al. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie F, Steven V, Ingram A, et al. Quantitation of biomolecules conjugated to nanoparticles by enzyme hydrolysis. Chem Commun. 2009;2009:2872–2874. doi: 10.1039/b823057a. [DOI] [PubMed] [Google Scholar]
- 23.Randeria PS, Jones MR, Kohlsted KL, et al. What controls the hybridization thermodynamics of spherical nucleic acids? J Am Chem Soc. 2015;137(10):3486–3489. doi: 10.1021/jacs.5b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouri FM, Hurley LA, Daniel WL, et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29(7):732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosi NL, Giljohann DA, Thaxton CS, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 26.Hill HD, Millstone JE, Banholzer MJ, et al. The role radius of curvature plays in thiolated oligonucleotide loading on gold nanoparticles. ACS Nano. 2009;3(2):418–424. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat Phys Sci. 1973;241:20–22. [Google Scholar]