Abstract
Rationale
The relative actions and synergism between distinct myocardial-derived stem cell populations remains obscure. Ongoing debates regarding optimal cell population(s) for treatment of heart failure prompted implementation of a protocol for isolation of multiple stem cell populations from a single myocardial tissue sample to develop new insights for achieving myocardial regeneration.
Objective
Establish a robust cardiac stem cell isolation and culture protocol to consistently generate three distinct stem cell populations from a single human heart biopsy.
Methods and Results
Isolation of three endogenous cardiac stem cell populations was performed from human heart samples routinely discarded during implantation of a left ventricular assist device (LVAD). Tissue explants were mechanically minced into 1 mm3 pieces to minimize time exposure to collagenase digestion and preserve cell viability. Centrifugation removes large cardiomyocytes (CMs) and tissue debris producing a single cell suspension that is sorted using magnetic-activated cell sorting (MACS) technology. Initial sorting is based upon c-Kit expression that enriches for two c-kit+ cell populations yielding a mixture of cardiac progenitor cells (CPCs) and endothelial progenitor cells (EPCs). Flow through c-Kit− mesenchymal stem cells (MSCs) are positively selected by surface expression of markers CD90 and CD105. After one week of culture the c-Kit+ population is further enriched by selection for a CD133+ EPC population. Persistence of respective cell surface markers in vitro is confirmed both by flow cytometry and immunocytochemistry.
Conclusions
Three distinct cardiac cell populations with individualized phenotypic properties consistent with CPCs, EPCs and MSCs can be successfully concurrently isolated and expanded from a single tissue sample derived from human heart failure patients.
Keywords: Adult stem cells, human myocardium, cardiac cell isolation, methodology
Subject Terms: Stem Cells, Basic Science Research, Myocardial Biology, Cardiovascular Disease, Cell Therapy, Cell Biology/Structural Biology
INTRODUCTION
Identification and selection for optimal stem cell type(s) remains a critical issue for successful therapeutic implementation of myocardial regeneration in patients suffering from heart failure. Cell populations from a variety of tissue sources, including bone marrow mononuclear cells, skeletal myoblasts, hematopoietic and endothelial progenitors, and induced pluripotent or embryonic stem cells have been extensively tested for their ability to regenerate lost myocardium1, 2. However, clinical trial outcomes repeatedly fall short of expectations raised by preclinical animal studies3–8 prompting concern regarding the translational impact of experimental models. Nevertheless, there is growing acceptance of the new dogma that the adult mammalian heart is capable of cellular replacement throughout life and in response to pathologic injury, including the cardiomyocyte population (CM)9. Myocardial regeneration is facilitated by resident stem cells that activate endogenous tissue repair by both direct and indirect contribution to cellular replacement of CMs, smooth muscle cells, and endothelial cells. Considering the heart as a self-renewing organ opens up exciting possibilities for therapeutic intervention by cellular activation to promote regenerative processes.
Effective tissue regeneration necessitates not only replacement of CMs, but also vasculature comprised of smooth muscle and endothelial cells. Relative contributions of divergent resident cardiac stem cell types must be appreciated and delineated to maximize therapeutic potential of cell-mediated repair. Coordinated action of multiple cardiac stem cell types forms the basis for normal myocardial biology and therefore will likely be essential to achieve clinically meaningful restoration of tissue structure and function in the wake of pathologic damage. Regardless of whether the ultimate effectors of repair are derived from an adoptively donated cell population or recruited from the endogenous pool(s) of resident cells, it is imperative to establish a working understanding of interactions and contributions of the distinct stem cell types participating in mediation of myocardial homeostasis and repair. Toward that goal, a simple and cost-effective protocol to separate and enrich multiple cardiac stem cells into distinct subsets is essential. Based upon established precedents, three such resident adult cardiac stem cells include cardiac progenitor cells (CPCs) as well as supportive cell types comprised of mesenchymal stem cells (MSCs) and endothelial progenitor cells (EPCs) that can be differentially separated based upon surface marker expression profiling.
Cardiac stem cells were originally isolated in the adult rat heart on the basis of tyrosine-protein kinase Kit (c-Kit) or CD117 expression and were found to lack hematopoietic lineage markers10. A similar population of c-Kit+ CPCs was identified in the adult human heart11, 12, prompting clinical testing to assess their potential efficacy for enhancing myocardial regeneration13, 14. Early patient results have been encouraging showing improved heart function, however advanced age, comorbidities, and genetic factors in patients with heart failure constrain the regenerative capacity of CPCs. Rejuvenation of senescent CPCs, such as with genetic modification15, 16, repeated cell administrations17, or in conjunction with additional supportive cell types18 may improve outcomes for a substantial patient population possessing functionally impaired stem cells.
Stromal MSCs are a supportive cell that contributes to regeneration by secretion of paracrine factors that activate endogenous stem cells, promote angiogenesis, protect CMs and reduce scar formation19, 20, 21. MSCs are multipotent stem cells that give rise to skeletal myoblasts, chondrocytes, and adipose tissue19, 22. Adherent MSCs express cell surface markers CD73, CD105, CD29, CD44, and CD90 while lacking CD45 that is expressed by hematopoietic stem cells (HSCs)23. Lack of major histocompatibility complex II expression allows MSCs to evade host immune responses and overcome host rejection19, 24 leading to their suggested use for allogeneic transplantation into patients25, 26.
EPCs were formerly defined as progenitor cells positive for both the HSC marker CD34 and the endothelial receptor known as vascular endothelial growth factor receptor 2 (VEGFR2)27. Because CD34 is not exclusively expressed on progenitor cells, but also on mature endothelial cells, enrichment with an additional early lineage stem cell marker CD13328 demonstrated that purified CD133+ EPCs differentiate into endothelial cells in vitro29. CD133, also known as prominin or AC133, is a highly conserved antigen with unknown biological activity expressed on EPCs but absent on mature endothelial cells30. EPCs promote paracrine-dependent vasculogenesis and angiogenesis, and form microvessels upon transplantation in vivo31. However, these microvessels regress without support from MSCs to allow vessel maturity32. For this reason combinatorial cell therapies have been suggested for the treatment of heart failure in hopes of synergistic effects,18 but isolation and examination of multiple enriched stem cell subpopulations from a single human heart has not been previously performed.
Knowledge and understanding of myocardial regenerative mechanisms will be facilitated by adoption of the protocol to derive three resident cardiac stem cell populations from adult heart biopsies procured during left ventricular assist device (LVAD) implantation described herein. Our sorting protocol allows for isolation of c-Kit+ CPCs, c-Kit+CD133+ EPCs, and c-Kit−CD90+CD105+ MSCs from patients of varying age and gender. Here we show these three cell populations maintain their unique phenotypic properties during ex vivo cell culture. Phenotypically, these cells show distinct morphology, growth kinetics, cell surface marker and gene expression profiles, and cardiac lineage potential. Isolation of multiple cells types from a single tissue source will allow for concurrent study of cell interactions, empower studies using cells derived from the target human heart failure population that will be involved in regenerative therapy, and expand the repertoire of possibilities for manipulation and modification of stem cells to treat cardiovascular disease. Therefore, the protocol and initial characterizations in this report represent an important and valuable technical advance in the development of novel techniques to facilitate understanding and implementation of regenerative medicine.
METHODS
Human cardiac stem cell isolation
Cardiac biopsies were obtained from patients undergoing LVAD implantation. NIH guidelines for human subject research are consistent with Institutional Review Board (IRB) exemption based upon the use of tissues that are waste discards from normal and routine clinical procedures of LVAD surgery (45 CFR 46.101). After excision, cardiac tissue remained on ice in cardioplegic solution until processed. Fatty tissue was excised and remaining cardiac tissue was suspended in Basic Buffer (15 mL) and minced into 1 mm3 pieces. After mincing, tissue and Basic Buffer were collected in 50 mL Falcon tube. Digestive solution containing collagenase, type II 225 U/mg dry weight (Worthington, #LS004174, Bio Corp, Lakewood, NJ) was dissolved in Basic Buffer (2–2.5 mg/mL) and incubated with tissue pieces for 1.5–2 hours at 37°C with continuous shaking. Digestion solution was refreshed at the one-hour time point and resulting suspensions were centrifuged at 350 g for 5 minutes and resuspended in CPC media (see Table 1). Final suspension was filtered through a 100-μm filter (Corning, #352360) followed by a 40-μm filter (Corning, #352340) and centrifuged at 150 g for 2 minutes to collect CMs. The supernatant was collected and centrifuged at 350 g for 5 minutes and resuspended in CPC media and incubated overnight at 37°C in CO2 incubator.
Table 1.
List of Media
| Component | Catalog Number | |
| Cardiac Stem Cell Medium | F12 HAM’s (1×) | SH30026.01, HyClone |
| 10% ES FBS | 16141079, Gibco | |
| 1% Penicillin-Streptomycin-Glutamine (100×) | 10378016, Gibco | |
| 5 mU/mL human erythropoietin | E5627, Sigma-Aldrich | |
| 10 ng/mL human recombinant basic FGF | HRP-0011, Biopioneer | |
| 0.2 mM L-Glutathione | 66013-256, Sigma-Aldrich | |
| Endothelial Progenitor Cell Medium | EBM-2 Basal Medium | CC-3156, Lonza |
EGM-2 Kit Supplements and Growth Factors:
|
CC-4176, Lonza | |
| Mesenchymal Stem Cell Medium | 10.1 g/L Minimum Essential Medium Eagle, Alpha Modification | M0644, Sigma-Aldrich |
| 20% FBS | FB-01, Omega Scientific, inc. | |
| 1% Penicillin-Streptomycin-Glutamine (100X) | 10378-016, Gibco | |
| Cell Culture Grade Water | ||
| Basic Buffer | 11 g/L Minimum Essential Medium Eagle, Joklik Modification | M0518, Sigma-Aldrich |
| 3 mM HEPES | H3375, Sigma-Aldrich | |
| 1% Penicillin-Streptomycin-Glutamine (100X) | 10378-016, Gibco | |
| 10 mM Taurine | T0625, Sigma-Aldrich | |
| Insulin, solvate in 3% Acetic Acid/PBS | I-5500, Sigma-Aldrich | |
| 1% Amphotericin B | 15290-018, Invitrogen | |
| 50 mg Gentamicin | G1397, Sigma-Aldrich | |
| Cell Culture Grade Water |
The following day, cells in suspension were collected in 50 mL Falcon tube. Any cells attached were dissociated using a 1:1 mixture of Cellstripper (Corning, #25-056-CI) and TrypLE Express (1X) (Thermo Fisher Scientific, #12604-013). Resulting suspension was filtered through a 40-μm filter, centrifuged at 350 g for 5 minutes, and resuspended in wash buffer (PBS plus 0.5% bovine serum albumin). To isolate c-Kit+ cells, suspension was incubated with c-Kit–labeled beads (Miltenyi Biotec, #130-091-332) and sorted according to the manufacturer’s protocol. The c-Kit+ fraction was divided as such: half the population was suspended in CPC media (see Table 1) and the other half was suspended in EPC media (see Table 1). The c-Kit− population was further incubated with CD90/CD105–labeled beads and sorted according to the manufacturer’s protocol (Miltenyi Biotec, #130-096-253/130-051-201). Cells positive for CD90/CD105 were suspended in MSC media (see Table 1). To isolate an EPC population, at 1 week the c-Kit+ population plated in EPC media was further sorted using CD133–labeled beads and sorted according to the manufacturer’s protocol (Miltenyi Biotec, #130-097-049). All cells were cultured at 37°C in CO2 incubator in their respective growth media. CPC and EPC were split 1:2 when they reached 60–70% confluency. MSC were split 1:2 when they reached 90% confluency. Patient information for the five cardiac samples used in this study can be found in Online Table III. All cells used in this study were mid-passage (passages 5–9).
Quantitative Reverse-Transcriptase Polymerase Chain Reaction (qRT-PCR) and bioinformatics
Total RNA was isolated from cardiac stem cell populations using Quick-RNA MiniPrep kit (Zymo Research, #R1055) according to manufacturer’s protocol. 500 ng of RNA were used to generate complementary DNA (cDNA) using an iScript cDNA Synthesis kit (Bio-Rad Laboratories, #170-8891). The amplified cDNA was diluted at a ratio of 1:10 in DNase- and RNase- free water. qRT-PCR were completed using iQ SYBER Green (Bio-Rad Laboratories, #170-8882) on a CFX Real-Time PCR Detection System (Bio-Rad). cDNA was amplified using primers specific to genes of interest (listed in Online Table II). The comparative or ΔΔCt method of qRT-PCR data analysis was used to analyze samples; variability in cDNA concentration was normalized using 18S. Hierarchical clustering and supervised clustering for gene expression profiling were performed using Expander 7.1 software33.
Immunocytochemistry
Cardiac stem cell populations were plated on 2-well chamber glass slides (10,000 cells/well) in their respective growth media (see Table 1) for a minimum of 24 hours. After incubation, slides were washed with PBS and fixed in 4% paraformaldehyde for 5 minutes at 4°C. Following fixation, the slides were washed twice with PBS and permeabolized in PBS plus 0.1% Triton X-100, 0.1 M Glycine for 3 minutes, then washed once with PBS and blocked with TNB (1X TN (Tris-HCl, NaCl) Buffer, 5 μg/mL blocking reagent (PerkinElmer, #FP1012)) for 30 minutes. Primary antibodies were diluted in TNB (see Online Table I) and incubated overnight at 4°C. The following day slides were washed twice with PBS. Fluorescently conjugated secondary antibodies were diluted in TNB (1:200) and incubated 1.5 hours at room temperature. For c-Kit staining a horseradish peroxidase (HRP)-linked secondary antibody (1:500) was used, followed by tyramide signal amplification (1:50) (PerkinElmer, #NEL753001KT). After washing twice with PBS, DAPI was included in a final wash to fluorescently label the nuclei, and slides were coverslipped with Vectashield® mounting reagent (Vector Laboratories, #H-1000). All slides were imaged using a Leica TCS SP8 confocal microscope. A table of antibodies and dilution ratios is available in Online Table I.
Cell morphology measurement
Cardiac stem cell populations were imaged using a Leica DMIL inverted tissue culture phase contrast microscope. Cell morphology was measured by tracing the outline of the cells using Image J software. The three measurements analyzed were Area, Roundness, and Length-to-Width (L/W) ratios. L/W ratios were calculated by dividing Feret/Min Feret measurements. A minimum of 30 cells was measured per cell line.
Cell proliferation assay
Cell populations were plated in quadruplicate (1,000 cells/well) in a 96-well black flat bottom plate with 100 μL/well of their respective growth media. Cell proliferation rate was determined using a CyQUANT Direct Cell Proliferation Assay (Thermo Fisher Scientific, #C35011) on days 0, 1, 3 and 5. Doubling times were calculated based on periods of exponential growth using a population doubling time online calculator (http://www.doubling-time.com/compute_more.php).
Flow cytometry
For live cell analysis, single cells were suspended in 100 μL wash buffer (PBS plus 0.5% bovine serum albumin) and incubated with primary antibody (see Online Table I for dilutions) on ice for 30 minutes. Following, cells were washed with wash buffer and incubated with secondary antibody (1:100) for 20 minutes on ice. For fixed cell analysis, cells were suspended in 4% paraformaldehyde for 5 minutes at room temperature and then washed twice with wash buffer. For c-Kit analysis requiring permeabilization, cells were washed twice and resuspended in PBS plus 0.1% Triton X-100, 0.1 M Glycine for 3 minutes, then washed once. Fixed cells were suspended in 100 μL wash buffer and incubated with primary antibody on ice for 1 hour. Following, cells were washed twice and incubated with secondary antibody (1:100) for 30 minutes on ice. For both fixed and live cells a total of 300 μL wash buffer was added post secondary incubation and the cells were analyzed by flow cytometry with a BD FACS Canto instrument. Unstained and isotype controls were used to establish baseline fluorescence levels. Data was analyzed by Flow Jo software (BD Biosciences). A minimum of 10,000 cell counts was analyzed. Due to low cell count for H13-066 MSC, the c-Kit count was 5,000.
Matrigel tube formation
Growth factor reduced matrigel (Corning, #356231) was used to coat a 96-well flat bottom plate (50 μl/well) and incubated for 30 minutes at 37°C. Cardiac stem cell populations were plated in duplicate (5,000 cells/well) suspended in 100 μL/well of EPC basal medium (see Table 1) and incubated at 37°C in CO2 incubator. Cell tube formation was imaged using a Leica DMIL inverted tissue culture phase contrast microscope 12–16 hours after plating.
MSC- colony-forming unit-fibroblast (CFU-F) assay
Cells were suspended in CFU-F assay medium: DMEM-low glucose (Thermo Fisher Scientific, #11054-020) with 10% FBS, 2 mM L-Glutamine and Gentamicin (10 mg/mL), and plated at 200 cells per 100 mm. Medium changed every 3 days and after 14 days of growth, dishes were washed with PBS and incubated in crystal violet at room temperature for 30 minutes. Solution removed by 4 washes of PBS. After dishes were dry, colonies were enumerated for each plate. A minimum of 3 dishes plated per cell line.
Multilineage mesenchymal differentiation potential
The potential for osteogenesis, adipogenesis, and chondrogenesis differentiation was assessed for the three cardiac stem cell populations using StemPro Differentiation Kits following manufacturer’s protocol (Thermo Fisher Scientific, #A1007201, #A1007001, and #A1007101). For osteocyte differentiation cells were stained with Alizarian-Red Staining Solution (Millipore, #TMS-008-C), for adipocyte differentiation cells were stained with Oil Red O (Sigma-Aldrich, #O0625), and for chondrocyte differentiation cells were embedded in optimal cutting temperature (OCT) compound, cryosectioned at 5–10 μm, and stained with Alcian-Blue Staining Solution (Millipore, #TMS-010-C).
Statistical analysis
Data expressed as mean±SEM. Statistical analyses of multiple groups were assessed by 1-way ANOVA with Bonferroni post hoc test. Multiple groups over time were analyzed by 2-way ANOVA. Statistical analysis was performed using GraphPad Prism version 5.0 software. The Pearson product-moment correlation coefficient was calculated using Microsoft Excel 2010. Experiments were performed in triplicate unless stated otherwise. A p-value of less than 0.05 was considered statistically significant.
RESULTS
Isolation of three distinct cardiac stem cell populations
The three different cardiac stem cell populations were isolated and expanded as described in the Methods section: Human Cardiac Stem Cell Isolation (Figure 1). At the time of extraction, the c-Kit− MSC population comprises 90–95% of our isolated stem cell pool, whereas c-Kit+ cells comprise 5–10% of the isolated cells. One week later, roughly half of the c-Kit+ cells are also positive for the endothelial progenitor marker CD133+.
Figure 1. Cardiac stem cell isolation protocol.
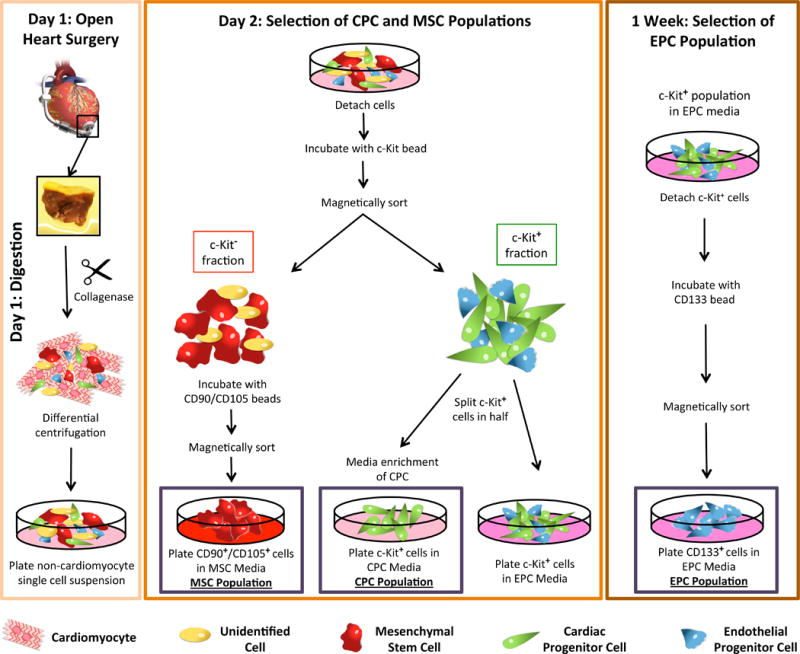
Schematic representation of the process used to isolate cardiac cells from LVAD tissue. Following open-heart surgery the tissue plug is digested to the single cell level and, following centrifugation to remove the cardiomyocytes, plated overnight at 37°C in CO2 incubator. Day 2, the single cell suspension is incubated with microbeads conjugated to c-Kit and magnetically sorted. C-Kit+ cells are split in half with a portion being plated in CPC media and the other half being plated in EPC media. The c-Kit− fraction is further sorted for CD90 and CD105 with positive cells being plated in MSC media. 1 week later, the c-Kit+ cells plated in EPC media are further enriched for CD133. Red cells indicate mesenchymal stem cells (MSC); green cells indicate cardiac progenitor cells (CPC); blue cells indicate endothelial progenitor cells (EPC); yellow cells indicate non-specific cell population.
Distinctive cell morphology among cardiac stem cell populations
After expansion ex vivo, mid-passage cells were assessed for morphometrics. Phase contrast imaging (Figure 2A and Online Figure I) measured parameters of Area, Roundness, and L/W Ratio. MSC area is significantly larger (23,301±1,018) relative to both CPC (7,435±358) and EPC (4,738±202) (Figure 2B). EPCs are significantly more round (EPC, 0.57±0.016; CPC, 0.26±0.012; MSC, 0.38±0.016) (Figure 2C), while CPCs show increased L/W ratio (CPC, 4.2±0.16; EPC, 2.0±0.081; MSC, 3.0±0.12). Representative examples of cultures of the three cell types show close clustering of morphometric parameters (Online Figure II) with minor variation between individual patients.
Figure 2. Isolation of three distinct cardiac stem cell populations from LVAD patients.
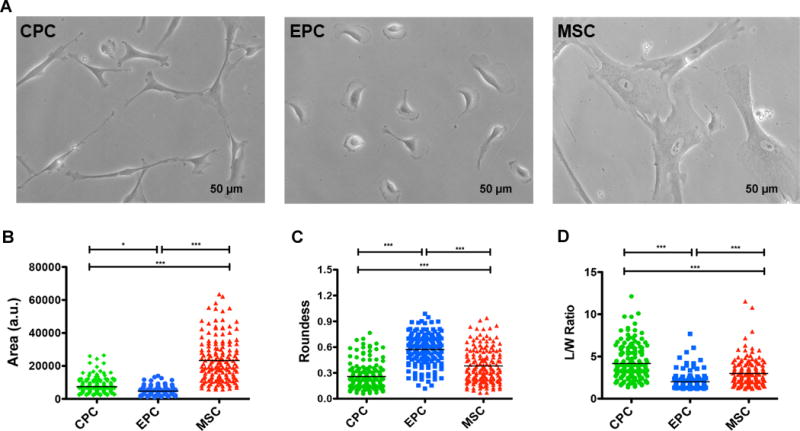
A, Representative phase contrast images for the three isolated cardiac cell populations. B-D, Cell morphometric parameters measuring area (B), roundness (C), and length-to-width (L/W) ratio (D) (n=4–5 patients, minimum of 30 cells traced per cell type per patient). Data are presented as 1 way ANOVA, *p<0.01, **p<0.001, ***p<0.0001. Scale bar, 50 uM.
Cell population kinetics vary by cell type
Population growth kinetics was determined by CyQuant proliferation assay (Figure 3A–3C). The MSC population exhibited slowest proliferation rate (doubling time: 119±35 hours) in agreement with prior studies using mid-passage MSC populations34–36. CPCs and EPCs show markedly faster proliferation rates (doubling times: CPC, 33±2 hours; EPC, 35±7 hours) (Figure 3D and 3E). Cell type growth kinetics varies by patient indicative of heterogeneity in cell biology between patient isolates (Online Figure III).
Figure 3. Cardiac stem cell populations exhibit distinct growth kinetics.
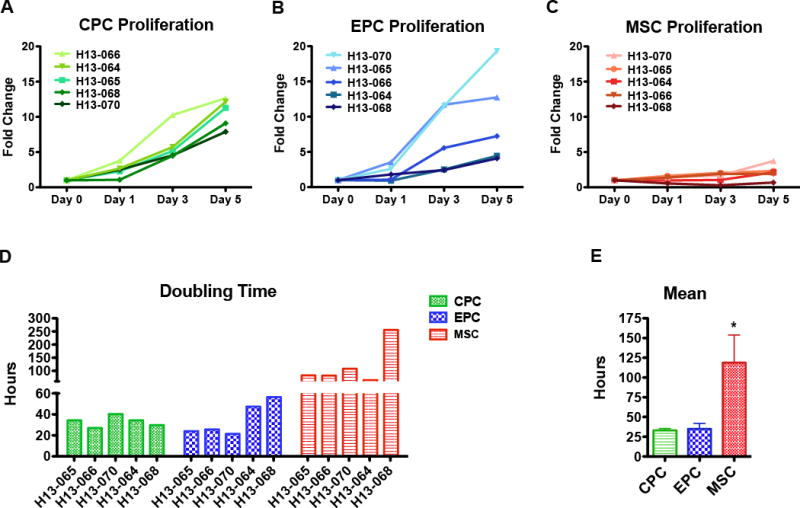
A-C, Cardiac cell proliferation measured using CyQuant assays at day 0, day 1, day 3, and day 5 for CPC (A), EPC (B), and MSC (C). D, Bar graph of doubling times calculated using periods of exponential cell growth for each cell line. E, Bar graph of mean doubling time for each cell type (n=5 patients). Data are presented as 1 Way ANOVA, *p<0.05.
Flow cytometry analysis of markers expressed upon in vitro culture
Persistence of markers used to isolate the cardiac subpopulations was analyzed by flow cytometric analysis of single cell suspensions with fluorescence signal comparison between the differentially enriched cell types (Figure 4 and Online Figure IV) and variance among patients (Online Figure V). All cells were negative for HSC marker CD45 (CPC, <1%; EPC, ~3%; MSC, <1%)(Online Figure V) as expected for cardiac-derived cell populations. CPCs and EPCs were initially isolated for c-Kit surface expression, however the extent of c-Kit expression and internalization varied among the three populations: CPC were ~97% positive for c-Kit, while EPC were ~43% and MSC were ~27% (Figure 4A). CPCs internalized c-Kit, while c-Kit expression did not change upon permeabilization for EPCs and MSCs (Online Figure VI). These observations are consistent with prior studies showing surface c-Kit expression is variable37–39, a characteristic of rapidly cycling receptors.
Figure 4. Flow cytometry analysis of markers expressed upon in vitro culture.
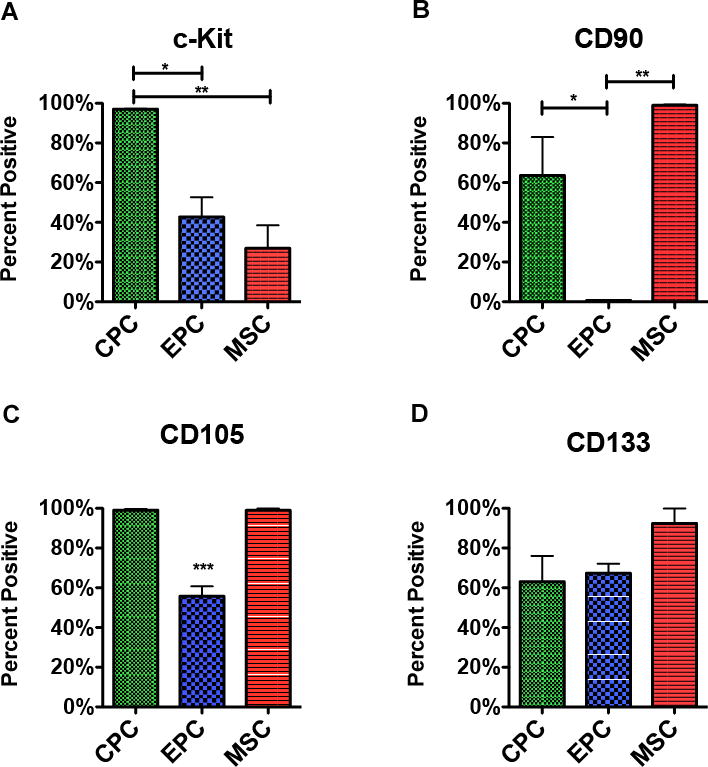
A-D, Single cell suspensions stained with antibodies for the markers used in cell isolation. Flow cytometry analysis of cardiac cell populations sorted for the presence of c-Kit (A), CD90 (B), CD105 (C), and CD133 (D) (n=3 patients per marker). Data are presented as 1 way ANOVA, *p<0.05, **p<0.01, ***p<0.001.
MSC populations were ~100% positive for both mesenchymal markers CD90 and CD105, with insignificant change between live (Figure 4B and 4C) and fixed cells (Online Figure V). While ~99% of CPCs expressed CD105, only a little over half (~64%) expressed CD90 (Figure 4B and 4C). Of the three cell populations, EPCs had the lowest parentage of cells positive for mesenchymal markers (CD105: ~56%; CD90: <1%)(Figure 4B and 4C). Whereas fixation did not alter the percent of MSCs positive for CD90 and CD105, fixation did decrease the percent of CPCs and EPCs positive for these two mesenchymal markers (CD90: fixed CPC, ~40%; fixed EPC, <1% and CD105: fixed CPC, ~28%; fixed EPC ~11%)(Online Figure V). CD133 could not be detected on live cells; with fixation CD133 could be identified on all cell populations (EPC, ~67%; CPC, ~63%; MSC, ~92%)(Figure 4D and Online Figure V).
Immunocytochemistry corroboration of flow cytometry data
Immunofluorescence microscopy was utilized to assess endogenous expression levels of the panel of markers used for stem cell isolation. Expression of c-Kit in CPCs was uniformly high, whereas c-Kit expression was comparatively low in MSCs. While a subset of the EPC population (marked by asterisks) expressed high levels of c-Kit, the majority expressed low to undetectable levels (Figure 5A). CD133 was prominently expressed by all three cells types (Figure 5B), consistent with flow cytometry data. Antibody labeling confirmed clear expression of mesenchymal markers CD90 and CD105 by the MSC population. Both mesenchymal markers were barely detectable above background in the EPC population with very low immonolabeling for CD105. While CPCs expressed CD105, CD90 level in CPCs was relatively low in comparison to the MSC population (Figure 5C).
Figure 5. Immunofluorescence imaging of stem cell markers.
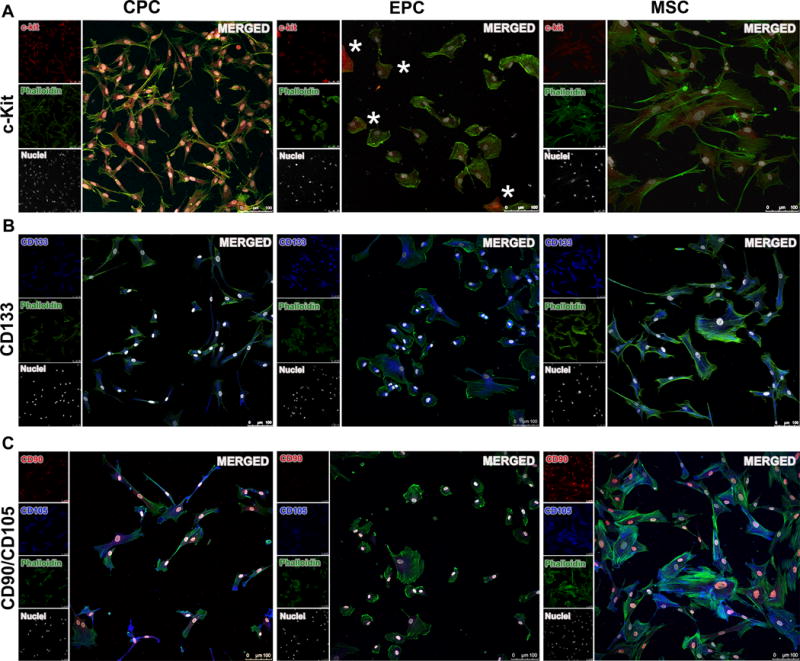
A, Representative florescence microscopy showing c-Kit localization in CPC, EPC, and MSC (red, c-Kit; green, phalloidin; white, nuclei (DAPI)). B, Immunofluorescence labeling of the mesenchymal markers CD90 and CD105 showing varying degree of co-localization for these two markers in the three cell populations (red, CD90; blue, CD105; green, phalloidin; white, nuclei (DAPI)). C, CD133 expression revealed by immunofluorescence in the cardiac cells (blue, CD133; green, phalloidin; white, nuclei (DAPI)). Scale bar, 100 um. Asterisks (*) indicate cells positive for c-Kit.
Gene expression profiles show Epcs diverge from Cpcs and Mscs
Transcriptional signatures for each cell type from various patients was performed using quantitative qRT-PCR focused upon three groups of genes: 1) growth factors and cytokines 2) extracellular matrix (ECM) proteins and 3) inflammatory factors (Online Table II). Results of qRT-PCR were subjected to bioinformatic analyses by hierarchical clustering for 45 samples (5 patients × 3 cell types × 3 replicates). Supervised clustering analysis revealed that CPCs and MSCs have closely related transcriptional profile, whereas EPCs are divergent. Two groups of genes differentiate EPCs from CPCs and MSCs: one group showed elevated gene expression (ANGPT2, PECAM1, COL3A1, HGF, IGF2, IRF1, TIMP1 and TNF) whereas another showed diminished gene expression only in EPCs (COL1A1, FGF2, HBEGF, IL1B, IL6, MMP1, NRG1 and CXCL12) (Figure 6A and 6B). Pearson product-moment correlation coefficient heat map matrix of individual patients revealed that four out of five patients showed high gene expression correlation (>0.82), meaning although there is inherent heterogeneity among patient samples, individual cell types display characteristic profiles (Online Figure VII).
Figure 6. Supervised cluster analysis of gene expression levels in cardiac stem cells.
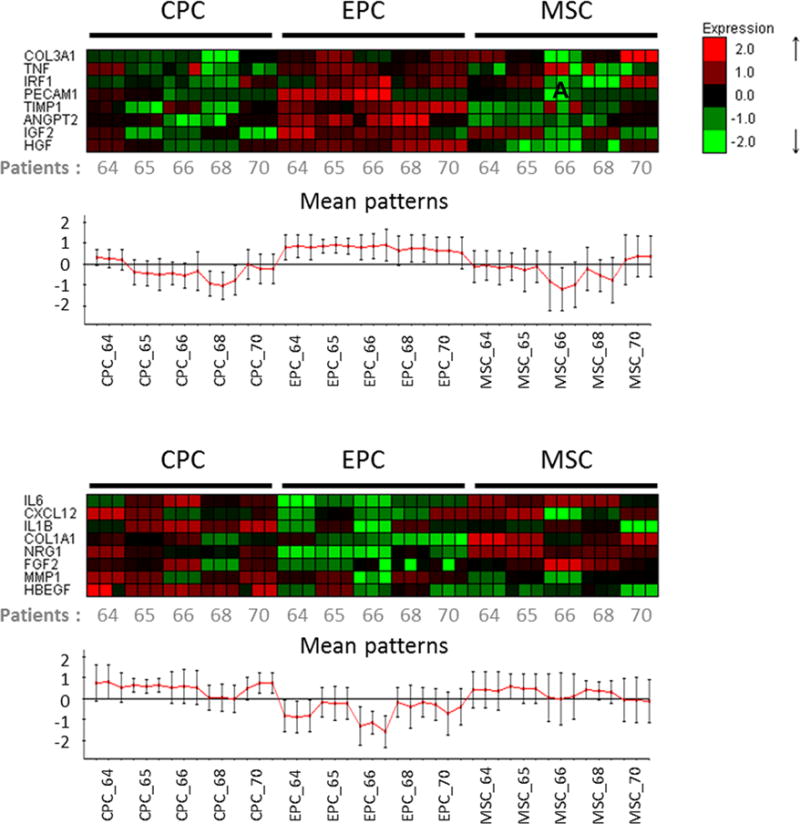
A-B, Heatmaps representing differentially expressed genes in three different cell types. A, Cluster of genes showing specific up-regulation in EPC, but not CPC and MSC. B, Second cluster of genes showing specific down-regulation in EPC compared to CPC and MSC (n=5 patients).
In vitro lineage assessment and comparison to non-cardiac controls
Tube formation assay using growth factor-reduced Matrigel demonstrates angiogenic potential of the three cardiac stem cells in vitro. Ability to form tubular networks varied among patients as well as cell types, with only a few CPC lines being able to form rudimentary tubules (Figure 7A), EPCs were able to form robust tubular networks within 15 hours (Figure 7B) similar to HUVEC control (data not shown), whereas MSCs could not form tubular structures, instead producing “star-burst” structures (Figure 7C).
Figure 7. In vitro lineage assessment and comparison to established cell lines.
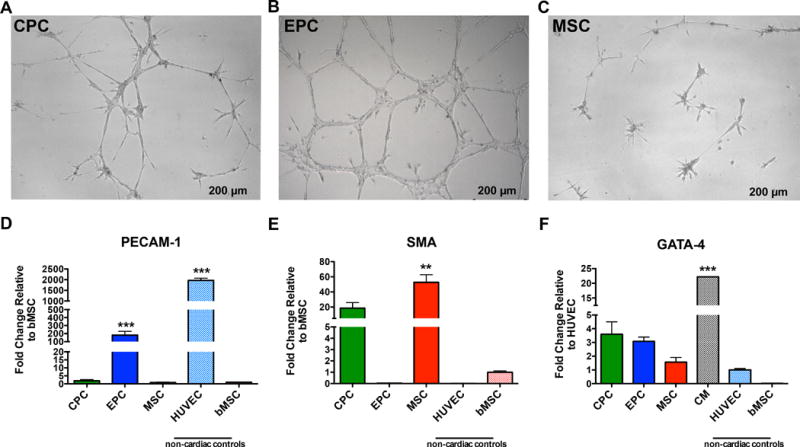
A-C, Representative images of tubular network formation when plated on growth factor reduced matrigel for CPC (A), EPC (B), and MSC (C). D–F, Bar graphs using established cell lines, HUVECs, bMSCs and adult CM, to assess the potential of cardiac stem cells to commit to an angiogenic (D), smooth muscle (E), and cardiogenic fate (F) (n=3 patients). Data are presented as 1 Way ANOVA, **P<0.001, ***p<0.0001, versus cell type used for normalization. Scale bar, 200 um. CM indicates Cardiomyocytes; GATA4, GATA binding protein 4; PECAM-1, Platelet endothelial cell adhesion molecule; and SMA, α-smooth muscle actin (SMA).
Multilineage mesenchymal differentiation assays to determine adipocyte, chondrocyte, and osteocyte potential were performed using MSC cultures (Online Figure VIII). Similar to bone marrow derived MSCs (bMSCs), cardiac derived MSCs differentiate into adipocyte, chondrocyte, and osteocyte lineages, whereas EPCs and CPCs did not exhibit phenotypic characteristics of fully committed cell types. Cardiac derived MSCs showed comparable levels of major histocompatibility complex (MHC) molecules, both MHC Class I, Class II, as well as co-stimulatory molecules to bMSC (Online Figure IX) and produce colony forming units (data not shown).
HUVECs and bMSCs were used to assess the potential of cardiac stem cell commitment toward angiogenic or smooth muscle fate at the transcript level. HUVECs and EPCs expressed the highest levels of PECAM-1 (HUVEC, 1,967±106; EPC 181±47) with respect to either CPCs (1.9±0.69) or MSCs (0.85±0.25) (Figure 7D). SMA is expressed by both bMSCs (1.0±0.12) and cardiac MSCs (52.67±10.06), and to a lesser extent CPCs (18.60±7.27). Both the EPCs and HUVECs expressed near undetectable levels of SMA (EPC, 0.04±0.01; HUVEC, 0.01±0.01) (Figure 7E). GATA4 is expressed by the adult CMs (22.19±0.06) and to a lesser degree by the CPCs (3.6±0.91). The other cell types showed lower expression of GATA4 (EPC, 3.09±0.31; MSC, 1.57±0.34) (Figure 7F).
DISCUSSION
Unbiased sampling of tissue biopsies from heart failure patients undergoing LVAD implantation represents an ideal source for tissue to isolate and study characteristics of CPCs, EPCs, and MSCs in the context of decompensated heart failure. LVAD recipients on “destination therapy” are desperately in need of regenerative therapy as the only other available option for treatment is cardiac transplantation. In an effort to improve the rather modest outcomes of current cell-based regenerative medicine intervention, the use of multiple cell types in combination or sequentially could enhance efficacy. Indeed, experimental animal studies support the premise of combining CPC and MSC to enhance beneficial effect18 and ongoing clinical trials are moving this concept into patients. However, these studies are focused upon allogeneic approaches that limit survival, engraftment, and persistence of the donated cell population. The protocol described in this report allows for simultaneous isolation of three distinct cell populations from a single tissue sample: CPC, MSC, and EPC. Each cell type exhibits characteristics that render them desirable to promote regenerative repair. EPCs promote vasculogenesis and angiogenesis and differentiate into mature endothelial cells. In vivo, MSCs have been shown to contribute to endogenous regeneration by secretion of paracrine factors that activate endogenous stem cells, promote angiogenesis, protect CMs and reduce scar formation19, 20, 21. In animal models, transplanted CPCs give rise to CMs, smooth muscle cells, and endothelial cells40, 41, but lack the power of MSCs to activate and recruit endogenous stem cells. For this reason combinatorial cell therapies have been advanced in hopes of initiating synergistic effects in myocardial repair, but no one to date has attempted to inject multiple stem cells expanded solely from the human heart. With this protocol, combinatorial studies can be performed using three cardiac cell populations isolated from a single heart. These cells can be studied combinatorial or individually to better elucidate how these distinct cell populations regulate and/or contribute to cardiac regeneration following ischemic injury.
Cardiac stem cell populations play intrinsically distinct roles in cardiac regeneration. Insight into what makes each cell type unique will translate to superior clinical application depending upon disease state of the patient. For example, for chronic ischemia the primary intent may be to salvage the CMs and improve blood supply. Consequently, using a cell type that induces vascular regeneration such as EPCs would be important. On the other hand, if the goal were to replace lost CMs, perhaps CPCs supported by MSCs would favor activation of endogenous cardiomyocytes as well as exhibiting inherent cardiomyogenic potential. Alternatively, co-injection of all three cardiac stem cell types may provide the most robust reparative and regenerative outcome. Achieving long-lasting myocardial benefits likely requires the interaction of multiple cardiac cell types and testing this hypothesis is the focus of ongoing experimental studies.
The CPC, MSC, and EPC populations can be reproducibly isolated using samples obtained from multiple patients with varying age and disease etiologies up to 84 years of age. Patients presented with a range in cardiomyopathies and comorbidities including diabetes and coronary artery disease, and/or heavy smokers and drinkers. Success rate of isolating the three cell populations was 80–90%. For a minority of patient samples received the ex vivo culture resulted in outgrowth from only two populations, which we attribute to a number of possible factors including tissue size, culture error, or absence of a particular cell type in the biopsy sample received. Not to be overlooked, another potential explanation for inability to expand a particular cell type could be correlated patient etiology and will be interesting to examine in future studies. We were unable to establish such relationships owing to the relatively small number of patients assessed in our isolation sampling. While showing some variation, consistent trends are evident with regard to cell morphology (Figure 2), growth kinetics (Figure 3), and gene profiles (Figure 6). Specifically, gene expression levels for cytokines, extracellular matrix proteins, paracrine and inflammatory factors clustered based on the three cardiac stem cell lineages. EPCs were transcriptionally distinct from CPCs and MSCs that possessed more similar profiles (Figure 6). Transcript profiles of EPCs isolated from the heart were comparable to observed profiles in HUVECs (an established endothelial cell line), and cardiac EPCs also retain the capacity to form tubular networks on Matrigel, consistent with endothelial cell phenotypic properties (Figure 7).
This protocol provides detailed culture procedures that will allow for stem cell maintenance evidenced by preservation of stem cell markers (Figure 4 and Figure 5). Key factors for successful propagation and expansion ex vivo are culture conditions and growth medium. Each cell type has its own preferential plating density and depending on cell type, differentiation and/or senescence can occur with prolonged culture15, 34 or due to cell-cell contact if cultured to confluency42, 43. CPCs and EPCs are successfully cultured at 50–70% confluency. On the other hand, MSCs prefer close contact with neighboring cells (70–90% confluency), appear cytopathic, and grow extremely slow when cultured below 60% confluency.
Although the initial relative abundance of MSCs is higher compared to either CPCs or EPCs following isolation, the latter two populations proliferate quickly (Figure 3) and senesce at a later passage than MSCs. The MSC population in most patient samples slows down proliferation around passages 8–10 and will completely senesce a few passages later. Considering the demands of cell manufacturing for clinical utilization, approximately 100 million cells can be produced by passage 10. The majority of CPC and EPC lines continue to proliferate until passages 15–20, and thus these two cell types can readily produce hundreds of millions of cells.
The protocol described herein is a highly reproducible and straightforward method independently conducted by multiple individuals at varied training levels ranging from graduate to post-doctoral level. Additionally, the procedure is amenable to being performed under a wide variety of experimental conditions. An example already initiated in our laboratory includes: splitting a single cardiac biopsy in half and conducting initial isolation under normoxic versus hypoxic conditions to study altered oxygen concentration and resulting phenotypic changes that occur in the isolated cardiac stem cell populations. Other applications could include variation in media formulation, such as glucose or growth factor supplementation, or examining differences caused by extracellular matrices and biologically coated surfaces.
Desperate unmet need to alleviate the suffering and burden of heart failure has understandably prompted a rapid progression into clinical trials while necessarily foregoing a deeper fundamental understanding of cardiac stem cell biology or identification of which cell or combination of cells yields the most efficacious outcome to mediate repair and regeneration in vivo. Initial clinical trials performed using bone marrow-derived stem cells were quickly pursued, in part owing to relative ease of isolation and established protocols, but yielded generally disappointingly modest myocardial recovery.44 The challenging frontier for myocardial regenerative medicine with the ever-expanding tapestry of potential interventional strategies requires careful analysis of critical factors in cell biology such as self-renewal potential, survival, and mechanisms that allow particular cell population(s) to repopulate the damaged myocardium more effectively than others. Each individual cell type may be specialized to perform in a specific context, and combinations of cell types likely will exert actions through concerted cooperative networking that any one single cell type cannot provide. Equipped with this basic understanding, secondary considerations of cell dosage, timing and delivery approach will need to be optimized. These concepts represent a small sampling of the nearly limitless conceptual possibilities that lie ahead for myocardial regenerative research. All journeys begin with a single step, and this protocol paves the way by isolating multiple cardiac cell populations that can be studied individually or combinatorial so that the field of cardiac stem cell therapy can come to a better understanding on which stem cell population(s) hold the most promise for cardiac regeneration.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Stem cell transplantation for heart repair shows efficacy in both animal studies and clinical trials, but relative actions and synergism between distinct myocardial-derived stem cell populations remains obscure.
Cardiac Progenitor Cells (CPCs) resident within the human myocardium promote replacement of cardiomyocytes (CMs) and vasculature (smooth muscle and endothelial cells) following injury.
Mesenchymal Stem Cells (MSCs) contribute to regeneration primarily through paracrine secretion.
Endothelial Progenitor Cells EPCs) contribute to microvessels and vascular structure.
What New Information Does This Article Contribute?
We describe a simple and cost-effective protocol to isolate and enrich for multiple human cardiac stem cell populations from a single myocardial sample that maintain their individual unique phenotypic properties during ex vivo cell culture.
Isolation of multiple stem cell types from a single human heart sample allows for concurrent study of cell interactions and development of novel techniques to facilitate understanding and implementation of cardiac regenerative medicine.
Desperate unmet need to alleviate the suffering and burden of heart failure has understandably prompted rapid progression to clinical trial while foregoing deeper fundamental understanding of cardiac stem cell biology or identification of which cells most effectively mediate repair and regeneration. Our protocol paves the way for greater understanding and improved outcomes through isolation of multiple cardiac cell populations from a single tissue sample. This approach facilitates studies to determine which stem cell population(s) hold the most promise for cardiac regeneration studied either individually or in combination. Our sorting protocol allows for isolation of c-Kit+ CPCs, c-Kit+CD133+ EPCs, and c-Kit− CD90+CD105+ MSCs from patients that vary in both age and gender. Phenotypically, these cells show distinct morphology, growth kinetics, cell surface marker and gene expression profiles, and cardiac lineage potential. Each cell type exhibits characteristics desirable to promote regenerative repair, with further careful analysis leading to elucidation of the mechanistic basis underlying the ability of selected cell population(s) to repair damaged myocardium.
Acknowledgments
We gratefully acknowledge Dr. Roland Wolkowicz and Cameron Smurthwaite from the San Diego State University flow cytometry core facilities for allowing us to use their equipment and Dr. Walter Dembitsky from Sharp Memorial Hospital for providing the Sussman lab with the human heart biopsy samples used for our cell isolations.
SOURCES OF FUNDING
M. Monsanto is supported by NIH grant R01HL122525, Rees Stealy Research Foundation, Achievement Rewards for College Scientists (ARCS), and Elliott Family Fund Scholarship. T. Kim is supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1A6A3A03019855). M. Sussman is supported by NIH grants: R01HL067245, R37HL091102, R01HL105759, R01HL113647, R01HL117163, P01HL085577, and R01HL122525, as well as an award from the Fondation Leducq.
Non-standard Abbreviations and Acronyms
- ANGPT2
angiopoietin-2
- bMSC
bone marrow derived mesenchymal stem cell
- cDNA
complementary DNA
- CM
cardiomyocyte
- COL1A1
collagen type I alpha 1
- COL3A1
collagen type III alpha 1
- c-Kit
tyrosine-protein kinase Kit or CD117
- CPC
cardiac progenitor cells
- CXCL12
C-X-C motif chemokine 12 or stromal cell-derived factor 1 (SDF1)
- ECM
extracellular matrix
- EPC
endothelial progenitor cell
- FGF2
basic fibroblast growth factor
- GATA4
GATA binding protein 4
- HBEGF
heparin-binding EGF-like growth factor
- HGF
hepatocyte growth factor
- HRP
horseradish peroxidase
- HSC
hematopoietic stem cell
- HUVEC
human umbilical vein endothelial cell
- IGF2
insulin-like growth factor 2
- IL1B
interleukin 1 beta
- IL6
interleukin 6
- IRF1
interferon regulatory factor 1
- LVAD
left ventricular assist device
- MACS
magnetic-activated cell sorting
- MHC
major histocompatibility complex
- MMP1
matrix metalloproteinase-1
- MSC
mesenchymal stem cell
- NRG1
neuregulin 1
- OCT
optimal cutting temperature
- PECAM1
platelet endothelial cell adhesion molecule or CD31
- qRT-PCR
quantitative reverse-transcriptase polymerase chain reaction
- SMA
smooth muscle actin
- THY1
thymocyte differentiation antigen 1 or CD90
- TIMP1
TIMP metallopeptidase inhibitor 1
- TNF
tumor necrosis factor
- VEGFR2
vascular endothelial growth factor receptor 2
Footnotes
AUTHOR CONTRIBUTIONS
M. Monsanto and M. Sussman designed experiments. M. Monsanto, K. White, T. Kim, B. Wang, K. Fisher, F. Khalafalla, and S. Mohsin performed experiments and analyzed data. A. Casillas, K. Broughton, T. Kim and M. Monsanto helped in the schematic design. M. Monsanto, K. Ilves, and M. Sussman wrote the article. All authors read and approved the final article.
DISCLOSURES
M. Sussman is Founder/Chief Scientific Officer for CardioCreate.
References
- 1.Sanganalmath SK, Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circulation research. 2013;113:810–34. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixit P, Katare R. Challenges in identifying the best source of stem cells for cardiac regeneration therapy. Stem Cell Res Ther. 2015;6:26. doi: 10.1186/s13287-015-0010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assmus B, Schachinger V, Teupe C, Britten M, Lehmann R, Dobert N, Grunwald F, Aicher A, Urbich C, Martin H, Hoelzer D, Dimmeler S, Zeiher AM. Transplantation of Progenitor Cells and Regeneration Enhancement in Acute Myocardial Infarction (TOPCARE-AMI) Circulation. 2002;106:3009–17. doi: 10.1161/01.cir.0000043246.74879.cd. [DOI] [PubMed] [Google Scholar]
- 4.Clifford DM, Fisher SA, Brunskill SJ, Doree C, Mathur A, Watt S, Martin-Rendon E. Stem cell treatment for acute myocardial infarction. Cochrane Database Syst Rev. 2012:CD006536. doi: 10.1002/14651858.CD006536.pub3. [DOI] [PubMed] [Google Scholar]
- 5.Hare JM, et al. Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. JAMA. 2012;308:2369–79. doi: 10.1001/jama.2012.25321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills JS, Rao SV. REPAIR-AMI: stem cells for acute myocardial infarction. Future Cardiol. 2007;3:137–40. doi: 10.2217/14796678.3.2.137. [DOI] [PubMed] [Google Scholar]
- 7.Traverse JH, et al. Effect of intracoronary delivery of autologous bone marrow mononuclear cells 2 to 3 weeks following acute myocardial infarction on left ventricular function: the LateTIME randomized trial. JAMA. 2011;306:2110–9. doi: 10.1001/jama.2011.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menasche P, Alfieri O, Janssens S, McKenna W, Reichenspurner H, Trinquart L, Vilquin JT, Marolleau JP, Seymour B, Larghero J, Lake S, Chatellier G, Solomon S, Desnos M, Hagege AA. The Myoblast Autologous Grafting in Ischemic Cardiomyopathy (MAGIC) trial: first randomized placebo-controlled study of myoblast transplantation. Circulation. 2008;117:1189–200. doi: 10.1161/CIRCULATIONAHA.107.734103. [DOI] [PubMed] [Google Scholar]
- 9.Leri A, Kajstura J, Anversa P. Role of cardiac stem cells in cardiac pathophysiology: a paradigm shift in human myocardial biology. Circulation research. 2011;109:941–61. doi: 10.1161/CIRCRESAHA.111.243154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 11.Bearzi C, et al. Human cardiac stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyngbaek S, Schneider M, Hansen JL, Sheikh SP. Cardiac regeneration by resident stem and progenitor cells in the adult heart. Basic Res Cardiol. 2007;102:101–14. doi: 10.1007/s00395-007-0638-3. [DOI] [PubMed] [Google Scholar]
- 13.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marban E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolli R, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Mohsin S, Khan M, Nguyen J, Alkatib M, Siddiqi S, Hariharan N, Wallach K, Monsanto M, Gude N, Dembitsky W, Sussman MA. Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circulation research. 2013;113:1169–79. doi: 10.1161/CIRCRESAHA.113.302302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulandavelu S, Karantalis V, Fritsch J, Hatzistergos KE, Loescher VY, McCall F, Wang B, Bagno L, Golpanian S, Wolf A, Grenet J, Williams A, Kupin A, Rosenfeld A, Mohsin S, Sussman MA, Morales A, Balkan W, Hare JM. Pim1 Kinase Overexpression Enhances ckit+ Cardiac Stem Cell Cardiac Repair Following Myocardial Infarction in Swine. Journal of the American College of Cardiology. 2016;68:2454–2464. doi: 10.1016/j.jacc.2016.09.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokita Y, Tang XL, Li Q, Wysoczynski M, Hong KU, Nakamura S, Wu WJ, Xie W, Li D, Hunt G, Ou Q, Stowers H, Bolli R. Repeated Administrations of Cardiac Progenitor Cells Are Markedly More Effective Than a Single Administration: A New Paradigm in Cell Therapy. Circulation research. 2016;119:635–51. doi: 10.1161/CIRCRESAHA.116.308937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams AR, Hatzistergos KE, Addicott B, McCall F, Carvalho D, Suncion V, Morales AR, Da Silva J, Sussman MA, Heldman AW, Hare JM. Enhanced effect of combining human cardiac stem cells and bone marrow mesenchymal stem cells to reduce infarct size and to restore cardiac function after myocardial infarction. Circulation. 2013;127:213–23. doi: 10.1161/CIRCULATIONAHA.112.131110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein HS, Srivastava D. Stem cell therapy for cardiac disease. Pediatr Res. 2012;71:491–9. doi: 10.1038/pr.2011.61. [DOI] [PubMed] [Google Scholar]
- 20.Miyahara Y, Nagaya N, Kataoka M, Yanagawa B, Tanaka K, Hao H, Ishino K, Ishida H, Shimizu T, Kangawa K, Sano S, Okano T, Kitamura S, Mori H. Monolayered mesenchymal stem cells repair scarred myocardium after myocardial infarction. Nature medicine. 2006;12:459–65. doi: 10.1038/nm1391. [DOI] [PubMed] [Google Scholar]
- 21.Gnecchi M, Zhang Z, Ni A, Dzau VJ. Paracrine mechanisms in adult stem cell signaling and therapy. Circulation research. 2008;103:1204–19. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malgieri A, Kantzari E, Patrizi MP, Gambardella S. Bone marrow and umbilical cord blood human mesenchymal stem cells: state of the art. Int J Clin Exp Med. 2010;3:248–69. [PMC free article] [PubMed] [Google Scholar]
- 23.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815–22. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 25.Kuraitis D, Ruel M, Suuronen EJ. Mesenchymal stem cells for cardiovascular regeneration. Cardiovasc Drugs Ther. 2011;25:349–62. doi: 10.1007/s10557-011-6311-y. [DOI] [PubMed] [Google Scholar]
- 26.Hare JM, Traverse JH, Henry TD, Dib N, Strumpf RK, Schulman SP, Gerstenblith G, DeMaria AN, Denktas AE, Gammon RS, Hermiller JB, Jr, Reisman MA, Schaer GL, Sherman W. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. Journal of the American College of Cardiology. 2009;54:2277–86. doi: 10.1016/j.jacc.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–7. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 28.Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M, Oz MC, Hicklin DJ, Witte L, Moore MA, Rafii S. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95:952–8. [PubMed] [Google Scholar]
- 29.Gehling UM, Ergun S, Schumacher U, Wagener C, Pantel K, Otte M, Schuch G, Schafhausen P, Mende T, Kilic N, Kluge K, Schafer B, Hossfeld DK, Fiedler W. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood. 2000;95:3106–12. [PubMed] [Google Scholar]
- 30.Handgretinger R, Gordon PR, Leimig T, Chen X, Buhring HJ, Niethammer D, Kuci S. Biology and plasticity of CD133+ hematopoietic stem cells. Annals of the New York Academy of Sciences. 2003;996:141–51. doi: 10.1111/j.1749-6632.2003.tb03242.x. [DOI] [PubMed] [Google Scholar]
- 31.Jujo K, Ii M, Losordo DW. Endothelial progenitor cells in neovascularization of infarcted myocardium. Journal of molecular and cellular cardiology. 2008;45:530–44. doi: 10.1016/j.yjmcc.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee WY, Wei HJ, Wang JJ, Lin KJ, Lin WW, Chen DY, Huang CC, Lee TY, Ma HY, Hwang SM, Chang Y, Sung HW. Vascularization and restoration of heart function in rat myocardial infarction using transplantation of human cbMSC/HUVEC core-shell bodies. Biomaterials. 2012;33:2127–36. doi: 10.1016/j.biomaterials.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 33.Sharan R, Maron-Katz A, Shamir R. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19:1787–99. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- 34.Ridzuan N, Al Abbar A, Yip WK, Maqbool M, Ramasamy R. Characterization and Expression of Senescence Marker in Prolonged Passages of Rat Bone Marrow-Derived Mesenchymal Stem Cells. Stem Cells Int. 2016;2016:8487264. doi: 10.1155/2016/8487264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaer A, Azarpira N, Aghdaie MH, Esfandiari E. Isolation and characterization of Human Mesenchymal Stromal Cells Derived from Placental Decidua Basalis; Umbilical cord Wharton’s Jelly and Amniotic Membrane. Pak J Med Sci. 2014;30:1022–6. doi: 10.12669/pjms.305.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aliborzi G, Vahdati A, Mehrabani D, Hosseini SE, Tamadon A. Isolation, Characterization and Growth Kinetic Comparison of Bone Marrow and Adipose Tissue Mesenchymal Stem Cells of Guinea Pig. Int J Stem Cells. 2016;9:115–23. doi: 10.15283/ijsc.2016.9.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H, Drummond CA, Fan X, Haller ST, Liu J, Malhotra D, Tian J. Hiding inside? Intracellular expression of non-glycosylated c-kit protein in cardiac progenitor cells. Stem Cell Res. 2016;16:795–806. doi: 10.1016/j.scr.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CL, Faltusova K, Molik M, Savvulidi F, Chang KT, Necas E. Low c-Kit Expression Level Induced by Stem Cell Factor Does Not Compromise Transplantation of Hematopoietic Stem Cells. Biol Blood Marrow Transplant. 2016;22:1167–72. doi: 10.1016/j.bbmt.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Babina M, Rex C, Guhl S, Thienemann F, Artuc M, Henz BM, Zuberbier T. Baseline and stimulated turnover of cell surface c-Kit expression in different types of human mast cells. Exp Dermatol. 2006;15:530–7. doi: 10.1111/j.1600-0625.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 40.Fischer KM, Cottage CT, Wu W, Din S, Gude NA, Avitabile D, Quijada P, Collins BL, Fransioli J, Sussman MA. Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing Pim-1 kinase. Circulation. 2009;120:2077–87. doi: 10.1161/CIRCULATIONAHA.109.884403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohsin S, Khan M, Toko H, Bailey B, Cottage CT, Wallach K, Nag D, Lee A, Siddiqi S, Lan F, Fischer KM, Gude N, Quijada P, Avitabile D, Truffa S, Collins B, Dembitsky W, Wu JC, Sussman MA. Human cardiac progenitor cells engineered with Pim-I kinase enhance myocardial repair. Journal of the American College of Cardiology. 2012;60:1278–87. doi: 10.1016/j.jacc.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sitte N, Saretzki G, von Zglinicki T. Accelerated telomere shortening in fibroblasts after extended periods of confluency. Free Radic Biol Med. 1998;24:885–93. doi: 10.1016/s0891-5849(97)00363-8. [DOI] [PubMed] [Google Scholar]
- 43.Butura A, Johansson I, Nilsson K, Warngard L, Ingelman-Sundberg M, Schuppe-Koistinen I. Differentiation of human hepatoma cells during confluence as revealed by gene expression profiling. Biochem Pharmacol. 2004;67:1249–58. doi: 10.1016/j.bcp.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 44.Nowbar AN, Mielewczik M, Karavassilis M, Dehbi HM, Shun-Shin MJ, Jones S, Howard JP, Cole GD, Francis DP, group Dw Discrepancies in autologous bone marrow stem cell trials and enhancement of ejection fraction (DAMASCENE): weighted regression and meta-analysis. BMJ. 2014;348:g2688. doi: 10.1136/bmj.g2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


