Abstract
The ability to label individual cells is useful for single-cell-level studies of complex cellular interactions and heterogeneity. Optically readable cell labeling is attractive as it can be interrogated noninvasively and repeatedly at high speeds. Here we demonstrate the feasibility of large-scale cell barcoding and identification using fluorescent polystyrene microbeads loaded into cells. Intracellular beads with different diameters in a range of 5 to 12 µm produce spectrally distinguished features or barcodes. A microfluidic chip was used to measure fluorescence resonance peaks emitted from individual cells. An algorithm comparing the peak wavelengths to a reference barcode library allowed barcode identification with high accuracy. This work provides guideline to increase the number of unique identifiers and reduce various false-positive, false-negative errors.
Graphical abstract
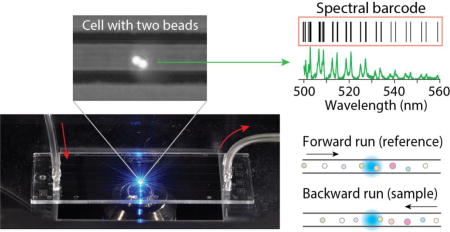
We demonstrate the feasibility of large-scale cell barcoding and identification using intracellular micro-resonators with different diameters.
Introduction
Barcoding in biomedical sciences is a labeling technique to identify objects from otherwise non-distinguishable population. DNA barcoding was developed for the identification of species and tracing the migratory behaviors of animals, such as birds, but its applications have been extended to biological cells for tracing their lineage, genetic mutations, and migration.1,2 Since DNA barcoding is encoded in the genome, its readout is typically done by DNA sequencing, which requires cell lysis. Possibly, DNA barcodes can be detected by using fluorescence in situ hybridization,3 but this method requires multiple stages of staining and washing, making measurements slow and not readily applicable to cells in living organisms. In mass cytometry, rare-earth metals with distinct atomic weights are widely used for labeling target molecules and cells.4,5 Multiplexing up to 96 labels have been demonstrated,6 and the number of identifiable labels may be further increased.
In contrast to the DNA and rare-earth metal barcodes, optically accessible barcodes allow noninvasive, rapid readouts and offer the possibility of real-time and time-lapse analyses. Among the variety of optical barcoding schemes developed to date,7 graphical encoding relies on micro- or nano-patterns that are distinguished by using optical microscopy or nanoscopy.8–10 However, this method is prone to error unless the barcode patterns are clearly visualized and so may not be applicable to in vivo applications. Information can also be encoded by the magnitudes of forward and side scattering of specifically engineered layered spheres11 and luminescence lifetimes.12 Spectrometric barcoding is based on the combination of non- or minimally-overlapping emission spectra from different organic dyes, quantum dots,13,14 Raman scattering molecules15, and lanthanides16. Stochastic expression of multiple fluorescent proteins17,18 can produce numerous colors distinguishable by ratiometric spectral analysis and can be combined with spatial mapping.19 Fluorescent beads that use three dyes and ten different concentration levels to generate 500 possible combinations have been commercially developed.20 While such beads can in principle be used for cellular barcoding, intensity-graded coding is less reliable than spectroscopic barcoding especially for intra-tissue and in vivo settings, where optical scattering and absorption in the tissue and photo-bleaching of the incorporated dyes may diminish the accuracy of intensity-based readout.
Among the various optical barcoding schemes, coupling of fluorescence with an optical microcavity is an attractive approach because it can generate narrowband spectral features defined by cavity resonance. In particular, fluorescent microbeads can support whispering-gallery modes (WGM) and generate feature-rich resonance spectra21 with good photo-stability, making them suited for cellular barcoding. Recently, we and others have shown that dye-containing microspheres can be incorporated into cells and generate stable WGM spectra in both spontaneous (fluorescence) and stimulated emission (lasing) regimes.22–24 In the event of cell division, beads can be passed down to daughter cell(s) over several generations with very small spectral shifts (< 30 pm) of their WGM peaks.25 Here, we report the feasibility of reading WGM-based spectral barcodes from cells in a microfluidic channel. Our experiments revealed several sources of readout errors and provide insights into how and to what extent the errors may be reduced for large-scale cellular barcoding and high-throughput readout with improved accuracy.
Results
Loading microspheres into cells
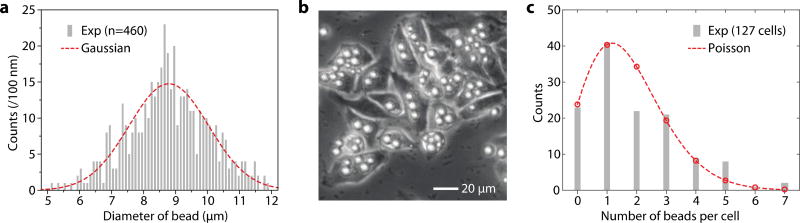
A commercial batch of fluorescent polystyrene beads (Thermo Scientific, Fluoro-Max, with a wide range of sizes from 5 to 12 µm, the coefficient of variation of the particle diameter, CV = 18%) was used for each labeling experiment. The bead sizes measured from their fluorescence microscope images exhibited a Gaussian distribution with a mean diameter of 8.79 µm and a full-width-half-maximum of 2.92 µm (standard deviation of 1.24 µm) (Fig. 1a), consistent with the manufacturer’s specification. After washing with ethanol and water, the microbeads were immersed in 1% w/v aqueous solution of poly-L-lysine hydrobromide (MW 30,000–70,000, Sigma) for 30 min to form poly-L-lysine coating on the surface. They were washed three times with water, centrifuged at 5000 g for 5 min, and transferred to phosphate buffered saline (PBS) solution. HeLa cells were grown at 37°C with 5% CO2 in full growth medium (DMEM medium with 10% fetal bovine serum and 1% pen-strep). The microbeads in PBS (1.84×106 beads/ml) were added to the cell culture medium (8.57×104 cells/ml) and incubated for 24 h, by which the cells reached ~70% confluency. After the 24-hr incubation, we typically found that about 80–85% HeLa cells contained at least one bead, and ~ 50% the bead-containing cells had more than one bead (Fig. 1b). The measured distribution of the number of beads fits well with a Poisson distribution with an average of 1.7 beads per cell (Fig. 1c). This indicates that internalization of beads can be described as a random process, independent of the bead size,24 and unaffected by the presence of other beads inside the cell. The cells were washed with PBS to remove non-internalized beads, detached by trypsin, centrifuged at 125 g for 5 min, and re-dispersed in PBS medium containing 1% w/v bovine serum albumin (BSA). BSA reduces sticking of cells to the surfaces in syringes, tubing and microchannel walls. The dispersion was filtered through a 35-µm mesh to remove clusters of cells and other debris before loading into 100-µl glass syringes (Gastight 1710).
Fig. 1.
(a) Histogram distribution of the bead diameter measured from fluorescence microscope images of 460 beads. Bin size: 100 nm; dashed curve (red): Gaussian distribution fit. (b) HeLa Cells grown with polystyrene beads in a dish. (c) Histogram of the number of beads per cell. Dashed curve (red): best-fit Poisson distribution (mean=1.7).
Spectral readout of resonance-encoded cells
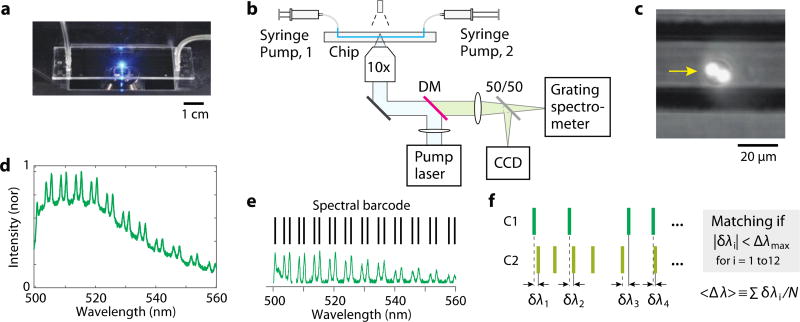
Microfluidic chips used were made of cyclic olefin copolymer (ChipShop) and had 58.5-mm-long channels on a 140 µm-thick bottom plate (Fig. 2a). Each channel had a square cross-section of 50 µm by 50 µm. The syringe filled with a suspension of cells (2.1×105 cells/ml) was connected to one end of the channel via 8-cm-long tubing with an inner diameter of 1.0 mm and Olive connectors. Another syringe filled with PBS was connected to the other end of the channel using the same type of tubing. The syringes were placed on two separate syringe pumps (KDS Legato 100). Air bubbles inside the syringes, tubes, and channel were carefully removed. The chip was mounted on a custom-built inverted microscope setup (Fig. 2b). For fluorescence excitation, the output of a continuous-wave 491-nm laser (Cobolt Dual Calypso) was delivered to the middle of the channel through an objective lens (10×, 0.25 NA, Nikon). The beam was slightly de-focused so that the beam size at the channel was 50 µm, matched to the width of the channel. The fluorescence emission from the channel at the focus of the objective lens was directed to a CCD camera (Mightex, MT9001) for bright-field and fluorescence imaging (Fig. 2c) and to a diffraction-grating spectrometer (Andor Shamrock 303i-B) with a spectral resolution of 0.05 nm. The fluorescence spectra were recorded at a rate of 28 spectra/s with an exposure time of 36 ms per measurement.
Fig. 2.
(a) The microfluidic chip mounted on a microscope. (b) Schematic of the optical setup. Pump laser: a continuous-wave laser at 491 nm. DM: Dichroic mirror. (c) CCD image of a cell (arrow) with two fluorescent beads, superimposed on a bright-field image of the channel. (d) A typical fluorescence spectrum from a cell containing a single bead, featuring a periodic double-peak WGM structure (TM and TM polarizations) on top of a broad fluorescence background. (e) The group of WGM peak wavelengths identified from a fluorescence-subtracted spectrum constitutes the “spectral barcode” of the cell. (f) Two barcodes, C1 and C2, are compared in terms of spectral differences, δλ, and deemed identical if |δλ| < Δλmax for at least 12 resonance peaks. The average of the spectral differences is denoted <Δλ>.
There is more than one method to convert the spectrum to a unique identifier of the cell. In principle, the WGM spectrum can provide sufficient information to determine the bead diameter with sub-nanometer resolution by curve fitting based on the WGM theory 22. This method can distinguish cells by the diameters of the beads. This fitting-based method was shown to be effective for individual single beads 22 but can be unreliable for spectra originating from multiple beads. Another approach is to use the spectral curve itself as an identifier and to perform some sort of cross-correlation integral to determine the identity between two spectra. This method is also effective for single beads but far less robust when cells have multiple beads because the overall spectral shape is sensitive to fluorescence intensity and the relatively intensities of individual beads within a cell can vary as the relative position of the two beads is changed, particularly with respect to the pump beam axis.
To enhance the robustness of tagging, we devised a simple, yet effective, protocol, as described in the following. The spectrum of a cell containing one or a few beads shows many WGM resonance peaks on top of a broad fluorescence spectrum over a spectral range of 500 to 580 nm (Fig. 2d). In post processing, the broadband background was subtracted from the measured spectrum, leaving only the WGM resonance peaks. The spectral peaks were fitted to Lorentzian line shape functions to locate the central positions of the peaks. The numerical array of the WGM peak wavelengths in an increasing order constitutes a “barcode” (Fig. 2e). We considered a WGM peak “clearly resolved” when the peak amplitude was at least 3-times larger than the root-mean-square (rms) value of the noise floor in the spectrum and the peak wavelength must be separated from its neighboring spectral peaks by at least 1 nm. Only the cells that exhibited 12 or more clearly-resolved spectral peaks were considered “barcoded” successfully and included in further analysis. A typical barcoded cell contained 12 to 24 clearly-resolved spectral peaks or barcode lines.
We used the following protocol to determine whether two barcodes are identical or different. First, the numbers of barcode lines are compared. If the difference in the number of peaks is greater than 6, the two barcodes would be determined to be different or non-matching, because this situation would occur when the barcodes are comprised of either different number of beads or same number of beads but very dissimilar sizes. On the other hand, when the difference is 6 or less, then their peak wavelengths are compared. For this comparison, between the two barcodes the one with smaller number of spectral peaks (C1) is selected. For each reference peak of the first barcode, the closest peak in the second barcode (C2) is found, and the spectral difference in wavelength, δλi, is calculated (Fig. 2f). The spectral difference is compared to a fixed parameter Δλmax, termed the maximum spectral shift. If δλi is smaller than Δλmax for at least 12 spectral peaks, the two barcodes would be regarded as identical or matching; if not, they would be deemed different. So, the criterion for barcode matching is |δλi| < Δλmax for i = 1 to 12. We optimized the parameter Δλmax experimentally to achieve the most desirable outcomes. The optimization step is described in more detail later. Since we compare only the central wavelength positions of peaks, this algorithm is relatively insensitive to the relative intensities of spectral peaks.
Verification of barcode reading with stationary cells
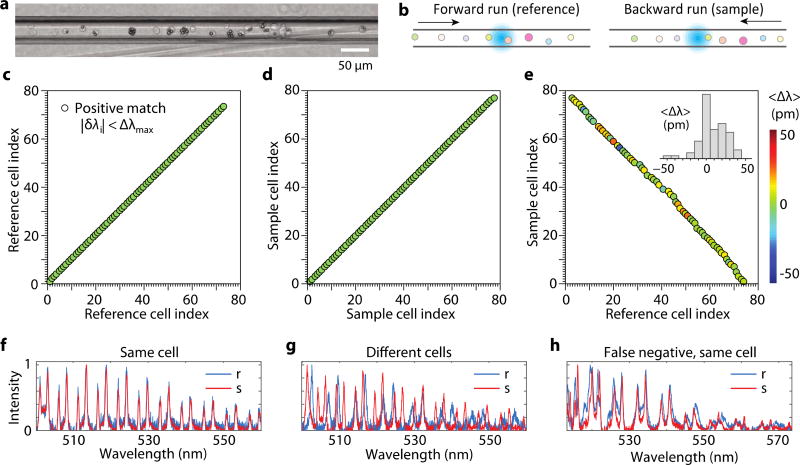
As an initial test of the readout scheme, we loaded a suspension of cells into a microfluidic chip and sealed both ends of the channel (Fig. 3a), so that the position of cells in the channel was not changed during measurements. The chip was mounted on a motorized stage, and the stage was translated in the forward and then backward directions at a speed of 0.9 mm/s while fluorescence spectra were continuously recorded (Fig. 3b). Between the two scans, the order of cells was retained. In the forward run, a total of 87 cells containing beads passed through the laser-excitation zone. From the typical bead-uptake efficiency of 80–85% we measured for this HeLa cell line, we estimated that about 18 cells without any beads passed through the excitation zone during the measurement without being detected in the fluorescence measurement. Out of 87 detected cells (containing beads), 72 generated emission spectra, each with at least 12 clearly-resolved spectral peaks, satisfying the detection criterion defined earlier. Almost all single-bead and most two-bead cells satisfied the criteria. However, almost all cells containing 3 or more beads did not satisfy the detection criterion because of the overlap of spectral peaks and, hence, were rejected (not “detected”). The detected 72 cells constitute a reference group, among which ~50 cells had single beads and ~22 had two or more beads. In the backward run, 75 barcoded cells met the criteria and were “detected”. The reason why there were 3 more cells detected in the backward scan is presumably because of the small intensity fluctuations between the forward and backward measurements. These cells were not detected in the forward scan because they had less than 12 clearly-resolved spectral peaks, but some of the undetected peaks exceeded the detection threshold — 3 times the rms intensity noise — in the backward scan. Considering the 75 cells as a test sample group, we applied our barcode matching protocol described earlier with respect to the reference cells.
Fig. 3.
(a) Micrograph of the channel loaded with cells. (b) Measurement strategy. Cells are scanned in the forward and then backward directions across the excitation beam (cyan). (c-d) Correlation maps obtained with Δλmax = 100 pm for (c) the reference (forward flowing) and (d) test sample (backward flowing), and (e) cross correlation between reference and sample cells. Inset, histogram of the measured mean spectral shift of identified WGM peaks between reference and sample measurements of matching cells. Color represents the mean spectral shift. (f) An example set of spectra of a cell in reference (r) and sample (s) dataset, showing high cross-correlation between them. Fluorescence background was subtracted from all spectra. (g) Example of reference and sample spectra of two different cells with apparently low cross-correlation. (h) An exemplary failure mode (false-negative error) that originates from spectral shifts between the reference and sample spectra of a cell containing two beads.
First, we analyzed correlation between reference cells for various Δλmax values and found Δλmax = ~100 pm was adequate to resolve all the 72 cells without any correlation hit between two different cells (all cells had a unique barcode). A correlation map for Δλmax = 100 pm was drawn (Fig. 3c), where the x- and y-axis represent cell indices in the temporal order they were measured, and a circular correlation hit mark is placed where the corresponding pair have identical barcodes according to the criterion for the given Δλmax. Along the diagonal axis, each barcode is compared to its own self, so this axis is trivial and always filled with circular marks. On the other hand, any correlation hit in the non-diagonal space of the map is non-trivial and could result from two scenarios: (1) Δλmax is too large so that barcodes from different beads satisfy the marching criterion, which leads to a false-positive error; or (2) the two cells have different beads but their diameters are too similar to be resolved with the given Δλmax. Reducing Δλmax may decrease these false-positive counts, but this comes at the expense of increasing false-negative errors failing to identify the same cells. At Δλmax = 100 pm, the analysis for the backward-scan dataset yielded no false-positive error as clearly shown in a correlation map (Fig. 3d).
The same protocol was applied to analyze cross-correlation between the sample and reference cells. 66 of 75 cells in the test sample had a match with different reference cells. This result is illustrated in a correlation map (Fig. 3e), where correlation marks appear along a negative-slope diagonal axis simply because of the order of cells was reversed in the backward scan. There was no double-matching error; that is, each test cell was matched to only one reference cell, and no two test cells were matched to a single reference cell. As noted above, 3 in 75 cells did not have matching references, and additional 6 cells failed to find their matching cells (false-negative errors). To better understand the causes of the errors, we manually examined the measured spectra. Any pairs of matching cells exhibited well-overlapped spectra (Fig. 3f). The spectra from non-matching cells showed apparent differences (Fig. 3g). However, the 6 cells which failed to find their matching cells (false-negative errors) had their spectra slightly shifted between the reference and sample scans (Fig. 3h). The magnitude of the shift was greater than 100 pm for some peaks, which reduced the number of matching peaks that satisfy |δλi| < 100 pm to less than 12. The spectral changes of >100 pm is presumably caused by changes in the effective refractive index of the surrounding of the beads.
We calculated a mean spectral shift, <Δλ>=Σδλi/N, where the summation is over the entire set of spectral lines (N) in the barcode that satisfied |δλi| < Δλmax, where typically N = 12 to 24. The color of each circle in the cross-correlation map (Fig. 3e) represents the mean spectral shift of the test barcodes with respect to their matching reference barcodes. Histogram of the calculated values (inset of Fig. 3e) ranged from −50 to 50 pm and showed some indication of redshift. Possible phenotype changes or temperature changes of the cells in the microfluidic channel may have caused the asymmetric shift, changing the surrounding refractive index of beads.
Test with flowing cells in microfluidic channels
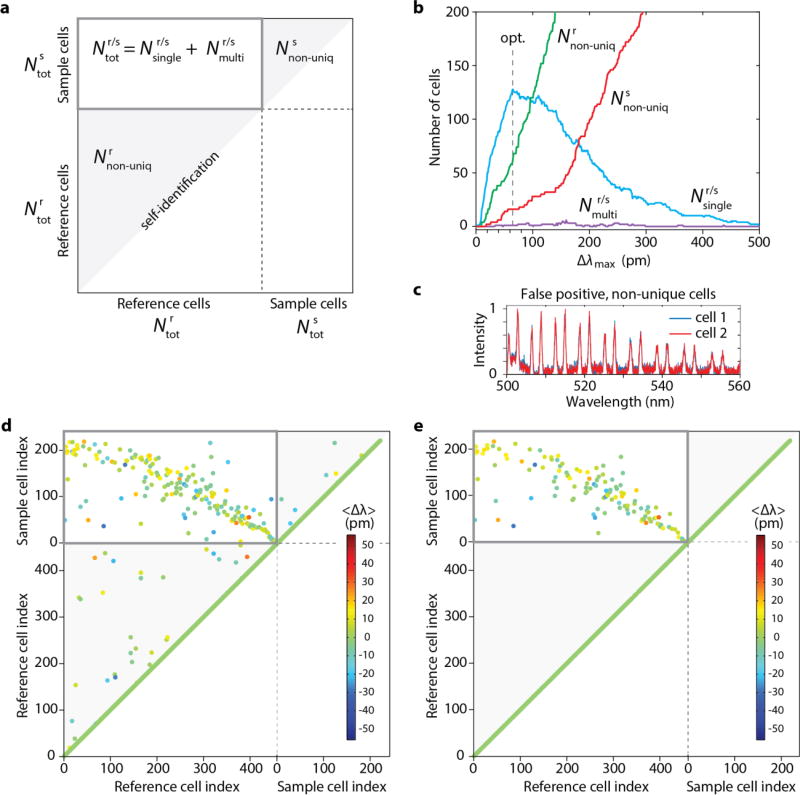
The emission spectra from stationary cells and beads therein were measured to be very stable over repeated measurements. When cells are in motion, however, the intracellular positions of beads and the relative position of multiple beads in a single cell may change, and these movements affect the emission spectra and the barcode and, therefore, may cause errors in barcode identification. We tested the barcode readout scheme for hundreds of cells in a non-stationary condition by making the cells flow in one direction through a microfluidic channel and then flow in the backward direction. In the backward scan, the cells are not expected to be in the exact reverse order because they are mixed in the tubing before returning to the channel. After loading a dispersion of cells into the syringe, the syringe pump was set to a volume flow rate of 250 nl/min, so that the cells pass the 50-µm-long laser-excitation zone in the channel, one cell at a time with a speed of 1.67 mm/s. As the cells were flown through the channel, their emission spectra were acquired at a frame rate of 28 spectra/s. A total of 479 cells containing at least one bead passed through the excitation zone. Of these, a total 457 ( ) cells were detected (met the detection criterion) in the forward direction, 320 of which contained spectral fingerprint from one bead and 137 from more than one bead. An estimated additional 97 cells did not contain beads and were thus undetected. All detected cells represent a reference barcode library. Next, we reversed the flow direction and scanned a partial group of the reference cells in the backward flow at the same flow rate. A total of 222 ( ) cells were detected (i.e. met the detection criterion), which constitutes a test sample group.
With the total 679 ( ) measured spectra, we drew a full correlation map showing the correlations of 679 × 679 scan data for a given Δλmax (Fig. 4a; Supplementary Information). Each pair of non-unique cells with identical barcodes appear in the lower-left (or upper-right) shaded triangular regions and are counted into (or ). The optimum Δλmax value was chosen that maximizes the number ( ) of sample cells matched to single reference barcodes. With the measured spectra, the numbers of various types of correlation counts were calculated as a function of Δλmax ranging from 0 to 500 pm (Fig. 4b). At Δλmax = 0, there are no correlation hits between two different cells (Fig. 4b). The numbers of both uniquely identified and non-unique cells increase as Δλmax increases and criterion for identity is relaxed. However, as Δλmax increases beyond ~ 65 pm, the numbers of non-unique cells ( ) continued to increase, and as the non-unique cells are removed from the reference and sample, the number of unique identification ( ) begins to decrease. In principle, the number of non-unique cells, i.e. false positive errors, can be reduced by using a batch of beads with all dissimilar diameters. Indeed, some cells appeared to contain similar beads with almost indistinguishable spectra (Fig. 4c). We have chosen Δλmax = 65 pm that maximized . A full correlation map (Fig. 4d) obtained with Δλmax = 65 pm shows 403 unique and 54 ( ) non-unique barcodes in the reference and 206 unique and 16 ( ) non-unique cells in the sample. Without removing the non-unique cells from reference and sample, 20 sample cells had matching with more than one reference cells ( ).
Fig. 4.
(a) A full correlation map representation. (b) Dependence of uniquely defined cells and errors on the choice of Δλmax. An optimum value of 65 pm for Δλmax was determined, which maximizes the number of uniquely identified sample cells. (c) An example of two non-unique cells. (d) A correlation map obtained with Δλmax = 65 pm. (e) A modified correlation map when the non-unique cells in the reference and sample have been removed.
To have only uniquely barcoded cells, the non-unique cells could be physically removed from the reference group. In our experiment, we simulated this situation of presorting in post processing, where all the non-unique cells were removed from the reference and sample groups in silico (Supplementary Information). After the removal, a re-calculated correlation map (Fig. 4e) confirms zero false-positive errors in the reference and samples ( ). The cross-correlation map (boxed domain) displays 128 ( ) uniquely identified cells and one multiply identified cell ( ). The remaining 77 ( ) cells in the sample group did not find uniquely matched references (false-negative errors) (Supplementary Information).
Conclusions
The spectral barcode readout scheme is suitable for tagging a few hundreds of cells with reasonable error rates. Most errors stemmed from the overlapping size distribution of current polystyrene beads. Using similar spectral analysis, the beads can be pre-sorted to create a batch with non-overlapping sizes from 8 to 12 µm with a size increment of 2 nm. Theoretically, two beads that are different in diameter by 2 nm produce WGM barcode peaks that are shifted by |Δλi| = 100 pm and, therefore, are resolvable when Δλmax = 65 pm. A batch of such uniformly poly-dispersed beads can produce 2,000 unique beads and label 2,000 cells when each cell is loaded with a single bead. When each cell is loaded with two beads, theoretically, up to 2,000,000 (=2,000 × 2,000 / 2) cells can be uniquely barcoded22. But this possibility should wait for the development of a method to control the number of beads per cell precisely.
The spectral barcodes are subject to change when the surrounding of beads is altered. Identical beads in environments with different refractive indices can produce non-identical WGM spectra. From simple spectral measurements, we cannot distinguish whether the spectral difference is because of different diameters or same diameters but in different environments. From theory it can be calculated that the spectral sensitivity to external refractive index of polystyrene beads is ~90 nm/RIU. So a change in the refractive index of the cytoplasm of 0.001 would cause a spectral shift of Δλ≈ 90 pm. In principle, the sensitivity to environment could be reduced by coating beads with a thin layer of low-index material to shield the evanescent field of optical modes from the environment. In addition, the spectral peaks are sensitive to temperature through thermal expansion of beads and temperature-dependent refractive indices. These two factors tend to have opposite effects for most materials. For polystyrene beads, the temperature sensitivity of WGM peaks is calculated to be 3 pm/°C,22 which may be negligible in most applications. Optical cross-talk can arise between beads when they are in optical contact within the range of evanescent fields and/or when the emission from one bead is absorbed or refracted by its neighboring beads. Nonetheless, we have shown that such cross-talks in uncoated polystyrene beads are small enough to allow more than a hundred unique barcodes, and in principle the multiplicity of barcodes can be substantially increased by using multiple dyes and 2–3 distinct beads per cell. Further, beads with core-shell structures can minimize optical coupling.
It has been shown that cells containing polystyrene beads with diameters of 7–12 µm undergo cellular division and movement over 1–2 days 22–24, and long-term cell viability in the presence of micro- and sub-micron-scale beads have been well established. Nevertheless, the subtle effects of intracellular objects on cellular phenotypes and gene expression patterns are not fully understood and should be an important subject of research in the future. Biocompatibility is expected to improve as beads get smaller, for example, by using high-index materials, such as high-index glass and semiconductors, as well as plasmonics. Also, coating with biocompatible polymers could further enhance biocompatibility.
The polystyrene beads used in this study have a Q-factor of ~1000, a mode spacing of ~5 nm, and the finesse of ~10. With this modest finesse, it was proven to be difficult to distinguish cells containing three or more beads. Fluorescent beads made of materials, such BaTiO3, that have higher refractive indices than polystyrene could be used with an advantage of smaller sizes at the same finesse25 or enhanced Q-factor and finesse at the same size.
Microfluidic chips are a useful platform to scan the barcodes in cells rapidly both for the construction of reference barcode libraries and for the barcode identification of sample cells. In the present study, the barcode scan in the forward flow simulates the process to build a reference barcode library. Test sample sets were produced by sending these cells back to the channel with no or partial cell mixing, and the barcodes of the test cells were compared to the reference library. It is envisioned that in actual applications, sample cells are harvested from cell cultures, tissues, or live animals.
Beside microfluidic chips, the barcode readout method could also be integrated into the conventional flow cytometry platform with modifications to include pumping and high-resolution spectral measurements. State-of-the-art flow cytometers offer 3 to 12 spectral channels.26 The brightness of fluorescent beads is sufficient to enable high-spectral-resolution measurements with millisecond- and sub-millisecond integration times for high-throughput barcode identifications and cell sorting. WGM cellular barcoding may prove to be a useful tool for biomedical investigations at single-cell levels. Besides cell tagging, resonator-based barcodes have potential as luminescent probes for highly-multiplexed molecular assays on chip. Finally, the optical barcode technology has a potential for non-biological labeling and authentication for documents, work of arts, industrial materials and products.
Supplementary Material
Acknowledgments
The authors acknowledge discussions with Sheldon Kwok and Nicola Martino. This research was supported by the U.S. National Institutes of Health (R01-CA192878, P41-EB015903, P01-HL120839, DP1-OD022296), National Science Foundation (CBET-1264356, EEC-1358296, ECCS-1505569, CMMI-1562863), and MGH Research Scholar Award. M.H. was supported in part by the Marie Curie International Outgoing Fellowship N° 627274 within the 7th European Community Framework Programme.
References
- 1.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe D, Tanner SD, Nolan GP. Science (80-.) 2011;332:687–697. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik M, Tanner S. J. Immunol. Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, et al. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng Y, Sun K, Chen X, Li W. Chem. Soc. Rev. 2015;44:5552–5595. doi: 10.1039/c4cs00382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating CD, Natan MJ. Adv. Mater. 2003;15:451–454. [Google Scholar]
- 9.Lin C, Jungmann R, Leifer AM, Li C, Levner D, Church GM, Shih WM, Yin P. Nat Chem. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Rosas E, Gomez R, Ibanez E, Barrios L, Duch M, Esteve J, Nogués C, Plaza JA. Small. 2009;5:2433–2439. doi: 10.1002/smll.200900733. [DOI] [PubMed] [Google Scholar]
- 11.Lawrie G, Battersby BJ, Trau M. Adv. Funct. Mater. 2003;13:887–896. [Google Scholar]
- 12.Lu Y, Zhao J, Zhang R, Liu Y, Liu D, Goldys EM, Yang X, Xi P, Sunna A, Lu J, Shi Y, Leif RC, Huo Y, Shen J, Piper JA, Robinson JP, Jin D. Nat. Photonics. 2013;8:32–36. [Google Scholar]
- 13.Mattheakis LC, Dias JM, Choi Y-J, Gong J, Bruchez MP, Liu J, Wang E. Anal. Biochem. 2004;327:200–8. doi: 10.1016/j.ab.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 14.Han M, Gao X, Su JZ, Nie S. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Yan B, Chen L. Chem. Rev. 2013;113:1391–428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 16.White K, Chengelis D, Gogick K, Stehman J, Rosi NL, Petoud S. J. Am.Chem. Soc. 2009;131:18069–18071. doi: 10.1021/ja907885m. [DOI] [PubMed] [Google Scholar]
- 17.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 18.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Nat. Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 19.Xiong F, Obholzer ND, Noche RR, Megason SG. PLoS One. 2015;10:e0127822. doi: 10.1371/journal.pone.0127822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunbar SA. Clin. Chim. Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramiro-Manzano F, Fenollosa R, Xifré-Pérez E, Garín M, Meseguer F. Adv. Mater. 2011;23:3022–3025. doi: 10.1002/adma.201100986. [DOI] [PubMed] [Google Scholar]
- 22.Humar M, Yun S Hyun. Nat. Photonics. 2015;9:572–576. doi: 10.1038/nphoton.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert M, Steude A, Liehm P, Kronenberg NM, Karl M, Campbell EC, Powis SJ, Gather MC. Nano Lett. 2015;15:5647–5652. doi: 10.1021/acs.nanolett.5b02491. [DOI] [PubMed] [Google Scholar]
- 24.Schubert M, Volckaert K, Karl M, Morton A, Liehm P, Miles GB, Powis SJ, Gather MC. Sci. Rep. 2017;7:40877. doi: 10.1038/srep40877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humar M, Yun SH. Optica. 2017;4:222–228. doi: 10.1364/OPTICA.4.000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolan JP, Condello D. Curr. Protoc. Cytom. 2013;63:1–18. doi: 10.1002/0471142956.cy0127s63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.