Abstract
RAS proteins are binary switches, cycling between ON and OFF states during signal transduction. These switches are normally tightly controlled, but in RAS-related diseases, such as cancer, RASopathies, and many psychiatric disorders, mutations in the RAS genes or their regulators render RAS proteins persistently active. The structural basis of the switch and many of the pathways that RAS controls are well known, but the precise mechanisms by which RAS proteins function are less clear. All RAS biology occurs in membranes: a precise understanding of RAS’ interaction with membranes is essential to understand RAS action and to intervene in RAS-driven diseases.
RAS proteins play a causal role in human cancer: this has been recognized for many years and has inspired multiple attempts to find RAS inhibitors. Mutations in RAS regulators, such as neurofibromin and SPRED1, also make significant contributions to cancer. “RASopathies,” a collection of developmental conditions caused by germline mutations that activate RAS/MAPK signaling, affect more than 400,000 individuals in the United States alone. Abnormal RAS activity may also play a significant role in autism and other neurological disorders. Here, we will review biochemical and biophysical properties of RAS proteins and how they affect human disease.
RAS proteins are binary molecular switches that cycle between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound states. This switch mechanism has been highly conserved among GDP/GTP binding proteins as diverse as bacterial elongation factors, heterotrimeric G-proteins, and a myriad of small GTPases with diverse biological functions. The conversion from stable, inactive GDP-bound forms to the active GTP-bound form is stimulated by guanine nucleotide exchange-factors (GEFs). Conversion back to the inactive form is mediated by GTPase-activating proteins (GAPs) (Cherfils and Zeghouf, 2013). GEFs and GAPs are large, multi-domain proteins capable of an astonishing variety of interactions with other proteins, lipids, and regulatory molecules that control levels of active and inactive RAS (Bos et al., 2007).
Dependence of RAS and other GTPases on GEFs and GAPs to switch them on and off allows both processes to be highly regulated and responsive to multiple signal inputs. However, they are governed by one important principle: translocation. GEFs and GAPs turn RAS on and off when they are recruited to the plasma membrane and placed in direct proximity to RAS. This proximal positioning in a 2D surface is equivalent to five orders of magnitude increase in binding constant relative to free solution and is at the heart of understanding how RAS proteins are regulated. RAS proteins activate effectors by recruitment to the plasma membrane. Indeed, for RAF kinase, translocation appears to be sufficient to initiate the complicated activation process (Leevers et al., 1994; Stokoe et al., 1994). Recruitment of RalGDS to RAS in the plasma membrane leads to activation of RalA and RalB (reviewed in Ferro and Trabalzini, 2010). Recruitment of PI 3-kinase γ (PI3Kγ) to the plasma membrane through a myristoylation signal rescues a mutant that does not bind RAS, suggesting membrane recruitment is a critical step in the activation process (Castellano and Downward, 2011). However, as with other effectors, direct binding of RAS also contributes to activation. Furthermore, differential activation of RAS effectors may be determined by local lipid composition within the membrane (Zhou et al., 2017). Understanding RAS proteins in the context of the plasma membrane is, therefore, a fundamental aspect of RAS biology, and characterizing these interactions will be important for interventions aimed at RAS diseases.
RAS as a Binary Switch
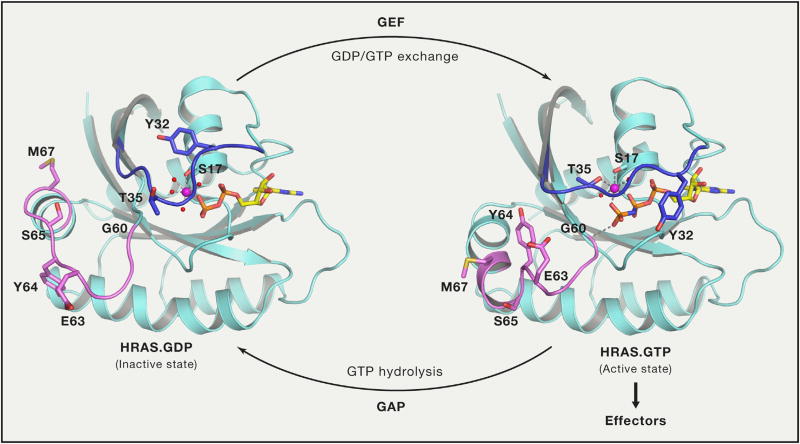
The crystal structure of the G-domain of HRAS in complex with GppNHp, a non-hydrolyzable GTP analog, provided the first insight into how RAS functions as a binary switch (Milburn et al., 1990; Pai et al., 1989). Subsequent structural studies on RAS in complex with GAP, GEF, and RAS-binding domains (RBDs) of effectors revealed two switch regions, switch-I (aa 30–38) and switch-II (aa 59–76) (Figure 1), and their critical role in protein-protein interactions. The two switch regions undergo conformational changes between the GDP and GTP states and have been described using a loaded-spring mechanism (Vetter and Wittinghofer, 2001). In the GTP-bound state, T35 and G60 form hydrogen bonds with the γ-phosphate and hold switch-I and switch-II regions in the active conformation, respectively. Upon GTP hydrolysis, the γ-phosphate is released and both switch regions return to the flexible conformation in the GDP-bound state.
Figure 1. Conformational Changes in Switch Regions when RAS Transitions from Inactive GDP-Bound State to Active GTP-Bound State.
Switch I and switch II regions are colored blue and violet, respectively, and side chain atoms of residues that undergo large conformation changes during transition are shown in stick representation. Interactions formed by γ-phosphate and magnesium ions are shown using dashed lines. HRAS.GDP and HRAS.GppNHp are from PDB: 4Q21 and 5P21, respectively. RAS is activated by GDP/GTP exchange stimulated by GEFs and inactivated by GTP hydrolysis stimulated by GAPs.
The very slow off-rate for GDP (t1/2 = 6 min, koff = 2 × 10−3 s−1 at 20°) (Hunter et al., 2015) allows RAS proteins to remain in their inactive states until signals provoke GDP/GTP exchange. This requires GEF activity to accelerate the exchange reaction by several orders of magnitude. GEF binding to RAS results in conformational changes in the switch regions and P loop (aa 10–17) that weakens the affinity of GDP, resulting in its release and replacement (Boriack-Sjodin et al., 1998). Because the affinity of RAS for GDP and GTP is similar, and the GEF does not favor rebinding of GDP or GTP, increased levels of GTP-bound RAS occur mainly due to the ~10-fold higher cellular concentration of GTP compared to GDP. Finally, binding of GTP dissociates the GEF and leads to the formation of an active GTP-bound RAS that can bind effectors.
The rate of intrinsic GTP hydrolysis for RAS proteins is very slow, (t1/2 = 16 min, koff = 6 × 10−4 s−1) (Hunter et al., 2015). Efficient GTP hydrolysis requires interaction with a GAP that accelerates the cleavage step by several orders of magnitude. Structural studies on GDP and AlF3 bound HRAS in complex with the GTPase activating domain (often referred to as the GAP-related domain, GRD; aa 718–1037) of RASA1/p120GAP provided first insights on how GAP stimulates GTP hydrolysis (Scheffzek et al., 1997). GAP interaction with the switch II region of HRAS stabilizes the position of Q61, which then coordinates the catalytic water molecule. Additionally, residue R789 of RASA1, called the arginine finger, protrudes into the active site of HRAS and interacts with the α- and β-phosphate groups of GDP and AlF3 and stabilizes the transition state by neutralizing a negative charge at the γ-phosphate (Ahmadian et al., 1997). Interaction of Q61 with catalytic water and main chain carbonyl group of the arginine finger enables it to extract a hydrogen atom from the attacking water molecule. This results in formation of a negatively charged hydroxyl ion that attacks the γ-phosphate of GTP to carry out the GTP hydrolysis reaction.
The GRDs of RAS GAPs are highly conserved (Ahmadian et al., 1997). However, RAS GAPs diverge dramatically outside of the GRDs, reflecting distinct roles in signal transduction and RAS regulation (Bos et al., 2007). For example, RASA1/p120 RasGAP contains SH2 and SH3 domains that bind to activated receptors, such as PDGFR (Kaplan et al., 1990). This is thought to enable RASA1/p120 RasGAP to downregulate RAS appropriately during signaling. On the other hand, neurofibromin (the NF1 gene product) is conserved organisms that lack receptor tyrosine kinase signaling, such as S. cerevisiae, and presumably regulates RAS in response to very different signals, the nature of which are unknown. In mammalian cells, neurofibromin depends on SPRED proteins to recruit it to RAS in the plasma membrane (Stowe et al., 2012), but the signals governing this critical interaction have not yet been identified.
RAS Effectors
In normal mammalian cells, RAF kinases (ARAF, BRAF, and CRAF) are major RAS effectors. In mouse embryo fibroblasts, RAS sustains proliferation, survival, and migration through activation of RAF kinases with no apparent role for other effectors in these processes (Drosten et al., 2010). RAS proteins in Drosophila melanogaster and Caenorhabditis elegans regulate pathways that are strikingly similar to the canonical RAS-MAPK pathway in mammalian cells (Lusk et al., 2017). Indeed, genetic analysis of RAS signaling in these organisms played an important role in piecing together the RAS pathway in mammalian cells, including roles for the regulatory proteins KSR, CBL, SHOC2, and SHP2 (Figure 2). Likewise, in Schizosaccharomyces pombe, Ras1 activates a MAP kinase cascade, although in this organism, the kinase that Ras1 activates directly, Byr2, does not resemble RAF. On the other hand, Saccharomyces cerevisiae RAS proteins (RAS1p and RAS2p) activate adenylate cyclase and regulate production of cyclic AMP (cAMP) in response to intracellular glucose. While their effectors are unrelated to RAS in mammalian cells, the GEFs (CDC25 and SDC25) and GAPs (IRA1 and IRA2) are clearly orthologs of their mammalian counterparts, underscoring that the GTP/GDP switch mechanism is highly conserved, even when the signal output is not. Mammalian RAS proteins expressed in S. cerevisiae can activate adenylate cyclase: this is surprising because S. cerevisiae RAS proteins interact at two distinct sites on adenylate cyclase, neither of which resemble the RAS-binding domains of mammalian RAS effectors. Also, unlike mammalian RAS effectors, activation of adenylate cyclase by S. cerevisiae RAS1p and RAS2p does not appear to involve effector recruitment to the plasma membrane, rather is based on allosteric relief of a negative regulatory domain (Powers, 1992). These examples illustrate that RAS effector interactions can be remarkably diverse, while the fundamental machinery governing the binary switch is highly conserved.
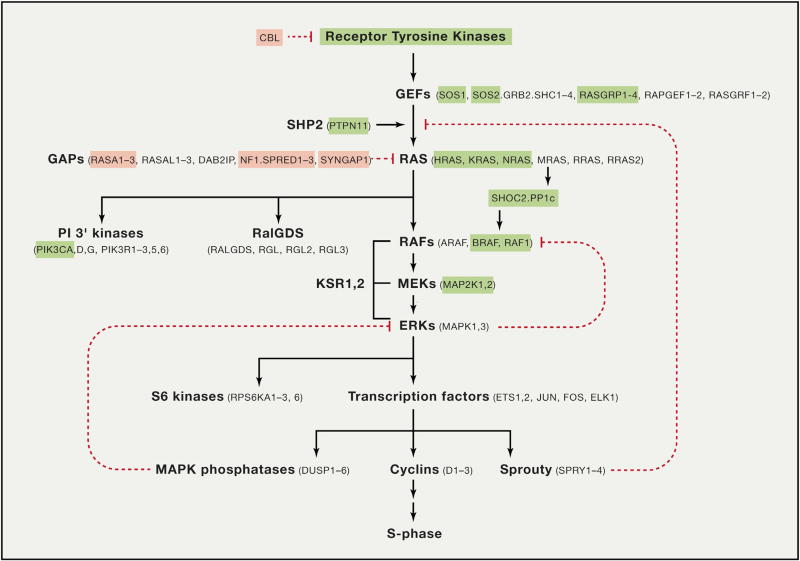
Figure 2. The RAS Pathway.
Genes highlighted in pink are frequently deleted in human cancers and RASopathies, as described in the text. Genes in green are frequently activated by mutation. This pathway is a simplified version of the pathways illustrated on http://www.cancer.gov compiled by the RAS Initiative and the RAS research community.
In humans, analysis of RASopathies (below) shows that germline-activated alleles of HRAS, NRAS, and KRAS exert similar phenotypes to those driven by activated BRAF, CRAF, MEK1, or MEK2, underscoring the central importance of the RAF/MAPK cascade in RAS biology. However, mammalian RAS proteins have other important effectors, of which PI 3-kinase (PI3K) is the best validated. For example, Catellano et al. (2013) and Gupta et al. (2007) generated mice expressing a mutant form of PI3Kα that fails to bind RAS: most of these mice did not survive to adulthood and those that did had defective lymphatic systems and were defective in angiogenesis and in macrophage function (Murillo et al., 2014). This interaction is, therefore, essential, albeit in specific tissue types. Other interactions between RAS and putative effectors have not yet been analyzed at this level in normal development.
More than seven distinct RAS effector proteins have been identified in cancer cells, in addition to well-studied RAF kinases, PI3K, RalGDS shown in Figure 2, including novel RAS effector 1A (NORE1A), Af6, phospholipase C (PLC), RAS and Rab interactor 1 (RIN1), T cell lymphoma invasion and metastasis-inducing protein (TIAM), and growth factor receptor 14 (Grb14) (summarized in Stephen et al., 2014).
Several crystal structures of GppNHp-bound RAS in complexes with effector proteins have been determined, all involving the G-domain of HRAS. No structures of RAS-effector complexes have been reported with KRAS or NRAS. Structures of HRAS-effector complexes include HRAS complexed with CRAF, PI3Kγ, RalGDS, NORE1A, PLCε, Grb14, and Bry2 (Mott and Owen, 2015). With the exception of a HRAS-PI3Kγ complex (Pacold et al., 2000), other HRAS-effector complex structures include only the effector RAS-binding domains (RBD) or effector RA-associating domains (RAs), thus providing limited information about the mechanism of RAS activation. Structural analysis reveals that the mode of interaction is highly similar in all these RAS-effector complexes. The RBDs of the effector proteins contain a ubiquitin-like fold and they interact with HRAS switch regions mainly via the β2 strand of RBD. Effector binding sites in HRAS vary for different effector proteins. RAF kinase primarily interacts via the β2 strand and switch I region whereas other effector proteins also interact with residues present in the switch II region.
The affinity of RAS for RAF kinases is in the 10–50 nM range, considerably higher than the affinity of RAS for other effectors or GAPs, which are in the low micromolar range. This has led to the suggestion that GAPs cannot compete effectively with RAF for binding to RAS, and GAPs cannot be responsible for terminating RAS signaling to RAF (discussed in Hunter et al., 2015). However, the mechanism by which GAPs (and GEFs) regulate RAS proteins is through recruitment to the plasma membrane in response to P-Tyr interactions or other signals. Once in the membrane, they achieve high local concentrations that are capable of interacting effectively with RAS. Likewise, the affinity of RAS for PI3Ks is relatively low, but these enzymes are recruited to the plasma membrane during receptor tyrosine kinase (RTK) signaling, again through P-Tyr interactions and other signals, again creating local concentrations that may be more compatible with direct interaction with RAS.GTP. Experimentally, complexes between RAS proteins and their effectors are hard to analyze when these complexes are released from membranes during cell lysis and analysis.
The effector binding regions of HRAS, NRAS, KRAS4a, and KRAS4b are identical, and all the effectors described above bind to all isoforms. Yet, clear differences between biological properties of these RAS proteins have been described (e.g., see Haigis et al., 2008). Some of these differences reflect differences in gene expression (To et al., 2008), others relate to specific interactions of proteins or membranes with the C-terminal hypervariable regions (HVRs) described below, but the possibility that there may be effectors that are specific for individual isoforms cannot be ignored.
RAS Proteins in the Membrane
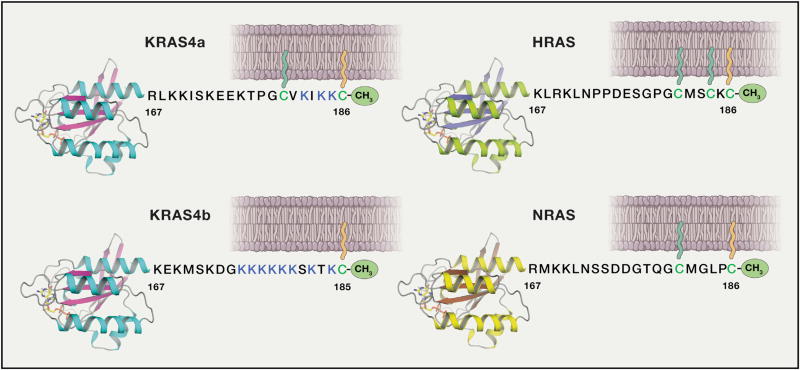
The subcellular localization of RAS proteins is determined by the specific lipid modification, composition of local membranes, and electrostatic nature of the isoform-specific HVRs formed by 19–20 residues present at the C-terminal end (Abankwa et al., 2010) (Figure 3).
Figure 3. Schematic of the Four Human RAS Proteins.
In humans, three RAS genes encode four distinct isoforms: HRAS, NRAS, and the two splice variants of KRAS gene, KRAS4a and KRAS4b, containing exons 4a and 4b, respectively. All four RAS isoforms contain identical residues in the first half of the GTPase domain (G-domain) and they share 82% sequence identity in the second half of the G-domain. The last 19–20 amino acids located at the C-terminal end exhibit significant sequence diversity among RAS isoforms and constitute the “hypervariable region (HVR)”. Unlike the other RAS isoforms, KRAS4b is not palmitoylated and contains a polybasic region (shown in blue color). In the figure, G-domains are shown in ribbon representation and membrane anchored farnesyl and palmitoyl lipid chains are shown in orange and green color.
Association with membranes is essential for activating downstream signaling pathways and this is facilitated by prenylation and palmitoylation of cysteine residues located in the HVR (Jackson et al., 1990; Willumsen et al., 1984). Prenylation of RAS proteins involves farnesylation of the cysteine residue present in the C-terminal CaaX motif (where “a” is an aliphatic amino acid whereas × can be any amino acid) by farnesyltransferase (Hancock et al., 1989). After prenylation, additional processing of CaaX motif involves cleavage of the -aaX residues by RCE1 (RAS converting CaaX endopeptidase 1) (Boyartchuk et al., 1997), followed by carboxymethylation of farnesylated cysteine residue by ICMT (isoprenylcysteine carboxyl methyltransferase) (Dai et al., 1998; Gutierrez et al., 1989). Almost two decades ago, farnesyltransferase inhibitors (FTIs) showed high preclinical anticancer activity in tumors driven by HRAS (Kohl et al., 1995). However, they failed to show the same efficacy against KRAS-driven cancers (Lobell et al., 2002). Later, it was shown that in the presence of FTIs, KRAS, and to a lesser extent NRAS, but not HRAS, undergoes alternative prenylation by geranylgeranyl transferase-I (Rowell et al., 1997; Whyte et al., 1997).
Trafficking between membranes requires additional targeting elements that HRAS and NRAS acquire by transiting to the Golgi where they are palmitoylated by DHHC9 and GCP16 (Swarthout et al., 2005) and subsequently undergo vesicular transport to the plasma membrane. KRAS4a is palmitoylated by an unknown palmitoylacyltransferase and apparently traffics to the plasma membrane without transiting the Golgi. Unlike other RAS isoforms, KRAS4b does not undergo palmitoylation but contains a poly-lysine region in the HVR that facilitates RAS-membrane association by interacting with the negatively charged head groups of membrane lipids (Hancock et al., 1991).
Differential HVR modification is also thought to drive the partitioning into distinct non-overlapping lipid microenvironments on the cytoplasmic face of the plasma membrane. For instance, HRAS and NRAS associate with cholesterol-rich liquid ordered (lo) lipid domains (rafts) (Vogel et al., 2009; Weise et al., 2009), while KRAS4a resides in liquid disordered (ld) domains (Weise et al., 2011). HRAS (Rotblat et al., 2004) and NRAS (Kapoor et al., 2012) move in opposite directions into and out of lipid rafts dependent on GDP/GTP-bound states that interact with lo or ld lipid components. KRAS4b clusters in “spatially distinct” nucleotide-dependent ld nanoclusters and the polybasic KRAS4b HVR orders lipids as part of a unique signaling complex. These distinct RAS micro-foci may contribute to isoform-specific signaling output as many of the effectors that interact with GTP-bound RAS have their own membrane interaction domains that display lipid specificity (Erwin et al., 2016).
The polybasic KRAS4b HVR can be phosphorylated at serine 181 by protein kinase C (Ballester et al., 1987) resulting in a decreased net charge, reduction in affinity for the PM, and accumulation of KRAS on endo-membranes (Zhang et al., 2017; Bivona et al., 2006; Sung et al., 2013). Ca2+/calmodulin is able to “extract” KRAS4b from the PM and redistribute it to endo-membranes (Fivaz and Meyer, 2005). A model to explain the Ca2+/calmodulin-mediated cycling of KRAS4b has been put forth (Sperlich et al., 2016): Ca2+/calmodulin releases KRAS4b from the plasma membrane where it is transported to endo-membranes by PDEδ (Schmick et al., 2014) that can deliver KRAS4b to a recycling endosome and transport it back to the plasma membrane. PDEδ displaces members of prenylated RAS family proteins from membranes and has been classified as a novel class of GDIs, proteins that extract and mobilize other small GTPases from membranes (Chandra et al., 2011). Depletion of PDEδ results in RAS mis-localization and consequently in attenuated signaling. Earlier deltarasin and recently deltazinone 1, which shows relatively less nonspecific cytotoxicity than deltarasin, were developed as inhibitors of the KRAS4b-PDEδ interaction (Papke et al., 2016; Zimmermann et al., 2013).
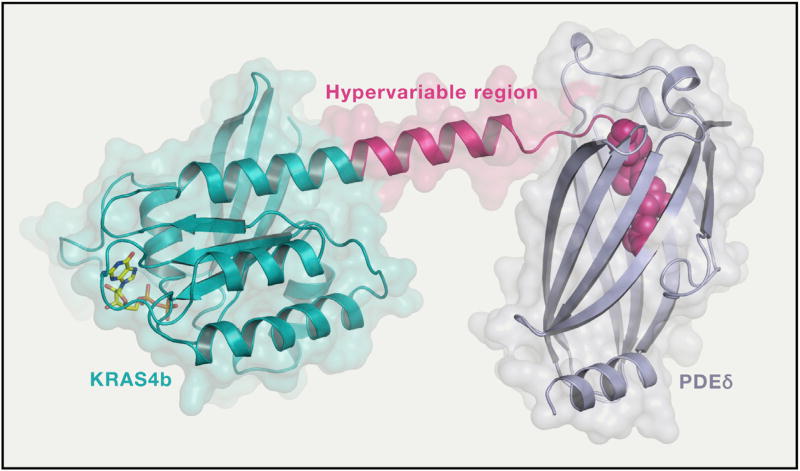
Our structural work on full-length KRAS4b in complex with PDEδ showed that unlike Rho and Rab GDIs that interact with both prenylated lipid and switch regions of G-domain, PDEδ only interacts with the C-terminal amino acids together with the farnesylated and methylated C185 of KRAS4b (Dharmaiah et al., 2016). Structure-based sequence analysis identified a 5-amino acid long sequence motif (Lys-Ser-Lys-Thr-Lys) in KRAS4b that may enable PDEδ to bind to both farnesylated as well as geranylgeranylated KRAS4b. Comparison of structures of PDEδ complexed with various prenylated proteins/peptides solved so far shows larger protein-protein interaction interface in KRAS4b-PDEδ complex that could be used for improving inhibitor design to minimize off-target effects (Figure 4).
Figure 4. Structure of Full-Length Farnesylated and Methylated KRAS4b in Complex with PDEδ.
This structure (PDB: 5TAR) provided atomic details of the hypervariable region for the first time. In this structure, HVR residues 166–180 are extend of helix 5. In the figure, KRAS4b and PDEδ are shown in ribbon and surface representations whereas prenylated C185 and GDP are shown in sphere and stick representations, respectively.
Computational approaches have been used to predict conformational dynamics of RAS on the plasma membrane and suggest that the G-domain determinants interact directly with the membrane and are dynamically regulated by activating mutations, as well as nucleotide-bound state (Jang et al., 2016; Prakash and Gorfe, 2016; Prakash et al., 2015, 2016). A combination of simulations and mutational analysis of signaling output have led to a model where GTP-bound HRAS G-domain helix 4 contacts the membrane while in the GDP-bound state, the protein adopts a conformation where helix 4 is away from the membrane and HVR residues contact the membrane (Abankwa et al., 2010). Similar computational studies predict that KRAS4b is subject to extensive conformational dynamics and interacts with membranes via two orientations (Prakash et al., 2016). NMR studies using chemically farnesylated full-length GDP-KRAS4b bound to nanodiscs also show G-domain membrane contacts via helices 4 and 5 (Mazhab-Jafari et al., 2015). Further simulations suggest two states for GTP-bound KRAS4b, one where the effector-binding domain of the protein is occluded and a second where it is solvent exposed and available for effector interaction (Jang et al., 2016).
In addition to effects on localization of KRAS4b, binding of KRAS4b to calmodulin (Villalonga et al., 2001) inhibits CaM kinase and affects downstream pathways involved in stem-like cell properties. In this way, oncogenic KRAS4b uniquely elicits a tumorigenic phenotype through downregulation of non-canonical Wnt/Ca2+ signaling (Wang et al., 2015). Binding of calmodulin to KRAS4b, but not to NRAS, HRAS, or KRAS4a, may be responsible for this major difference (Villalonga et al., 2001).
RAS May, or May Not, Be a Dimer
Bremner and Balmain (1990) reported the presence of a strong suppressor of HRAS-driven skin cancers in mice: this suppressor was wild-type HRAS. Likewise, wild-type KRAS had a clear suppressive effect on tumorigenesis by oncogenic KRAS (Zhang et al., 2001). More recently, To et al. (2013) showed that the suppressive effects of normal RAS alleles depends on the tissue of origin, the gene itself, and the strain of the host. Perhaps the most striking and baffling is the discovery that wild-type KRAS appears to suppress the Q61L mutant more effectively than Q61R (Westcott et al., 2015). These data suggest that RAS proteins form complexes, and wild-type proteins can act as sub-unit poisons in these complexes. Equally baffling, Xu et al. (2013) have shown that wild-type NRAS does not exert a suppressive effect on tumors induced by its oncogenic counterpart.
Because RAF activation by RAS is known to involve RAF dimerization, and each monomer has the potential to bind RAS an attractive model for the state of RAS in signaling complexes is that it is indeed present as a dimer (Freeman et al., 2013). Early evidence for an RAS dimer driving RAF activation comes from studies using single-lipid liposomes, HRAS isolated from insect cells, and BRAF complemented with extracts containing CRAF. Chemical cross-linking of RAS or forced protein:protein dimerization through tags resulted in RAF activation that was not seen in the presence of RAS monomers (Inouye et al., 2000). Additional support for a RAS dimer model came from work using fluorescently tagged RAS and visualization of single RAS molecules via super-resolution microscopy. The authors demonstrated the presence of RAS dimers in live cells and the coincident activation of MAPK signal at minimal RAS protein levels that result in dimers (Nan et al., 2015). This work also showed that forced RAS dimerization results in activation of the pathway.
The existence of RAS dimers in membranes has been hard to prove unequivocally (Güldenhaupt et al., 2012; Lin et al., 2014; Nan et al., 2015). RAS certainly forms clusters and higher order structures as approaches using higher-resolution electron microscopy have led to the observation that in fixed plasma membranes different isoforms of RAS localize to discrete membrane environments based on HVR modification (reviewed in Zhou and Hancock, 2015). These studies also suggest that RAS exists in “nanoclusters” comprised of five to ten monomers. Whether these nanoclusters are comprised of non-interacting monomers, dimers, or higher order protein:protein multimers and whether these are signaling foci that also mediate RAF dimerization is not known. A recently described synthetic binding protein “monobody” that interacts with helices 4 and 5 and occludes RAS:RAS dimers and nanoclusters may make it possible to modulate multimerization and determine the role of higher order structures in RAS signaling (Spencer-Smith et al., 2017).
RAS and Its Close Relatives
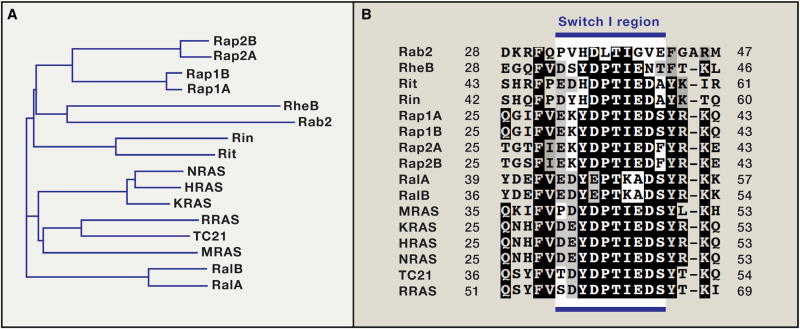
HRAS, NRAS and KRAS4a and 4b have several close relatives (Figure 5A). The binding regions (switch I) of these proteins are remarkably similar (Figure 5B), yet the biological functions of these relatives are completely different (Raaijmakers and Bos, 2009). For example, Rap1a can bind to the RBDs from the RAS effectors CRAF, PI3K, and RalGDS in vitro, but does not activate any of these effectors efficiently in cells (Rodriguez-Viciana et al., 2004). The major functions of Rap proteins relate to control of cell adhesion, cell junction formation, cell secretion, and cell polarity, all functions that are distinct from those of RAS and depend on distinct effectors, as well as specific GEFS and GAPs (Bos et al., 2007). These observations suggest that signaling specificity in vivo is determined by additional factors that may include specific points of contact between RAS proteins and their effectors, beyond the Switch I/RBD binding site, or contributions from other cellular proteins or membrane components, most of which are unknown.
Figure 5. Phylogenetic Analysis and Conservation of Switch I Region in Members of RAS Subfamily.
(A) Phylogenetic analysis of members of RAS subfamily proteins from humans. (B) Multiple sequence alignment showing conservation of residues in the switch I region in different members of RAS subfamily in humans.
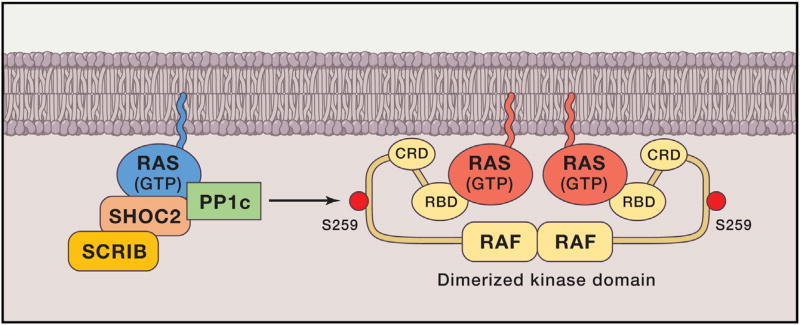
A clear, functional relationship has been demonstrated, however, between MRAS and other RAS proteins. MRAS is structurally similar to HRAS, NRAS, and KRAS and shares an almost identical effector binding sequence at switch I (Figure 5B). However, in contrast to other RAS proteins and 30 other GTPases, including the closest relatives RRAS and RRAS2/TC21, MRAS binds directly to SHOC2. SHOC2/SUR8 was first identified as a positive regulator of the RAS pathway in C. elegans. It is a scaffold protein composed almost entirely of leucine-rich repeats. The MRAS.SHOC2 complex also binds catalytic subunit of PP1 (PP1c) and recruits it to the plasma membrane, where it de-phosphorylates CRAF kinase at S259 (S365 in BRAF, S214 in ARAF), resulting in RAF activation (Figure 6). Remarkably, de-phosphorylation only occurs on RAF that is complexed to RAS proteins in the membrane. Therefore, MRAS cooperates with its cousins HRAS, NRAS, or KRAS to promote full activation of RAF kinase in the plasma membrane. This model was based on biochemical analysis of MRAS in cell-based systems (Rodriguez-Viciana et al., 2006), but was confirmed by analysis of gain-of-function mutations in SHOC2 that cause Noonan syndrome (NS) (discussed below): these mutations create a myristoylation site at the amino terminus so that SHOC2 is localized in the plasma membrane constitutively, leading to hyperactivation of MAPK signaling (Cordeddu et al., 2009). More recently, PP1 mutations and MRAS mutations have been identified in NS, adding more weight to the model (Gripp et al., 2016; Higgins et al., 2017). SHOC2 was recently identified as one of five genes necessary for proliferation of RAS mutant AML relative to RAS-wild-type AML (the others were RCE1, ICMT, CRAF, and PREX1) (Wang et al., 2017), further underscoring the importance of MRAS/SHOC2/PP1c in RAS signaling.
Figure 6. MRAS Recruits the Phosphatase PP1c to the Plasma Membrane and Dephosphorylates S259 on CRAF Bound to Active RAS.
Amongst all RAS subfamily members, only MRAS (shown in blue color) bind SHOC2.PP1c, and it does so in a GTPdependent manner. Recruitment of this complex to the plasma membrane de-phosphorylates the negative regulatory site on CRAF, S259, and equivalent sites on ARAF and BRAF. This step is essential for efficient signal transduction from RAS to the MAPK cascade.
MRAS has other interesting properties. It is part of an epithelial-to-mesenchymal transition (EMT) expression signature and can promote EMT itself. Its GAP is the semaphorin 4D receptor plexin-B1 that regulates invasion and migration. Furthermore, the SHOC2 complex that MRAS recruits to the plasma membrane also includes the protein SCRIB, to which SHOC2 binds directly. SCRIB has been implicated in several aspects of malignant growth, including altered cell polarity and migration, as well as regulation of ERK and Hippo signaling, and its participation in signaling from the MRAS/SHOC2/PP1c complex provides a direct link between RAF/ERK signaling and spatial cell organization and dynamics (Young et al., 2013b).
RAS and Human Disease
Cancer
The widespread prevalence of RAS mutations in human cancer has been recognized for many years: mutations in KRAS alone account for approximately one million deaths per year worldwide, a similar toll as malaria and tuberculosis. Analysis of the current data available at the cBioPortal database reveals that among the RAS isoforms, missense mutations are found most commonly in KRAS (85%) and much less in NRAS (12%) and HRAS (3%) (Table 1).
Table 1.
Frequency of Mutation in RAS Isoforms and Related Proteins in Selected Human Cancers.
| Tissue | KRAS (%) | NRAS (%) | HRAS (%) | RITI (%) | NFI (%) | SPRED1 (%) | SOS1 (%) |
|---|---|---|---|---|---|---|---|
| Pancreas | 91 | 0 | <1 | <1 | 2 | <1 | <1 |
| Colon | 42 | 9 | 0 | <1 | 4 | 3 | 3 |
| Lung | 33 | <1 | <1 | 2 | 12 | 1 | 3 |
| Skin (melanoma) | 2 | 27 | 1 | 1 | 13 | 2 | 3 |
| Endometrium | 21 | 4 | <1 | 1 | 8 | 1 | 5 |
| Testicular germ cell | 13 | 5 | <1 | 0 | 2 | 0 | 1 |
| Bone marrow | 4 | 8 | 0 | <1 | 1 | 0 | <1 |
| Thyroid | 1 | 8 | 3 | 0 | <1 | 0 | <1 |
| Thymus | <1 | 2 | 8 | 0 | 2 | 0 | 0 |
| Stomach | 9 | 1 | <1 | 0 | 8 | 3 | 3 |
| Adrenal glands | 0 | 0 | 10 | 0 | 8 | 0 | 0 |
| Bladder | 0 | 1 | 5 | 1 | 8 | 1 | 3 |
| Brain | 1 | 1 | 0 | 0 | 14 | 0 | 0 |
| Cervix | 6 | <1 | <1 | 2 | 4 | <1 | 2 |
| Bile duct | 6 | 0 | 0 | 3 | 0 | 3 | 3 |
Data were compiled from the TCGA datasets present in the cBioPortal database for cancer genomics.
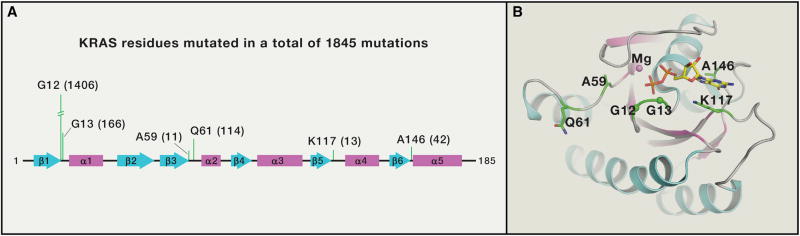
Most RAS mutations occur at codons 12, 13, or 61 (Figure 7). Replacement of glycine at codons 12 or 13 by any amino acid except proline are thought to present a steric block that prevents GAP’s arginine finger from entering the GTPase site and promoting hydrolysis (Scheffzek et al., 1997), although this has not yet been confirmed experimentally. Glutamine-61 is part of the GTP hydrolysis mechanism, so mutations at this residue destroy both intrinsic and GAP-mediated GTP hydrolysis. Mutations also occur that decrease affinity for nucleotide, such as A146 mutations (4% in colorectal cancer) (Edkins et al., 2006). A146 mutants and others (e.g., T158A, R164Q, and K176Q) allow GDP to dissociate rapidly, resulting in abnormal accumulation of RAS in the GTP-form without assistance from upstream signals and GEFs. The net result of all these activating mutations is increased GTP-loading without upstream input.
Figure 7. KRAS Residues with Higher Mutational Frequency in Cancer.
(A) KRAS residues with more than 10 mutations in cBioPortal database for cancer genomics are shown along with number of mutations (in a total of 1845 mutation) in parenthesis. All these residues are in the switch regions or in the G1-G5 sequence motifs that play a critical role in recognition and binding of guanine nucleotide. (B) Position of KRAS residues (shown in green color) with more than 10 mutations in cBioPortal database are mapped on the tertiary structure of KRAS.GDP (PDB: 4EPV).
Levels of GTP associated with oncogenic RAS proteins can be measured experimentally by labeling cells to equilibrium with 32-P, followed by immunoprecipitation and analysis of bound nucleotide by TLC (see Trahey and McCormick, 1987) or by pull-down experiments using the RAS-binding domain of RAF. The proportion of RAS in the GTP state can also be calculated from the GDP-off rate and the GTPase rate. RAS.GTP/RAS.GDP = Koff/Kcat assuming the cellular levels of GTP exceed GDP. For all G12, G13, and Q61 mutants, Kcat is much slower than Koff so that most of the RAS protein is in the GTP-state. Even G12C, which has a relatively high intrinsic GTPase rate for an oncogenic mutant, is ~75% GTP bound at steady state (Patricelli et al., 2016).
The RAS-related proteins Rit1 and Rit2 are activated in lung adenocarcinoma by mutations that inhibit GTP hydrolysis (F82V and T83P) or increase nucleotide exchange (S35T, A57G, and Y89H). However, the mutations that cause these shifts toward the GTP-bound form are distinct from those that activate RAS proteins, with the exception of one mutation corresponding to Q61 in RAS proteins (Q79 in Rit1). The lack of frequent mutations at codon 12, 13, and 61 equivalents is thought to be because substitutions in these codons create splice sites or termination codons (Zhu et al., 2016). Rit proteins activate RAF kinase, RalGDS, PI3Kα, and PI3Kγ (Rodriguez-Viciana et al., 2004). In these respects, they resemble HRAS, NRAS, and KRAS. Indeed, while Rit proteins are only 44% identical to RAS proteins, their effector binding site is nearly identical (Figure 5B). The GAPs and GEFs that regulate the Rit GTPase cycle have not been identified.
Activating mutations at codons 12, 13, and 61 render RAS proteins resistant to GAPs, as discussed above. Loss of GAPs through mutation or deletion might, therefore, phenocopy RAS activation. This may seem unlikely as there are at least 14 different forms of GAP in human cells (Bernards, 2003). Loss of any one of these may have less effect than mutations in RAS that are resistant to all of them. Unfortunately, this is not the case: loss of the RasGAP neurofibromin occurs frequently in human cancers, and tumors driven by loss of neurofibromin are often indistinguishable from those driven by mutant RAS. In these tumors, neurofibromin appears to be the major GAP that maintains RAS proteins in their inactive states.
Mutations in the RasGAPs were first reported in cell lines from sporadic cancers more than 25 years ago, but the high frequency of NF1 mutations in human cancer has only been recognized recently, facilitated by advances in DNA sequencing analysis. The NF1 gene spans 60 exons and >300 kb of genomic DNA with few recurrent mutations that would simplify the sequencing process. Furthermore, we understand very little about the function of the protein: the GAP domain is ~333 amino acids out of a total of protein of 2,818 amino acids. It is, therefore, hard to distinguish between mutations that cause loss of function from passenger mutations. Mutations associated with neurofibromatosis type 1 disease, of which over 1,400 have been published (Stenson et al., 2008), are informative, but calling true loss-of-function mutations in sporadic cancers is still complicated. Mutations in NF1 at high frequency (18%) were reported from one of the earliest TCGA projects, recurrent glioblastoma. In this analysis, very few RAS mutations (2%) were detected. However, the NF1-subset has features that distinguish this subset from other classes (defined by EGFR mutation, PDGFR overexpression) (Brennan et al., 2009), including increased expression of mesenchyme markers and high frequency of PTEN alterations (McGillicuddy et al., 2009). NF1 also plays a major role in lung adenocarcinoma, in which NF1 mutations occur at about the same frequency of mutations in EGFR (11%, TCGA). Loss of SPRED1, neurofibromin’s essential partner, occurs in another 4% of lung adenocarcinomas. In this disease, patients with KRAS mutations (32% in the same study) are excluded from treatment with EGFR inhibitors, as they derive no benefit. Whether NF1-deficient tumors respond to EGFR inhibitors is not clear, as this cohort has not yet been evaluated. However, low expression of neurofibromin in lung adenocarcinoma has emerged as a mechanism of resistance to the EGFR inhibitor erlotinib (de Bruin et al., 2014).
In sun-exposed melanoma, the NF1 gene is the third most frequently mutated gene after BRAF and NRAS. In the majority of NF1-mutated melanoma, other mutations occur in the RAS pathway, including SOS1, CRAF, SPRED1, and another RasGAP, RASA2 (Krauthammer et al., 2015). Further analysis of the role of RASA2 in melanoma revealed loss of expression, measured by immunohistochemistry, in ~33% of cases (Arafeh et al., 2015). In desmoplastic melanoma, the frequency of NF1 mutations is over 50% (Shain et al., 2015). Loss of neurofibromin is one of the most frequent recurrent mutations in myxofibrosarcoma and pleomorphic liposarcoma (Barretina et al., 2010). Similar studies have identified genomic loss of the NF1 locus in pediatric embryonal rhabdomyosarcomas (Paulson et al., 2017). Loss of SPRED1 also occurs in many sporadic tumors (cBioPortal).
The RASA1 gene encodes p120 RasGAP, which binds to activated RTKs, such as PDGFR, through its SH2 domains and presumably turns off RAS proteins associated with these receptors. p120 RasGAP also forms a complex with the RhoGAP p190 (Settleman et al., 1992), encoded by the ARHGAP5 gene, raising the possibility that loss of RASA1 leads activation of Rho proteins as well as RAS proteins. This gene is mutated quite frequently in human cancer (Campbell et al., 2016).
While somatic mutations and deletions in the RasGAPs neurofibromin/SPRED1, RASA1, and RASA2 reveal major roles in human cancer, loss of expression by epigenetic silencing of the RasGAPs DAB2IP and RASAL2 (McLaughlin et al., 2013) contributes directly to metastatic luminal B breast cancer and prostate cancer. These diseases have been notable for the absence of frequent mutations in RAS genes themselves. In prostate cancer, loss of DAB2IP expression leads to coordinated activation of RAS and nuclear factor κB (NF-κB) (Min et al., 2010), whereas in luminal B breast cancers, simultaneous loss of DAB2IP and RASAL2 drives invasion, EMT, and metastasis (Olsen et al., 2016).
RASopathies
The RASopathies are a group of clinical syndromes caused by hyperactivation of the RAS/MAPK pathway. One of the commonest is neurofibromatosis type 1, an autosomal dominant syndrome first described in detail in 1882 by Friedrich Daniel Von Recklinghausen. The incidence of neurofibromatosis type 1 is ~1 in 3,500, of which ~50% are inherited and 50% are due to de novo mutations. This translates to ~100,000 cases in the United States alone. Mosaic neurofibromatosis type 1 occurs at approximately one-tenth this frequency (García-Romero et al., 2016).
Neurofibromatosis type 1 is often first diagnosed by the presence of café-au-lait macules. Other features include Lisch nodules, optic gliomas, hypertension, short stature, and macrocephaly as well as skeletal abnormalities and learning disabilities (Boyd et al., 2010). When the NF1 gene was cloned, the predicted sequence revealed a region of homology to p120 RasGAP and S. cerevisiae IRA1 and IRA2 proteins, and it was quickly shown that the gene product, neurofibromin, is indeed a GAP that negatively regulates RAS (Martin et al., 1990; Xu et al., 1990). At that time, it seemed remarkable that that the multiple, complex phenotypes associated with this disease may simply be due to hyperactive RAS. However, the variety of phenotypes now known to be caused by mutations in other genes in the RAS pathway, described below, confirm that neurofibromatosis type 1 is indeed a RAS-driven disease in all its manifestations.
While it is clear that neurofibromin is a GAP that negatively regulates RAS, it is less clear which forms of RAS become active when neurofibromin is defective. This is because neurofibromin also acts on MRAS, RRAS, and RRAS2 (also known as TC21) (Ohba et al., 2000). RRAS proteins, unlike HRAS, KRAS, and NRAS activate PI3Kγ (Rodriguez-Viciana et al., 2004). MRAS, in its GTP-bound form, has the unique ability to recruit SHOC2/PP1c to the plasma membrane (Rodriguez-Viciana et al., 2006) and to regulate SCRIB activity (Young et al., 2013b), as described above (Figure 6). Therefore, loss of neurofibromin could have broader effects on cells than activation of HRAS, NRAS, and KRAS proteins themselves.
Neurofibromatosis type 1 patients may develop dermal or plexiform neurofibromas, ~10% of which progress to malignant peripheral nerve sheath tumors that can be metastatic. They are also at increased risk of developing breast cancer and other malignancies (Uusitalo et al., 2016). It is curious that the cancers frequently caused by sporadic loss of neurofibromin (e.g., lung adenocarcinoma, glioblastoma) are not especially prevalent in individuals who inherit a defective NF1 allele. This discordance is also observed in other RASopathies, as described below. Individuals with germline mutations in BRAF, for example, are not at higher risk for melanoma.
One important feature of neurofibromatosis type 1 is the extent of phenotypic variation between individuals with the same germline mutation pointing toward strong effects of modifier genes. Indeed, twin studies revealed that each of the major phenotypes associated with neurofibromatosis type 1 is affected by distinct modifiers (Easton et al., 1993), none of which have yet been identified. This feature, along with the natural history of this complex disease and the relatively small number of recurrent mutations has precluded identification of clear genotype-phenotype correlations (Li and Wu, 2016). Recent psychiatric analysis of neurofibromatosis type 1 patients revealed that the NF1 gene functions as a quantitative trait locus that is associated with autism spectrum disorders (ASD) (Morris et al., 2016). A strong association in quantitative autistic trait burden between first-degree relatives suggested a high degree of mutational specificity for ASD severity. Interestingly, mouse models of neurofibromatosis type 1 driven by either a nonsense mutation or a missense mutation have distinct phenotypes, providing the clearest evidence that different mutations have different consequences beyond simple loss-of-function activity.
Legius syndrome is a recently recognized, milder form of neurofibromatosis type 1, in which the NF1 gene is intact, but, instead, loss-of-function mutations occur in SPRED1, a previously known suppressor of RAS/MAPK signaling. SPRED stands for Sprouty-related EVH1-domain containing protein (Brems et al., 2007). By comparing proteins that bind to wild-type SPRED1 and to pathogenic mutants, we found that SPRED1 binds directly to neurofibromin through its EVH1 domain, and SPRED proteins are essential for neurofibromin to interact with RAS in the plasma membrane (Stowe et al., 2012). Loss of SPRED1, therefore, partially phenocopies loss of neurofibromin. We speculate that Legius syndrome is less severe than neurofibromatosis type 1 because SPRED2 and SPRED3 proteins can also bind neurofibromin and can compensate, to some extent, for loss of SPRED1. The regions of neurofibromin that binds SPRED proteins comprise a discontinuous domain that flanks the GAP-related domain. SPRED binding does not affect neurofibromin’s GAP activity (Dunzendorfer-Matt et al., 2016) but is essential for enabling neurofibromin to downregulate RAS through recruitment to RAS in the plasma membrane.
Noonan syndrome (NS) is the most genetically diverse RASopathy and is the most common, with estimates of incidence of 1 in 1,000–2,500. Clinical features of NS include craniofacial dysmorphic features and short stature, undescended testicles, bleeding and predisposition to cancer, and most notably, juvenile myelo-monocytic leukemia and heart disease. Indeed, NS is one of the leading congenital syndromes predisposing to cardiac disease, along with Down syndrome which occurs at about the same frequency. Most of the mutations are familiar components of the RAS pathway: PTPN11 (Shp2), SOS1 and SOS2, HRAS, KRAS, RRAS, NRAS, MRAS and RIT1, the GAP RASA2, the kinases CRAF, BRAF, MEK1, MEK2, and MAP3K8 as well as Sprouty 1 and CBL, SHOC2, and its binding partner PP1. Recent additions to the list of genes causing NS include the histone acetyltransferase MYST4, the adaptor protein LZTR1 and the protease inhibitor A2ML1. The role of these proteins in regulating the RAS/ MAPK pathway, if indeed this is how they function, is not yet known (Tidyman and Rauen, 2016).
SHOC2 mutations have been reported in combination with mutations in PTPN11 (SHP2) in NS, each contributing to the overall phenotype (Ekvall et al., 2011). Likewise, PTPN11 mutations have been found in combination with mutations in SOS2 (Brasil et al., 2010a), KRAS (Brasil et al., 2010b), and NF1 (Thiel et al., 2009) in Noonan-like syndromes. Clinically defined variants of NS include NS with multiple lentigines (also known as LEOPARD), caused by mutations in PTPN11, CRAF, or BRAF, and NS-like disorder caused by mutations in CBL (Aoki et al., 2016).
Cardiofaciocutaneous syndrome (CFC) shares many features with NS, such as distinctive facial appearance and sparse and brittle hair, and is characterized by thick scaly skin, delayed growth, and cardiac malformations (Rauen, 2007). CFC is caused by activating mutations in BRAF and less frequently by KRAS, MEK1, or MEK2. Some of these mutations have been reported in sporadic malignant melanoma, but the commonest melanoma allele V600E has not been reported in CFC. KRAS alleles in CFC are partially active relative to cancer-causing mutations, retaining some sensitivity to GAP inhibition (Schubbert et al., 2006). To date, CFC has not been associated with increased risk of cancer. In contrast, Costello syndrome, which is caused by germline mutations in HRAS, is associated with increased risk of rhabdomyosarcomas and neuroblastomas. Diseases associated with sporadic mutations in HRAS, such as thyroid cancer and bladder cancer, have not been associated with Costello syndrome (Rauen, 2007).
Comparison of RASopathies with germline diseases of the PI3K pathway, such as PIK3CA-related overgrowth spectrum, Proteus syndrome, Cowden syndrome, and tuberous sclerosis complex (Keppler-Noreuil et al., 2016) is also of great interest, because of the close and complicated relationship between these major signaling pathways. Some of their phenotypes overlap. Indeed, the disease suffered by the Elephant Man has been mistakenly associated with neurofibromatosis type 1, but is now thought more likely to be Proteus syndrome, a disease caused by activatingmutations in AKT (Huntley et al., 2015). Likewise, skin lentigines are characteristic of the RASopathy Noonan/LEOPARD syndrome but are also seen in Peutz-Jeghers syndrome, a disease with other distinctive features caused by germline loss of LKB1, a downstream component of PI3K signaling.
Capillary Malformations
RASA1-related disorders are characterized by multiple capillary malformations on the face and limbs and, in many cases, arteriovenous malformations and arteriovenous fistulas. These can lead to heart failure and other severe consequences. RASA1 mutations have also been associated with the related condition known as Parkes Weber syndrome (Bayrak-Toydemir and Stevenson, 1993).
Psychiatric and Neurodevelopmental Disorders
The potential effects of hyperactive RAS on cognitive function, revealed through analysis of individuals suffering from RASopathies, has been underscored by the recent discovery that the RasGAP, SYNGAP1, is one of the most frequently mutated genes in patients suffering from intellectual disorder and epilepsy with comorbid ASD in children (Mignot et al., 2016). The GAP domain of SYNGAP1 is closely related to the GAP domain from neurofibromin and p120 RasGAP encoded by the RASA1 gene, including the critical “arginine finger” that facilitates GTP hydrolysis. However, SYNGAP1 also has GAP activity against Rap1. This is surprising, as Rap1 proteins hydrolyze GTP through a different mechanism: they have threonine instead of glutamine at codon 61 and their GAPs utilize an asparagine residue to facilitate GTP hydrolysis on Rap proteins. Furthermore, SYNGAP’s GAP activity against Rap1 requires a C2 domain from SYNGAP1, and the specificity of SYNGAP1 for RAS versus Rap proteins is regulated by phosphorylation by CaM kinase II and CDK5. Loss of SYNGAP1 may, therefore, activate both RAS and Rap proteins to contribute to its effects on neurocognition (Jeyabalan and Clement, 2016).
In mice, loss of SPRED2, a neurofibromin binding partner, is associated with facial lesions from excessive grooming, a phenotype resembling obsessive compulsive disorder (OCD). In these mice, RAS.GTP and MAPK signaling were elevated, consistent with loss of neurofibromin function. The OCD phenotype could be reversed by antipsychotic drugs, or, remarkably, by MEK inhibitors (Ullrich et al., 2017).
Therapeutic Approaches
Targeting Upstream
Cancers expressing oncogenic RAS mutations are resistant to therapies targeting RTKs. This is not surprising, as RAS proteins are downstream of these receptors. However, RAS-driven cancers retain varying degrees of dependence on upstream signaling and on wild-type RAS proteins (Young et al., 2013a; Grabocka et al., 2014). RTKs activate several pathways that can contribute to growth and survival of cells expressing oncogenic RAS mutations: first, they recruit and activate GEFs that increase GTP-loading on wild-type HRAS, NRAS, and KRAS proteins, as well as other members of the RAS subfamily, such as RRAS proteins and MRAS, each of which can contribute to overall signal output in these cells. Second, growth factors activate PI3K. In cells in which RAS proteins do not activate PI3K efficiently, growth factors may provide PI3K that is essential for proliferation and survival. Third, RTKs activate PLC gamma, leading to mobilization of calcium, generation of diacylglycerol, and activation of PKC. Ca2+ can affect localization and function of KRAS4b by promoting calmodulin binding, and PKC can phosphorylate KRAS4b and can contribute to activation of RAF kinase (Bivona et al., 2006; Alvarez-Moya et al., 2010).
Direct Attack on RAS Proteins
Small molecules that bind tightly to RAS proteins have been difficult to find, mostly because RAS proteins lack a deep pocket to which small molecules could bind with high affinity. Several low affinity compounds have been reported but do not appear to have been developed beyond their initial characterization (summarized in Ostrem and Shokat, 2016). More promising compounds that emerged from two independent fragment-based screens were reported in recent years, one from the drug discovery group at Genentech (Maurer et al., 2012) and the other from Sun et al. (2012). These compounds bind at the same pocket and prevent GEF-mediated GDP/GTP exchange. Because oncogenic mutants are relatively independent of SOS, in contrast to their wild-type counterparts, SOS inhibitors are likely to block wild-type RAS selectively. Neither of these compounds have advanced toward clinical evaluation, but Burns et al. (2014) have pursued this approach further and identified another series of interesting compounds that bind to a new pocket on SOS1 and unexpectedly increase SOS activity while preventing cell proliferation.
In 2013, Ostrem et al. (2013) developed a series of compounds that irreversibly target cysteine residue present in the G12C mutant. These compounds bind to RAS preferentially in the GDP-bound state and prevent exchange of GDP by GTP. They also impair SOS-catalyzed nucleotide exchange and block RAS-RAF interaction in G12C mutant cells. At first sight, this seems problematic, as oncogenic RAS proteins are mostly GTP-bound at steady state. Fortunately, however, G12C mutants have relatively high intrinsic GTPase activity, with a t½ of ~25 min, similar to wild-type RAS proteins. This affords the opportunity for GDP-targeted drugs to bind and covalently modify the GDP-bound form and eventually to reduce the active, GTP-state to negligible levels. Structural studies showed that these compounds bind in an allosteric pocket formed by stabilizing the highly dynamic switch-II region named as switch-II pocket. Further optimization of this scaffold resulted in an improved inhibitor, ARS-853, that modifies KRAS-G12C at lower concentrations than initial compounds and prevents exchange of GDP for GTP with much higher efficacy. Modification of KRAS-G12C in cells showed blockade of downstream signaling pathways (Patricelli et al., 2016).
Drugs that target mutant alleles such as G12C, such as that described above, and another by Xiong et al. (2016) obviously offer the best opportunity for safety and efficacy. However, strategies for targeting mutant proteins such as G12D and G12V have not yet been forthcoming. Therefore, drugs that do not discriminate between wild-type and mutant forms may be an inevitable compromise. RAS proteins are essential for transducing signals from all RTKs, as well as many GPCRs. Therefore, we expect that pan-RAS inhibitors would be unacceptably toxic. Indeed, ablation of all three RAS genes in most adult tissues is fatal in mice. However, adult mice can survive for ~1 year following ablation of only KRAS (M. Barbacid, personal communication). Drugs that ablate KRAS, but spare HRAS and NRAS, are likely to be tolerated. While this appears to simplify strategies for treating KRAS driven cancers, the fact that the KRAS gene encodes two splice variants, KRAS4a and KRAS4b, creates another challenge. Developing compounds that inhibit both forms of KRAS without affecting HRAS and NRAS will be difficult. Targeting KRAS4b alone might be effective, as KRAS4b is expressed at higher levels than KRAS4a in most tumors and has unique biochemical properties. KRAS4b binds calmodulin and is a substrate for phosphorylation by PKC (Alvarez-Moya et al., 2010; Villalonga et al., 2001; Bivona et al., 2006). KRAS4b, unlike other RAS proteins, promotes expansion of endoderm stem cells (Quinlan et al., 2008) and promotes a stem-like program through its interaction with Ca/Calmodulin, referred to above (Wang et al., 2015). However, it is currently unclear how effective a drug would be if it only targeted KRAS4b. KRAS4a is clearly capable of causing tumors. Indeed, Kirsten sarcoma virus is an activated form of KRAS4a.
Targeting RAS Effectors
Targeting RAF kinase appeared to be a logical strategy for treating KRAS mutant cancers, but the first RAF kinase inhibitor to enter clinical trials, sorafenib, showed no benefit in KRAS-driven cancers. Small skin lesions referred to as keratocanthomas were observed during these trials (Williams et al., 2011). Later, patients being treated with another RAF inhibitor, vemurafenib, also developed these lesions, at higher frequency. These are now attributed to paradoxical activation of RAF kinase by RAF inhibitors in RAS mutant cancers (reviewed in Holderfield et al., 2014).
This unexpected effect of RAF inhibition shifted focus to MEK inhibitors, which do not provoke the same paradoxical pathway activation, as well as efforts to target RAF kinase without paradoxical activation (Zhang et al., 2015). However, targeting MEK also has serious problems: first, MEK inhibition relieves feedback suppression of upstream signaling, leading to increased RTK signaling that can off-set the effects of inhibiting MEK (Lake et al., 2016). Second, MEK inhibition leads to loss of expression of DUSPs, allowing ERK activity to increase dramatically, again off-setting the effects of MEK inhibition. Third, and perhaps most importantly, the therapeutic window between normal cells and KRAS cancer cells may be minimal. In normal cells, MEK activity is transiently activated during growth factor signaling. In a population of proliferating normal cells, only a small fraction of cells are likely have high levels of MEK, giving the impression that MEK activity is low in normal cells compared to KRAS cancer cells, in which every cell has elevated MEK because of chronic RAS activation.
Despite these concerns and disappointing clinical activity against cancers driven by mutant RAS, MEK inhibitors showed promise in mouse models of neurofibromatosis type 1 (Jessen et al., 2013; Chang et al., 2013), and more recently, selumetinib appeared to benefit children with neurofibromatosis type 1 and inoperable plexiform neurofibromas with tolerable side effects, even over the long term (Dombi et al., 2016). This is a remarkable result that gives us reason to be optimistic about the future of treating this disease as well as sporadic tumors driven by loss of neurofibromin or SPRED1.
Blocking the protein-protein interfaces of RAS-effector complexes with small molecules constitutes an attractive therapeutic approach, and targeting the RBDs of various effectors has been suggested to be an effective strategy against RAS-driven cancers. Recently, a small-molecule inhibitor, rigosertib, was shown to act as a RAS mimetic and interact with the RBDs of RAF kinases, RalGDS and PI3Ks, resulting in their inability to bind to activated RAS (Athuluri-Divakar et al., 2016). Recently, results from Ritt et al. (2016) suggested a different mechanism on how rigosertib mediates its effects. The effects of rigosertib were seen to be largely indirect, mediated by the stress-induced phosphor-regulatory circuit that involves TNK cascade activation. This results in hyper-phosphorylation of the RAFs and SOS1, which made components of RAS pathway unresponsive to upstream signaling.
Synthetic Lethal Screens
Screens for targets that RAS cancers depend on, but normal cells do not, is an attractive concept that has been investigated extensively. Early attempts to find new drug targets using this approach have not yet translated successfully into clinical candidates, for various reasons recently discussed by Downward (2015). However, new efforts are underway using better screening technologies (e.g., CRISPR-based libraries), more sophisticated biological systems, and a clearer understanding of tumor heterogeneity (Howard et al., 2016). In a recent example, Kim et al. (2016) discovered that subsets of KRAS mutant non-small cell lung cancers are strongly dependent on the nuclear export receptor XPO1, a therapeutically tractable target. Another orthogonal approach to targeting RAS cancers emerged from the discovery that RAS turns on an important regulator of redox control NRF2, and NRF2 is essential for pancreatic cancer maintenance through inhibition of oxidation of specific translation regulators (Chio et al., 2016).
Mechanisms of Resistance
In mouse models, ablation of RAS oncogenes causes dramatic tumor regression. However, RAS-independent clones can emerge, driven by amplification of the YAP1 gene (Kapoor et al., 2014). Likewise, overexpression of YAP1 rescues cells from KRAS depletion (Shao et al., 2014). To date, these are the best indicators of resistant mechanisms that may emerge in cancer patients treated with RAS inhibitors. Other clues are likely to emerge from clinical trials in which HRAS mutant cancers are treated with farnesyl transferase inhibitors. Based on prior experience with drugs targeting the MAPK pathway, we expect that RAS inhibitors will relieve upstream feedback and increase signaling from growth factor receptors. If so, combination therapy will be necessary. In addition, mutations in other genes in the RAS pathway are likely to emerge at some frequency, such as mutations in other RAS genes, BRAF or MEK.
Future Prospects for Targeting RAS
Direct and indirect approaches to targeting RAS cancers are underway in many biotechnology and pharmaceutical companies, in academia, and in other organizations such as the Frederick National Laboratory, in which the RAS Initiative is focused on a major effort to target RAS supported by the National Cancer Institute and in collaboration with an extensive network of private and academic groups. There are major challenges that relate to unique features of the RAS proteins, such as lack of a deep pocket to which drugs can bind, as described above. The complexity of RAS effector functions and downstream pathways, as well as feedback loops and signaling redundancy, add to the challenge of shutting down RAS signaling. The degree to which RAS tumors remain dependent on RAS is another issue that has been challenging to address. On the other hand, recent advances in technologies such as fragment-based screening, tethering, and computer-based docking make RAS seem like a more tractable target. From a broader perspective, the prospects that the immune system might be harnessed to attack RAS cancers (Rosenberg et al., 2017), or that delivery of small interfering RNA (siRNA) (Yuan et al., 2014) or related technologies might be employed to attack RAS-driven tumors, or that synthetic lethal screens will yield druggable targets, are all reasons to be optimistic.
Acknowledgments
This project was funded in part with federal funds from the National Cancer Institute, NIH, under contract number HHSN261200800001E. In addition, this project was funded in part by the Outstanding Investigator Award to F.M. (5R35CA197709). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
References
- Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Alvarez-Moya B, López-Alcalá C, Drosten M, Bachs O, Agell N. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene. 2010;29:5911–5922. doi: 10.1038/onc.2010.298. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Inoue S, Matsubara Y. Recent advances in RASopathies. J. Hum. Genet. 2016;61:33–39. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]
- Arafeh R, Qutob N, Emmanuel R, Keren-Paz A, Madore J, Elkahloun A, Wilmott JS, Gartner JJ, Di Pizio A, Winograd-Katz S, et al. Recurrent inactivating RASA2 mutations in melanoma. Nat. Genet. 2015;47:1408–1410. doi: 10.1038/ng.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MVR, Ueno L, Hart JR, et al. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell. 2016;165:643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester R, Furth ME, Rosen OM. Phorbol ester- and protein kinase C-mediated phosphorylation of the cellular Kirsten ras gene product. J. Biol. Chem. 1987;262:2688–2695. [PubMed] [Google Scholar]
- Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrak-Toydemir P, Stevenson D. RASA1-related disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, editors. GeneReviews(R) University of Washington, Seattle; 1993. [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Boyd KP, Gao L, Feng R, Beasley M, Messiaen L, Korf BR, Theos A. Phenotypic variability among café-au-lait macules in neurofibromatosis type 1. J. Am. Acad. Dermatol. 2010;63:440–447. doi: 10.1016/j.jaad.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil AS, Malaquias AC, Wanderley LT, Kim CA, Krieger JE, Jorge AA, Pereira AC, Bertola DR. Co-occurring PTPN11 and SOS1 gene mutations in Noonan syndrome: does this predict a more severe phenotype? Arq. Bras. Endocrinol. Metabol. 2010a;54:717–722. doi: 10.1590/s0004-27302010000800009. [DOI] [PubMed] [Google Scholar]
- Brasil AS, Pereira AC, Wanderley LT, Kim CA, Malaquias AC, Jorge AA, Krieger JE, Bertola DR. PTPN11 and KRAS gene analysis in patients with Noonan and Noonan-like syndromes. Genet. Test. Mol. Biomarkers. 2010b;14:425–432. doi: 10.1089/gtmb.2009.0192. [DOI] [PubMed] [Google Scholar]
- Bremner R, Balmain A. Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-h. [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MC, Sun Q, Daniels RN, Camper D, Kennedy JP, Phan J, Olejniczak ET, Lee T, Waterson AG, Rossanese OW, Fesik SW. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl. Acad. Sci. USA. 2014;111:3401–3406. doi: 10.1073/pnas.1315798111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Cancer Genome Atlas Research Network. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Grönroos E, et al. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- Chang T, Krisman K, Theobald EH, Xu J, Akutagawa J, Lauchle JO, Kogan S, Braun BS, Shannon K. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J. Clin. Invest. 2013;123:335–339. doi: 10.1172/JCI63193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Öhlund D, et al. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, Gettinger S, Walther Z, Wurtz A, Heynen GJ, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov. 2014;4:606–619. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaiah S, Bindu L, Tran TH, Gillette WK, Frank PH, Ghirlando R, Nissley DV, Esposito D, McCormick F, Stephen AG, Simanshu DK. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. USA. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, Whitcomb P, Martin S, Aschbacher-Smith LE, Rizvi TA, et al. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N. Engl. J. Med. 2016;375:2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clin. Cancer Res. 2015;21:1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, Castellano E, Guerra C, Santos E, Barbacid M. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29:1091–1104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunzendorfer-Matt T, Mercado EL, Maly K, McCormick F, Scheffzek K. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc. Natl. Acad. Sci. USA. 2016;113:7497–7502. doi: 10.1073/pnas.1607298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am. J. Hum. Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]
- Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol. Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekvall S, Hagenäs L, Allanson J, Annerén G, Bondeson ML. Co-occurring SHOC2 and PTPN11 mutations in a patient with severe/complex Noonan syndrome-like phenotype. Am. J. Med. Genet. A. 2011;155A:1217–1224. doi: 10.1002/ajmg.a.33987. [DOI] [PubMed] [Google Scholar]
- Erwin N, Sperlich B, Garivet G, Waldmann H, Weise K, Winter R. Lipoprotein insertion into membranes of various complexity: lipid sorting, interfacial adsorption and protein clustering. Phys. Chem. Chem. Phys. 2016;18:8954–8962. doi: 10.1039/c6cp00563b. [DOI] [PubMed] [Google Scholar]
- Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell. Signal. 2010;22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Fivaz M, Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 2005;170:429–441. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman AK, Ritt DA, Morrison DK. The importance of Raf dimerization in cell signaling. Small GTPases. 2013;4:180–185. doi: 10.4161/sgtp.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Romero MT, Parkin P, Lara-Corrales I. Mosaic neurofibromatosis type 1: a systematic review. Pediatr. Dermatol. 2016;33:9–17. doi: 10.1111/pde.12673. [DOI] [PubMed] [Google Scholar]
- Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25:243–256. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Aldinger KA, Bennett JT, Baker L, Tusi J, Powell-Hamilton N, Stabley D, Sol-Church K, Timms AE, Dobyns WB. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am. J. Med. Genet. A. 2016;170:2237–2247. doi: 10.1002/ajmg.a.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kötting C, Gerwert K. N-Ras forms dimers at POPC membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Magee AI, Marshall CJ, Hancock JF. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis. EMBO J. 1989;8:1093–1098. doi: 10.1002/j.1460-2075.1989.tb03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Marshall CJ. Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B) EMBO J. 1991;10:641–646. doi: 10.1002/j.1460-2075.1991.tb07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins EM, Bos JM, Mason-Suares H, Tester DJ, Ackerman JP, MacRae CA, Sol-Church K, Gripp KW, Urrutia R, Ackerman MJ. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight. 2017;2:e91225. doi: 10.1172/jci.insight.91225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TP, Vazquez F, Tsherniak A, Hong AL, Rinne M, Aguirre AJ, Boehm JS, Hahn WC. Functional genomic characterization of cancer genomes. Cold Spring Harb. Symp. Quant. Biol. 2016:031070. doi: 10.1101/sqb.2016.81.031070. [DOI] [PubMed] [Google Scholar]
- Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- Huntley C, Hodder A, Ramachandran M. Clinical and historical aspects of the Elephant Man: exploring the facts and the myths. Gene. 2015;555:63–65. doi: 10.1016/j.gene.2014.09.056. [DOI] [PubMed] [Google Scholar]
- Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proc. Natl. Acad. Sci. USA. 1990;87:3042–3046. doi: 10.1073/pnas.87.8.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Banerjee A, Chavan TS, Lu S, Zhang J, Gaponenko V, Nussinov R. The higher level of complexity of K-Ras4B activation at the membrane. FASEB J. 2016;30:1643–1655. doi: 10.1096/fj.15-279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME, Eaves D, Widemann B, Kim MO, Dombi E, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J. Clin. Invest. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyabalan N, Clement JP. SYNGAP1: mind the gap. Front. Cell. Neurosci. 2016;10:32. doi: 10.3389/fncel.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Morrison DK, Wong G, McCormick F, Williams LT. PDGF beta-receptor stimulates tyrosine phosphorylation of GAP and association of GAP with a signaling complex. Cell. 1990;61:125–133. doi: 10.1016/0092-8674(90)90220-9. [DOI] [PubMed] [Google Scholar]
- Kapoor S, Weise K, Erlkamp M, Triola G, Waldmann H, Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur. Biophys. J. 2012;41:801–813. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Noreuil KM, Parker VE, Darling TN, Martinez-Agosto JA. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am. J. Med. Genet. C. Semin. Med. Genet. 2016;172:402–421. doi: 10.1002/ajmg.c.31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales J, Villalobos P, Neggers JE, Mendiratta S, Wei S, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538:114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, Omer CA, Conner MW, Anthony NJ, Davide JP, deSolms SJ, Giuliani EA, Gomez RP, Graham SL, Hamilton K, et al. Inhibition of farnesyltransferase induces regression of mammary and salivary carcinomas in ras transgenic mice. Nat. Med. 1995;1:792–797. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, McCusker JP, Ma S, Cheng E, Straub R, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake D, Corrêa SA, Müller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci. 2016;73:4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- Li HF, Wu ZY. Genotype-phenotype correlations of amyotrophic lateral sclerosis. Transl. Neurodegener. 2016;5:3. doi: 10.1186/s40035-016-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Iversen L, Tu HL, Rhodes C, Christensen SM, Iwig JS, Hansen SD, Huang WY, Groves JT. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, Koblan KS, Lee Y, Mosser S, Motzel SL, et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol. Cancer Ther. 2002;1:747–758. [PubMed] [Google Scholar]
- Lusk JB, Lam VY, Tolwinski NS. Epidermal growth factor pathway signaling in Drosophila embryogenesis: tools for understanding cancer. Cancers (Basel) 2017;9:E16. doi: 10.3390/cancers9020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon RM, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 2015;112:6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, Beroukhim R, Polyak K, Brown M, Kuperwasser C, Cichowski K. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–378. doi: 10.1016/j.ccr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot C, von Stülpnagel C, Nava C, Ville D, Sanlaville D, Lesca G, Rastetter A, Gachet B, Marie Y, Korenke GC, et al. EuroEPINOMICSRES MAE working group. Genetic and neurodevelopmental spectrum of SYNGAP1-associated intellectual disability and epilepsy. J. Med. Genet. 2016;53:511–522. doi: 10.1136/jmedgenet-2015-103451. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Tong L, deVos AM, Brünger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat. Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM, Acosta MT, Garg S, Green J, Huson S, Legius E, North KN, Payne JM, Plasschaert E, Frazier TW, et al. Disease burden and symptom structure of autism in neurofibromatosis type 1: a study of the International NF1-ASD Consortium Team (INFACT) JAMA Psychiatry. 2016;73:1276–1284. doi: 10.1001/jamapsychiatry.2016.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott HR, Owen D. Structures of Ras superfamily effector complexes: What have we learnt in two decades? Crit. Rev. Biochem. Mol. Biol. 2015;50:85–133. doi: 10.3109/10409238.2014.999191. [DOI] [PubMed] [Google Scholar]
- Murillo MM, Zelenay S, Nye E, Castellano E, Lassailly F, Stamp G, Downward J. RAS interaction with PI3K p110α is required for tumor-induced angiogenesis. J. Clin. Invest. 2014;124:3601–3611. doi: 10.1172/JCI74134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Tamgüney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, Nagashima K, Matsuda M. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 2000;275:20020–20026. doi: 10.1074/jbc.M000981200. [DOI] [PubMed] [Google Scholar]
- Olsen SN, Wronski A, Castano Z, Dake B, Malone C, De Raedt T, Enos M, DeRose YS, Zhou W, Guerra S, et al. Loss of RasGAP tumor suppressors underlie the aggressive nature of luminal B breast cancers. Cancer Discov. 2016;7:202–217. doi: 10.1158/2159-8290.CD-16-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat. Rev. Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- Papke B, Murarka S, Vogel HA, Martín-Gago P, Kovacevic M, Truxius DC, Fansa EK, Ismail S, Zimmermann G, Heinelt K, et al. Identification of pyrazolopyridazinones as PDEδ inhibitors. Nat. Commun. 2016;7:11360. doi: 10.1038/ncomms11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- Paulson VA, Shivdasani P, Angell TE, Alexander EK, Cibas E, Krane JF, Lindeman NI, Barletta J. NIFTP accounts for over half of “carcinomas” harboring RAS mutations. Thyroid. 2017;27:506–511. doi: 10.1089/thy.2016.0583. [DOI] [PubMed] [Google Scholar]
- Powers S. Genetic analysis of ras homologs in yeasts. Semin. Cancer Biol. 1992;3:209–218. [PubMed] [Google Scholar]
- Prakash P, Gorfe AA. Membrane orientation dynamics of lipidmodified small GTPases. Small GTPases Aug. 2016;11:1–10. doi: 10.1080/21541248.2016.1211067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Sayyed-Ahmad A, Gorfe AA. pMD-membrane: a method for ligand binding site identification in membrane-bound proteins. PLoS Comput. Biol. 2015;11:e1004469. doi: 10.1371/journal.pcbi.1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Zhou Y, Liang H, Hancock JF, Gorfe AA. Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys. J. 2016;110:1125–1138. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol. Cell. Biol. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J. Biol. Chem. 2009;284:10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA. HRAS and the Costello syndrome. Clin. Genet. 2007;71:101–108. doi: 10.1111/j.1399-0004.2007.00743.x. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Abreu-Blanco MT, Bindu L, Durrant DE, Zhou M, Specht SI, Stephen AG, Holderfield M, Morrison DK. Inhibition of Ras/Raf/MEK/ERK Pathway Signaling by a Stress-Induced Phospho-Regulatory Circuit. Mol. Cell. 2016;64:875–887. doi: 10.1016/j.molcel.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol. Cell. 2006;22:217–230. doi: 10.1016/j.molcel.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Rosenberg SA, Tran E, Robbins PF. T-cell transfer therapy targeting mutant KRAS. N. Engl. J. Med. 2017;376:e11. [PMC free article] [PubMed] [Google Scholar]
- Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol. Cell. Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, Bastiaens PI. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157:459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Settleman J, Narasimhan V, Foster LC, Weinberg RA. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell. 1992;69:539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, Chung J, Wang NJ, Kakavand H, Mann GJ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet. 2015;47:1194–1199. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017;13:62–68. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlich B, Kapoor S, Waldmann H, Winter R, Weise K. Regulation of K-Ras4B membrane binding by calmodulin. Biophys. J. 2016;111:113–122. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball E, Howells K, Phillips A, Mort M, Cooper DN. Human Gene Mutation Database: towards a comprehensive central mutation database. J. Med. Genet. 2008;45:124–126. doi: 10.1136/jmg.2007.055210. [DOI] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernández H, Burlingame AL, McCormick F. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, Foskett JK, Philips MR. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc. Natl. Acad. Sci. USA. 2013;110:20593–20598. doi: 10.1073/pnas.1306431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Thiel C, Wilken M, Zenker M, Sticht H, Fahsold R, Gusek-Schneider GC, Rauch A. Independent NF1 and PTPN11 mutations in a family with neurofibromatosis-i syndrome. Am. J. Med. Genet. A. 2009;149A:1263–1267. doi: 10.1002/ajmg.a.32837. [DOI] [PubMed] [Google Scholar]
- Tidyman WE, Rauen KA. Pathogenetics of the RASopathies. Hum. Mol. Genet. 2016;25(R2):R123–R132. doi: 10.1093/hmg/ddw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To MD, Wong CE, Karnezis AN, Del Rosario R, Di Lauro R, Balmain A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat. Genet. 2008;40:1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To MD, Rosario RD, Westcott PM, Banta KL, Balmain A. Interactions between wild-type and mutant Ras genes in lung and skin carcinogenesis. Oncogene. 2013;32:4028–4033. doi: 10.1038/onc.2012.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Ullrich M, Weber M, Post AM, Popp S, Grein J, Zechner M, Guerrero González H, Kreis A, Schmitt AG, Üçeyler N, et al. OCD-like behavior is caused by dysfunction of thalamo-amygdala circuits and upregulated TrkB/ERK-MAPK signaling as a result of SPRED2 deficiency. Mol. Psychiatry. 2017 doi: 10.1038/mp.2016.232. http://dx.doi.org/10.1038/mp.2016.232. [DOI] [PMC free article] [PubMed]
- Uusitalo E, Rantanen M, Kallionpää RA, Pöyhönen M, Leppävirta J, Ylä-Outinen H, Riccardi VM, Pukkala E, Pitkäniemi J, Peltonen S, Peltonen J. Distinctive cancer associations in patients with neurofibromatosis type 1. J. Clin. Oncol. 2016;34:1978–1986. doi: 10.1200/JCO.2015.65.3576. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Villalonga P, López-Alcalá C, Bosch M, Chiloeches A, Rocamora N, Gil J, Marais R, Marshall CJ, Bachs O, Agell N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 2001;21:7345–7354. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Reuther G, Weise K, Triola G, Nikolaus J, Tan KT, Nowak C, Herrmann A, Waldmann H, Winter R, Huster D. The lipid modifications of Ras that sense membrane environments and induce local enrichment. Angew. Chem. Int. Ed. Engl. 2009;48:8784–8787. doi: 10.1002/anie.200903396. [DOI] [PubMed] [Google Scholar]
- Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, McCormick F. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell. 2015;163:1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise K, Triola G, Brunsveld L, Waldmann H, Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J. Am. Chem. Soc. 2009;131:1557–1564. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- Weise K, Kapoor S, Denter C, Nikolaus J, Opitz N, Koch S, Triola G, Herrmann A, Waldmann H, Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- Williams VL, Cohen PR, Stewart DJ. Sorafenib-induced premalignant and malignant skin lesions. Int. J. Dermatol. 2011;50:396–402. doi: 10.1111/j.1365-4632.2010.04822.x. [DOI] [PubMed] [Google Scholar]
- Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy DR. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Lu J, Hunter J, Li L, Scott D, Choi HG, Lim SM, Manandhar A, Gondi S, Sim T, et al. Covalent guanosine mimetic inhibitors of G12C KRAS. ACS Med. Chem. Lett. 2016;8:61–66. doi: 10.1021/acsmedchemlett.6b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Xu J, Haigis KM, Firestone AJ, McNerney ME, Li Q, Davis E, Chen SC, Nakitandwe J, Downing J, Jacks T, et al. Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation. Cancer Discov. 2013;3:993–1001. doi: 10.1158/2159-8290.CD-13-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013a;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, et al. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol. Cell. 2013b;52:679–692. doi: 10.1016/j.molcel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace D, Carver JO, Zuber J, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, Anderson MW, Sills RC, Hong HL, Devereux TR, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G, Zhang J, Lin J, Ewing T, Matusow B, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526:583–586. doi: 10.1038/nature14982. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Sperlich B, Li FY, Al-Ayoubi S, Chen HX, Zhao Y, Li YM, Weise K, Winter R, Chen YX. Phosphorylation weakens but does not inhibit membrane binding and clustering of K-Ras4B. ACS Chem. Biol. 2017 doi: 10.1021/acschembio.7b00165. http://dx.doi.org/10.1021/acschembio.7b00165. [DOI] [PubMed]
- Zhou Y, Hancock JF. Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Prakash P, Liang H, Cho KJ, Gorfe AA, Hancock JF. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell. 2017;168:239–251. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Xu J, Mei X, Feng Z, Zhang L, Zhang Q, Zhang G, Zhu W, Liu J, Zhang C. Biochemical and structural insights into the aminotransferase CrmG in Caerulomycin biosynthesis. ACS Chem. Biol. 2016;11:943–952. doi: 10.1021/acschembio.5b00984. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PIH, Waldmann H. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]