By the use of comparative BioID of nuclear envelope (NE) proteins lamin A and Sun2, as well as a minimal inner nuclear membrane targeting motif, VRK2 is identified as a novel constituent of the NE. A-type lamins retain the transmembrane kinase VRK2 at the NE, where it phosphorylates and regulates the nuclear mobility of BAF.
Abstract
The nuclear envelope (NE) is critical for numerous fundamental cellular functions, and mutations in several NE constituents can lead to a heterogeneous spectrum of diseases. We used proximity biotinylation to uncover new constituents of the inner nuclear membrane (INM) by comparative BioID analysis of lamin A, Sun2 and a minimal INM-targeting motif. These studies identify vaccinia-related kinase-2 (VRK2) as a candidate constituent of the INM. The transmembrane VRK2A isoform is retained at the NE by association with A-type lamins. Furthermore, VRK2A physically interacts with A-type, but not B-type, lamins. Finally, we show that VRK2 phosphorylates barrier to autointegration factor (BAF), a small and highly dynamic chromatin-binding protein, which has roles including NE reassembly, cell cycle, and chromatin organization in cells, and subtly alters its nuclear mobility. Together these findings support the value of using BioID to identify unrecognized constituents of distinct subcellular compartments refractory to biochemical isolation and reveal VRK2A as a transmembrane kinase in the NE that regulates BAF.
INTRODUCTION
The nuclear envelope (NE) surrounds the nucleus during interphase, performing a wide variety of critical roles, including nucleoplasmic–cytoplasmic separation and transport (Knockenhauer and Schwartz, 2016), organization and regulation of the genome (Talamas and Capelson, 2015; Czapiewski et al., 2016) nuclear integrity (Lusk and King, 2017; Shah et al., 2017), DNA repair (Bermejo et al., 2012), and the positioning and mechanical stability of the nucleus (Kim et al., 2015a; Meier, 2016). Formed by a specialized extension of the endoplasmic reticulum (ER), the NE is composed of two membranes—the outer and inner nuclear membranes (ONM and INM, respectively), which are separated by the perinuclear space, which is continuous with the ER lumen. Annular junctions within the NE house the large proteinaceous nuclear pore complexes (NPCs), which, most notably, regulate nucleocytoplasmic transport (reviewed in Knockenhauer and Schwartz, 2016). Within the nuclear compartment and adjacent to the INM, the nuclear lamina is a scaffold composed of lamins—type V intermediate filaments (reviewed in Burke and Stewart, 2013; Gruenbaum and Foisner, 2015). With roles in chromatin organization, nuclear integrity, and gene expression, the nuclear lamina is also a vital structural feature of the NE.
The importance of the nuclear envelope extends beyond its role in various cellular functions to its role in human disease. Mutations in genes that encode constituents of the NE, including nuclear lamins, INM transmembrane proteins, NPC proteins, and soluble NE-associated proteins, are responsible for numerous rare diseases, including skeletal and cardiac diseases, progeria syndromes, peripheral neuropathy, lipodystrophies, and CNS diseases (reviewed in Worman et al., 2010; Schreiber and Kennedy, 2013; Burke and Stewart, 2014; Reddy and Comai, 2016). Although the etiology of these diseases is clear, the underlying cellular mechanisms largely remain unknown. Given its profound role in a myriad of cell functions and association with diverse diseases, there remains a clear value in identifying protein constituents of the NE and their network of functional protein–protein associations.
RESULTS AND DISCUSSION
BioID analysis of the INM with a minimal targeting motif
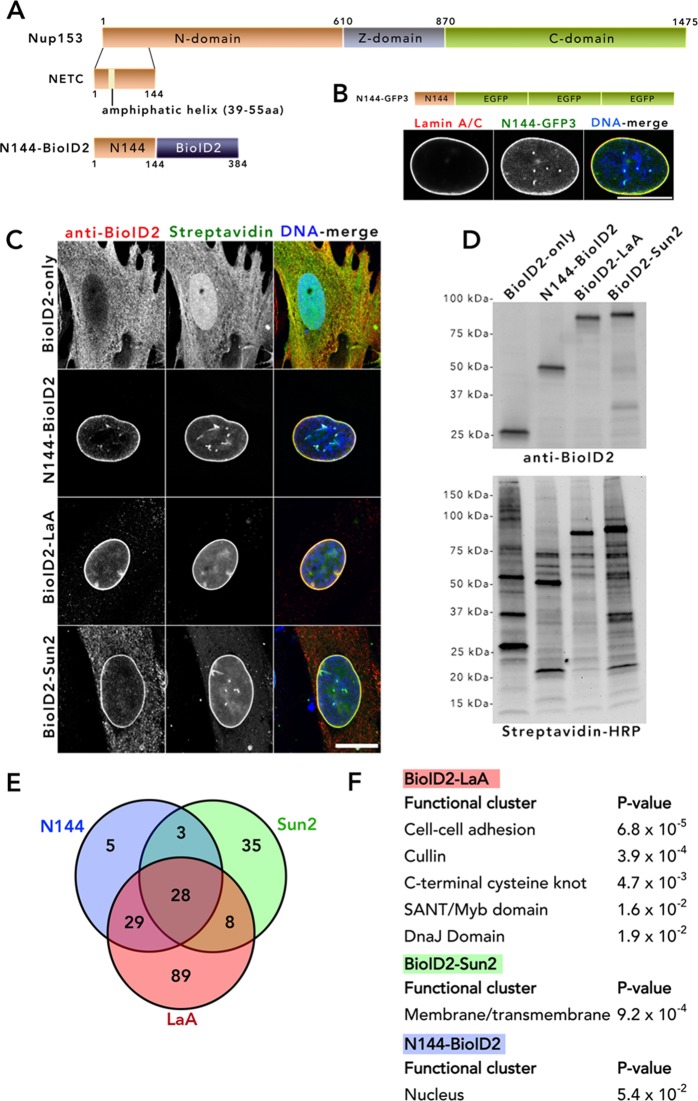
To map the protein constituency of the INM, we took advantage of the BioID method, which uses a promiscuous biotin ligase to covalently biotinylate proximate proteins and enable their isolation and identification. In our prior studies using BioID, we fused the ligase to full-length proteins of interest in order to identify associations specific to those proteins. However, here we sought to apply BioID to map the protein constituency of this distinct subcellular region by fusing the BioID ligase to a minimal targeting motif (zip code motif). We refer this application of BioID as zip code BioID. Ideally, zip code BioID imparts minimal specific protein interactions, thus permitting an “unbiased” screening of the constituent in a given subcellular region. To restrict BioID to the INM, we used a fragment of Nup153 to generate an INM-zip code motif. One of three nucleoporins, together with Nup50 and TPR, that localize on the nuclear basket (Sukegawa and Blobel, 1993; Cordes et al., 1997; Guan et al., 2000), Nup153 has a tripartite structure (Sukegawa and Blobel, 1993; Figure 1A) composed of an N-terminal domain (N-domain), central zinc-finger–containing domain (Z-domain), and C-terminal phenylalanine glycine–repeat containing region (C-domain). Contained within the N-domain is a minimal NE-targeting cassette (NETC). The NETC is crucial for the targeting of Nup153 to the NE (Enarson et al., 1998) and binding to the INM via an amphipathic helix (amino acids [aa] 39–55; Vollmer et al., 2015). The construct of three tandem green fluorescent protein (GFP) tags fused to the first 144 aa of Nup153 (N144-GPF3) clearly targets to the INM (Figure 1B) but is not enriched at, and appears largely excluded from, the NPCs (Supplemental Figure S1). We sought to use this NETC motif to serve as a zip code motif to target BioID to the INM and identify its constituents. We generated N144-BioID2 by fusing N144 to the N-terminus of BioID2, a smaller, second-generation promiscuous biotin ligase (Kim et al., 2016). To accurately gauge the efficacy of N144-BioID2 in its ability to detect INM proteome relative to the conventional targeted BioID at the NE, we used BioID2-lamin A (BioID2-LaA) and BioID2-Sun2 (Kim et al., 2016) for comparative purposes. LaA is an isoform of A-type lamins and a well-characterized constituent of the nuclear lamina; it is intimately associated with the INM. Sun2 is single-pass, type II INM protein with N- and C-termini facing the nucleoplasm and NE lumen, respectively.
FIGURE 1:
Harnessing a NE-targeting fragment of Nup153 to generate INM-BioID for comparison with BioID2-LaA and BioID2-Sun2. (A) The NETC domain (aa 1–144) of Nup153 was fused to BioID2 to generate N144-BioID2 as a means to detect NE constituents. (B) N144-GFP3 (green) was transiently expressed in BJ cells and its localization at the NE observed by fluorescence colocalization with LaA/C. (C) IF analysis of BJ cells stably expressing BioID2-only, N144-BioID2, BioID2-LaA, and BioID2-Sun2. Fusion proteins were detected by antibodies against BioID2 (red). Streptavidin (green) was used to label biotinylated proteins. For all IF images, DNA was labeled with Hoechst dye 33342 (blue in DNA merge). Scale bar, 10 µm. (D) By IB analysis, BioID2 fusion proteins were detected with anti-BioID2, and proteins biotinylated by BioID2-only, N144-BioID2, BioID2-LaA, and BioID2-Sun2 were detected with streptavidin-HRP. (E) Venn diagram showing the number of proteins differentially detected by the three NE baits (see Materials and Methods for inclusion criteria). (F) Functional annotation clustering using DAVID analysis of unique or enriched (see Materials and Methods for inclusion criteria) proteins differentially identified by the three BioID2 baits.
Identification and comparison of biotinylated proteins detected by each of the three NE baits
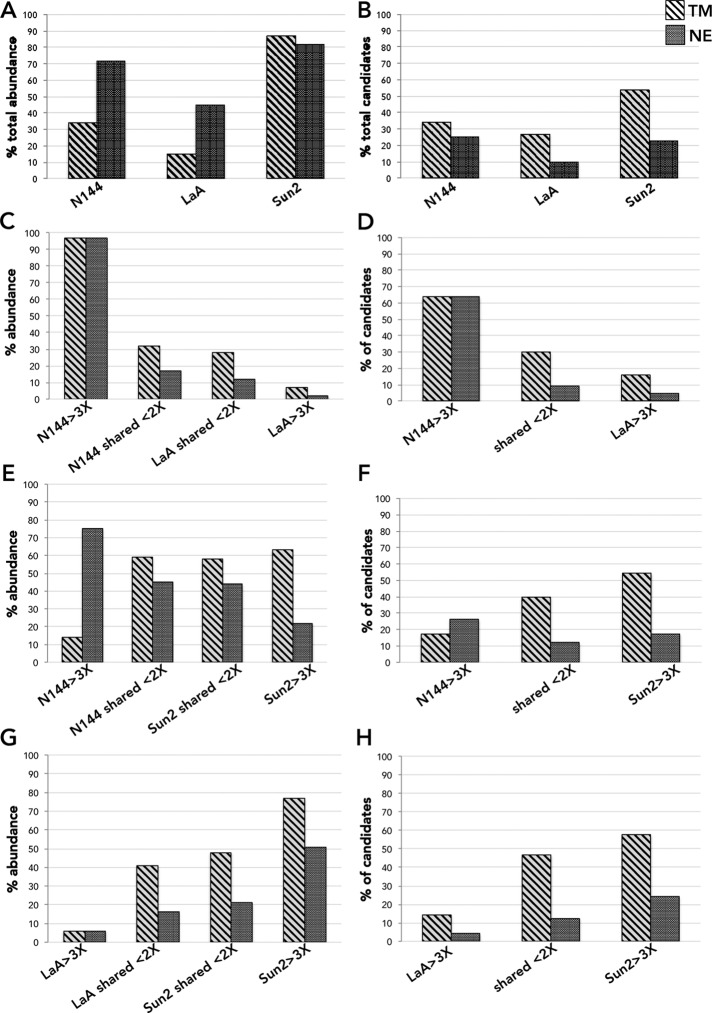
To identify NE constituents, we generated BJ cells, diploid human foreskin fibroblasts that stably express each of the N144-BioID2, BioID2-LaA, BioID2-Sun2, or BioID2-only. By immunofluorescence (IF) analysis, we observed that BioID2-only targeted to the nucleoplasm and cytoplasm, with biotinylation especially pronounced in the nucleus (Figure 1C). In contrast, N144-BioID2 was almost exclusively NE, as was the biotinylation, with some evidence of NE invaginations, a phenotype often seen in these cells, especially upon expression of N144. BioID2-LaA was predominantly detected at the NE, although a nucleoplasmic population can be observed, and biotinylation can be detected in both places. The targeting and biotinylation by BioID2-Sun2 are both predominantly at the NE, although there is obvious endoplasmic reticulum (ER) localization, common for ER-synthesized INM transmembrane proteins like Sun2, and some nucleoplasmic biotinylation. By immunoblot (IB) analysis, we observed similar levels of expression and appropriate migration of the BioID2-fusion proteins (Figure 1D). The pattern of biotinylated proteins, although unique, showed some overlap between the NE-associated BioID2 fusion proteins. To identify proteins biotinylated by the BioID2-fusion proteins, we analyzed streptavidin-based pull downs of denatured whole-cell lysates by protein mass spectrometry (MS) after 18 h of biotinylation by biotin supplementation. Only proteins detected uniquely or with relative peak intensity measurements at least three times greater than those detected by BioID2-only were considered as candidates (Supplemental Table S1). In addition, to obtain a higher level of confidence in the proteins identified, only proteins with the relative peak intensities greater ≥5 × 107 were included. The MS results revealed a similar number of candidates (65 and 74) detected for N144-BioID2 and BioID2-Sun2, with ∼30% of them shared between the two baits (Figure 1E). On the other hand, BioID2-LaA identified 154 candidates. The candidates that are shared between N144, LaA, and Sun2 BioIDs (∼15% of total identified proteins) likely reflect their close proximity within the two-dimensional (2D)–restricted plane of the NE. We next analyzed the relative number and abundance of TM and NE proteins identified by the BioID baits (Figure 2 and Supplemental Table S2). In comparison to BioID2-LaA, N144-BioID2 and BioID2-Sun2 detected a substantial amount of NE and TM candidates by abundance (Figure 2A). Of special note was the extent of NE protein detection by BioID2-Sun2. A similar trend was observed for the relative number of identified TM and NE candidates (Figure 2B). On comparing N144-BioID2 to BioID2-LaA, the vast majority of unique or enriched (>3× abundance) candidates, by relative abundance or number, detected by N144-BioID2 are transmembrane (TM) and/or NE proteins (Figure 2, C and D), likely a reflection of the predominant restriction of N144 to the INM and the functional role of lamins in the nucleoplasm. Compared with N144-BioID2, the majority of detected candidates by relative abundance unique or enriched in BioID2-Sun2 are TM but not exclusively NE proteins (Figure 2, E and F), likely reflecting that, although concentrated at the NE, Sun2 is synthesized in and traffics through the ER. For BioID2-LaA relatively few of the unique or enriched candidates are TM or NE, by relative abundance or number, as compared BioID2-Sun2 (Figure 2, G and H), again likely reflecting its partial nucleoplasmic localization and function. If we take a closer look at those proteins unique or enriched for each bait as compared with the others by Database for Annotation, Visualization, and Integrated Discovery (DAVID) analysis, a gene functional annotation clustering tool, we find significant enrichment of membrane/transmembrane proteins for BioID2-Sun2, whereas clusters of protein domains, families, or functional roles were identified for BioID2-LaA, and for the few N144-BioID2–specific candidates, a nuclear localization cluster was identified by DAVID analysis (Figure 1F). Because Sun2 and LaA both impart their BioID-fusions with a bias toward ER and nucleoplasmic compartments, respectively, we focused on the unique or enriched candidates of N144-BioID2 as potential novel constituents of the NE. MCM3AP (GANP) is reported to be NPC associated (Wickramasinghe et al., 2011), Sun1 and lamin B2 are known NE proteins, GNL3L shuttles between the nucleus and cytoplasm, SPCS2 is a transmembrane peptidase, and vaccinia-related kinase-2 (VRK2) is reported to be an ER-tethered or nucleoplasmic kinase, depending on the isoform. Of particular interest for further studies was the kinase VRK2, as there is a scarcity of reported resident NE kinases. Although the stringent exclusion criteria in this study classified VRK2 as a unique candidate for N144-BioID2, it was detected by the other two baits, for which it was excluded solely based on low intensity levels. We also previously detected low levels of VRK2 in BioID-LaA from various cell types (Roux et al., 2012; Kim et al., 2014a; unpublished data).
FIGURE 2:
The relative abundance and number of TM or NE candidate proteins identified by BioID2 baits. Percentage of total relative peak intensity (abundance; A) and relative number of TM or NE candidate proteins (B) identified by N144-BioID2, BioID2-LaA, or BioID2-Sun2. Percentage of identified TM or NE candidate proteins by abundance (C, E, G) and percentage of TM or NE candidate proteins among identified candidates (D, F, H) for comparative analysis of N144-BioID2 and BioID2-LaA (C, D), N144-BioID2 and BioID2-Sun2 (E, F), and BioID2-LaA and BioID2-Sun2 (G, H).
Vaccinia-related kinase 2A is a resident protein of the NE
The serine-threonine kinase VKR2 is a member of the vaccinia-related kinase protein family and shares homology with vaccinia virus B1 kinase (Nichols and Traktman, 2004). Two major splice isoforms exists for VRK2—VRK2A and VRK2B (Figure 3A). The predominant isoform A is a 508-aa C-terminal anchored transmembrane protein reported to reside predominantly in the ER; the less abundant isoform B is a 397-aa protein that lacks a TM anchor and is nucleoplasmic (Blanco et al., 2006). Both isoforms contain a functional serine/threonine kinase domain. A variety of functions have been reported for VRK2, including regulation of mitogen-activated protein kinase signaling (Blanco et al., 2007; Fernandez et al., 2010, 2012), apoptosis (Acar, 1982; Monsalve et al., 2013), tumor cell invasion (Vazquez-Cedeira and Lazo, 2012), expression of cyclooxygenase-2 (Vazquez-Cedeira and Lazo, 2012), and chaperonin function (Kim et al., 2014a,b). Of obvious relevance to the NE, members of the vaccinia kinase family, including VRK1, VRK3, and vaccinia kinase B1, are reported to phosphorylate barrier-to-autointegration factor (BAF), a small and highly dynamic chromatin-binding protein with roles including NE reassembly, cell cycle, and chromatin organization, in vivo (Lee and Craigie, 1998; Nichols et al., 2006). VRK2 has been shown to be capable of BAF phosphorylation in in vitro assays but likely discounted for this role in vivo due to its reported ER localization. To test the possibility that VRK2 is a NE constituent, we expressed human GFP-VRK2A and -VRK2B in BJ cells and C2C12 cells (Supplemental Figure S2B) and observed that GFP-VRK2A is clearly enriched at the NE and also localizes to the ER, especially when expression levels increase. In contrast, GFP-VRK2B is predominantly nucleoplasmic, with no obvious enrichment at the NE (Figure 3B). By IF, anti-VRK2 readily detected exogenous VRK2 (GFP-VRK2A or untagged VRK2A) but failed to detect endogenous VRK2 (Supplemental Figure S2A), suggestive of low expression levels in all cell types studied. These findings suggest that VRK2A is a novel NE-resident protein and the major isoform identified by the BioID2 baits.
FIGURE 3:
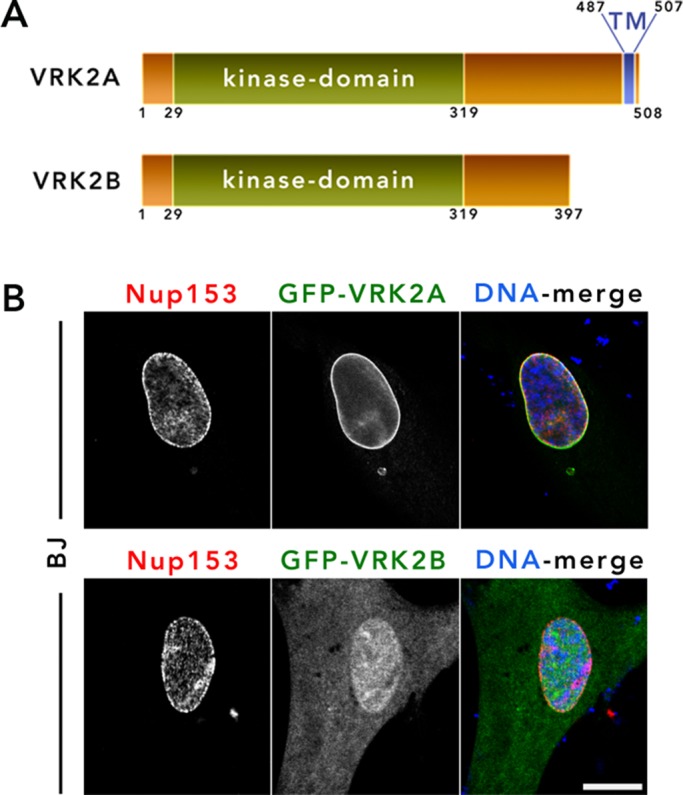
VRK2A is a novel constituent of the NE. (A) Schematic showing two predominant isoforms of VRK2—VRK2A and VRK2B—with TM and kinase domains labeled. (B) GFP-tagged VRK2A and VRK2B were transiently expressed in BJ cells and their localization monitored by IF. Anti-Nup153 (red) was used as the NE marker. DNA was labeled with Hoechst dye 33342 (blue). The experiment was repeated at least three times. Scale bar, 10 µm.
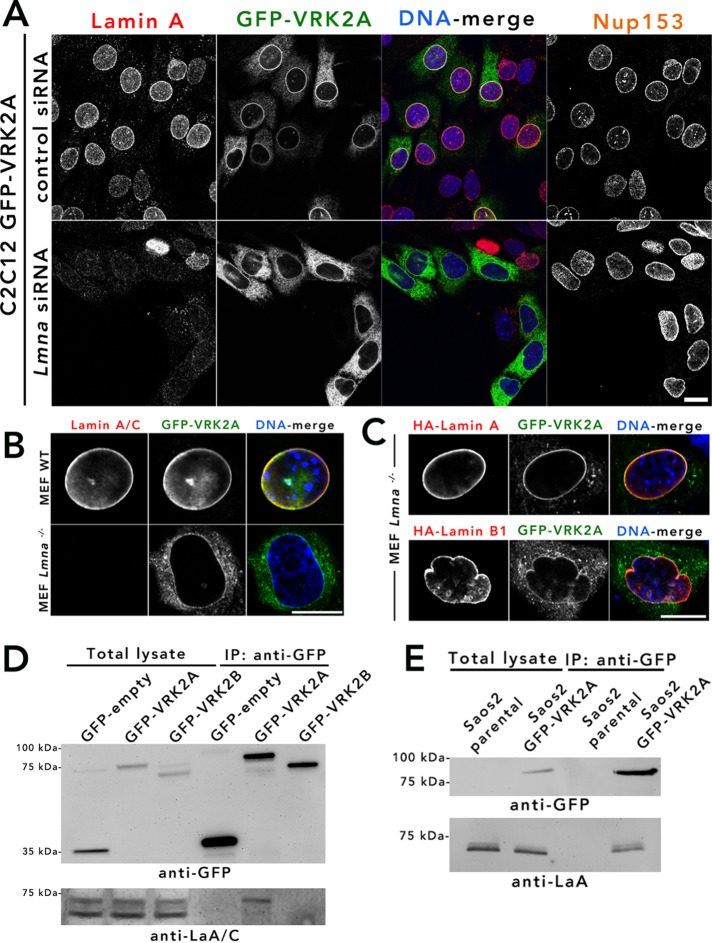
NE retention of VRK2A is dependent on A-type lamins
A-type lamins are required for the appropriate NE retention of some INM proteins, including emerin (Sullivan et al., 1999), SAMP1 (Borrego-Pinto et al., 2012), and Lap1C (Powell and Burke, 1990). Although BioID2-LaA detected only low levels of VRK2 relative to N144 (below our threshold for candidacy), we sought to determine whether A-type lamins help retain VRK2A at the NE. A reduction of Lmna levels in C2C12 cells stably expressing GFP-VRK2A led to a significant reduction of VRK2A localization at the NE, with redistribution throughout the ER (Figure 4A and Supplemental Figure S2B). A similar dependence on Lmna for NE localization of VRK2 was observed in wild-type (WT) and Lmna-deficient mouse embryonic fibroblasts (MEFs; Kim and Zheng, 2013; Guo et al., 2014) transiently expressing GFP-VRK2A (Figure 4B). The NE localization of VRK2 in Lmna-deficient cells could be rescued by coexpression of A-type but not B-type lamins (Figure 4C). Using MEFs that are deficient for emerin (Lammerding et al., 2005), we confirmed that loss of emerin does not affect the NE localization of VRK2A (Supplemental Figure S2D). Together these observations suggest that A-type lamins are responsible for retaining VRK2A at the NE.
FIGURE 4:
A-type lamins retain and physically interact with VRK2A at the NE. (A) siRNA-mediated knockdown of Lmna was performed on C2C12 cells stably expressing GFP-VRK2A. Lmna knockdown was confirmed by anti-lamin A (red). Anti-Nup153 (not in merged) was used as an additional NE marker. The experiment was repeated twice. (B) Transient expression of GFP-VRK2A in WT Lmna and Lmna-gene deleted (KO) MEF cells. (C) Coexpression of GFP-VRK2A with A-type lamins (HA–lamin A; top) and B-type lamins (HA–lamin B1; bottom) in Lmna KO MEF cells. DNA was labeled with Hoechst dye 33342 (blue). The experiment was repeated three times. Scale bar, 10 µm. (D) Anti-GFP coIP from total lysates of transiently expressed GFP empty vector and GFP-VRK2A and -VRK2B in HeLa cells, followed by immunoblotting with anti–lamin A/C. (E) Anti-GFP coIP in parental Saos-2 and Saos-2 cells stably expressing GFP-VRK2A followed by immunoblotting with anti-LaA.
VRK2A physically interacts with A-type lamins
Because VRK2A is retained at the NE by A-type lamins, we reasoned that A-type lamins might physically interact with VRK2A, either directly or indirectly. To investigate this hypothesis, we transiently transfected HeLa cells with GFP-VRK2A, GFP-VRK2B, or GFP-only for anti-GFP immunoprecipitation (IP) and probed for endogenous A-type lamin coIP. We observed that only GFP-VRK2A and not GFP-VRK2B or GFP was able to coimmunoprecipitate A-type lamins, predominantly isoform A (Figure 4D). Furthermore, anti-GFP coIP in parental Saos-2 cells or Saos-2 cells stably expressing GFP-VRK2A also revealed that VRK2A efficiently pulls down endogenous lamin A (Figure 4E). Together these findings indicate a physical interaction, either direct or indirect, between A-type lamins and VRK2A.
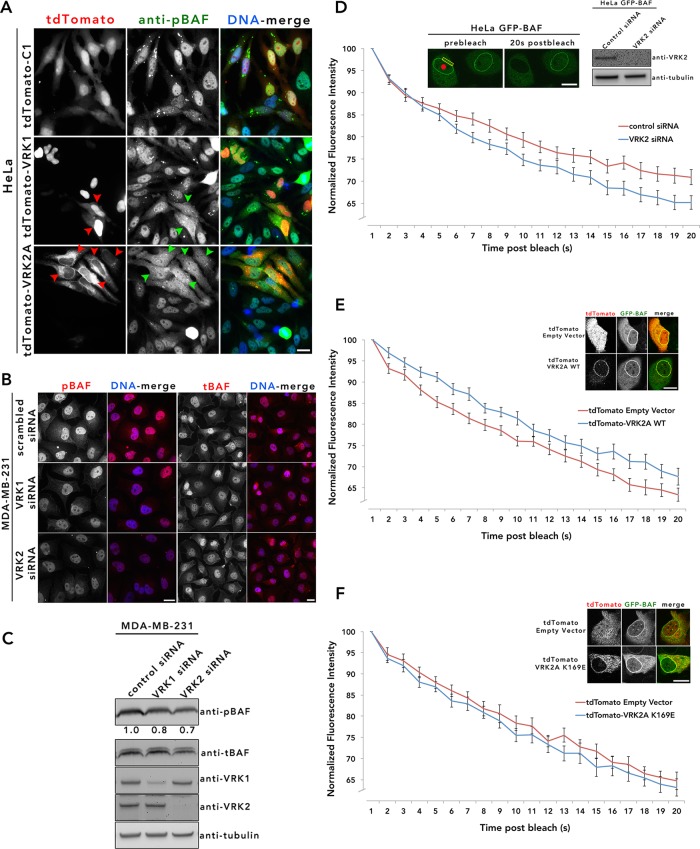
VRK2A phosphorylates cellular BAF and subtly regulates NE BAF during interphase
Knockdown or overexpression of VRK2 did not appreciably alter the nuclear or NE morphology nor the distribution of any NE constituents that we tested (unpublished data). We did detect an increase in G1/G0 in cells overexpressing VRK2A and a decrease of G1/G0 in cells with reduced VRK2 expression (Supplemental Figure S3). We turned our attention to the possibility that VRK2 regulated BAF phosphorylation similar to other VRK family members (Nichols et al., 2006; Park et al., 2015). Because it was unclear whether VRK2 can phosphorylate BAF in cells, we first investigated this possibility by transiently overexpressing tdTomato-only, tdTomato-VRK1, and tdTomato-VRK2A in HeLa cells and monitoring changes in BAF phosphorylation by IF. In cells that expressed elevated levels of VRK1 and VRK2A such that there was noticeable accumulation in the cytoplasm or ER, respectively, we observed abnormally phosphorylated BAF in the cytoplasm as detected by anti-pBAF (Zhuang et al., 2014; Figure 5A). Depletion of VRK2 in MDA-MB-231 cells by small interfering RNA (siRNA) reduced detectable BAF phosphorylation by IF and IB (Figure 5, B and C). This reduction in BAF phosphorylation is similar to that observed upon depletion of VRK1 (Figure 5C). Together these data establish BAF as a cellular substrate of VRK2. Although enriched in the nucleus, especially at the nuclear envelope, distinct nuclear and cytoplasmic populations of BAF exists during interphase (Shimi et al., 2004; Haraguchi et al., 2008; Molitor and Traktman, 2014). Nuclear BAF was shown to be highly mobile, likely due dynamic interaction with binding partners, including LEM-domain proteins (Cai et al., 2001; Shimi et al., 2004; Mansharamani and Wilson, 2005) and lamins (Montes de Oca et al., 2009; Capanni et al., 2010) at the NE, histones and DNA damage repair proteins (Montes de Oca et al., 2009), transcription factors (Wang et al., 2002; Huang et al., 2011), and sequence-nonspecific DNA (Lee and Craigie, 1998; Zheng et al., 2000). To investigate how VRK2 affects the mobility of BAF in the nucleus, we performed fluorescence loss in photobleaching (FLIP) studies in interphase HeLa cells stably expressing GFP-BAF, in which we photobleached a region of the nucleoplasm and recorded loss of GFP-BAF from a region of the NE. Compared to the control cells, the loss of GFP-BAF from the NE was increased in VRK2 depleted cells, albeit only slightly (Figure 5D). Overexpression of a tdTomato-VRK2A led to nominal reduction in BAF dissociation from the NE (Figure 5E), but a kinase-dead version (K169A; Blanco et al., 2006) did not (Figure 5F). None of these observed differences in BAF mobility resulting from modulation of VRK2 expression reached clear statistical significance. It is noteworthy that, in contrast to the dramatic accumulation of nucleoplasmic BAF in cells depleted of VRK1 (Molitor and Traktman, 2014), knockdown of VRK2 caused no substantial changes in the distribution of the BAF population. VRK1 knockdown was reported to reduce the mobility of BAF in the nucleus (Molitor and Traktman, 2014), likely due to enhanced nucleoplasmic DNA binding by nonphosphorylated BAF, whereas knockdown of VRK2, which, being NE tethered, would only phosphorylate the NE-associated BAF population, actually enhanced BAF dissociation from the NE. These findings suggest that whereas VRK1 may be the predominant kinase regulating the localization of nuclear BAF during interphase, VRK2 modulates a population of BAF enriched at the NE, with VRK1 and VRK2 acting to displace BAF from the nucleoplasm or NE, respectively. It has been reported that VRK1 functions early in mitosis to dissociate BAF from the condensing chromatin, and loss of VRK1 prevents the normal progression of mitosis (Gorjanacz et al., 2007; Lancaster et al., 2007). In contrast, we observed no obvious defects during mitosis or postmitotic envelope reassembly with depletion of VRK2 (unpublished data), suggesting that VRK1 is the primary kinase that phosphorylates BAF during late G2/M.
FIGURE 5:
VRK2 phosphorylates BAF in cells and subtly alters its mobility at the NE. (A) tdTomato-tagged empty vector and -VRK1 and -VRK2A were expressed transiently in HeLa cells and labeled with anti-pBAF (red). Red arrowheads depict cells overexpressing cytoplasmic VRK1 or NE/ER/cytoplasmic VRK2A; green arrowheads show cells with enriched pBAF labeling. (B) siRNA-mediated knockdown of VRK1 and VRK2 was performed in MDA-MB-231 cells and labeled for pBAF and tBAF (red) for IF analysis (B) or immunoblotted with VRK1, VRK2, pBAF, tBAF, and tubulin as a loading control (C). Band intensities for pBAF were quantified using ImageJ, normalized relative to tubulin, and expressed as ratios relative to control. For the IF images in A and B, DNA was labeled with Hoechst dye 33342 (blue). The experiments were repeated twice. Scale bar, 10 µm. (D) FLIP analysis was performed on GFP-BAF HeLa cells 48 h after VRK2 siRNA–mediated depletion; 44 control and VRK2-depleted cells from two independent FLIP experiments were analyzed. IF analysis (inset) shows a HeLa GFP-BAF cell before and after photobleaching. The red circle represents the bleached ROI, and the yellow rectangle denotes the ROI analyzed for the loss of GFP signal. VRK2 depletion is observed by IB analysis (inset). (E, F) FLIP analysis was performed on HeLa GFP-BAF cells stably expressing tdTomato-empty and -VRK2A WT (E) or -VRK2A K169E (F). For E and F, FLIP analysis was performed on 30 cells from three independent FLIP experiments. Data are presented as the normalized average intensity ± SEM. None of these FLIP studies reached clear statistical significance, but they did exhibit consistent trends for independent experiments.
Collectively the results of these studies illustrate the value of using a zip code BioID to assist in identification of novel constituents of a subcellular domain and for comparative studies to reveal candidates of specific BioID2-baits that are unique or enriched. The findings that N144-BioID2 but not BioID2-LaA predominantly detected VRK2 despite LaA being a binding partner, albeit not necessarily a direct binding partner, and retention mechanism highlights the limitations of this method and the risk of overinterpreting BioID results. The identification of VRK2A as an A-type lamin–retained constituent of the NE that phosphorylates BAF and subtly regulates its nuclear mobility adds to the repertoire of NE constituents and opens the door for the potential of yet-to-be-discovered roles for this membrane-tethered kinase in the regulation of the nuclear envelope and its cellular functions.
MATERIALS AND METHODS
Plasmids
The NETC fragment was amplified by PCR from human Nup153 in a pcDNA 3.1 and inserted into the previously generated BioID2-HA pBabe.puro vector using EcoRI and AscI. pBabe.puro constructs containing BioID2-only, BioID2-LaA, and BioID2-Sun2 were previously generated by Kim et al. (2016). VRK2A, VRK2B, and BAF were amplified by PCR from human cDNA. Digested PCR products (by XhoI and AflII) were inserted into GFP pcDNA 4T/O (Thermo Fisher Scientific, Waltham, MA). All GFP constructs used in this work are enhanced GFP. To generate untagged VRK2A construct, amplified VRK2A was digested by NheI and AflII and inserted into pcDNA 3.1 vector. Kinase-dead VRK2A was generated via K169E substitution. Amplified VRK2A WT and VRK2A K169E were digested by NheI and BamHI and inserted into tdTomato-C1 (Addgene plasmid 54653). GFP-BAF in pcDNA 4T/O vector was subcloned to pBabe.puro using EcoRI and SalI. The N144-GFP3 construct was generously provided by Brian Burke (Institute of Medical Biology, Singapore).
Cell culture
BJ cells were obtained from the American Type Culture Collection (ATCC; CRL-2522™). Stable cell lines for all BioID2 constructs and GFP-BAF construct were generated using retroviral transduction. HEK293 Phoenix cells (National Gene Vector Biorepository, Indianapolis, IN) were transfected with BioID2 or GFP-BAF constructs using Lipofectamine 2000 (Thermo Fisher Scientific) per manufacturer's recommendation. The transfected cells were incubated at 37°C overnight. After overnight incubation, the transfected cells were replenished with fresh medium and further incubated at 32°C for 36 h. The culture media was collected and centrifuged at 1000 × g for 5 min. The supernatant of the centrifuged medium was filtered through a 0.45-μm filter and added to BJ cells (target cells for BioID2 constructs) or HeLa (CCL-2; target cells for GFP-BAF construct) along with Polybrene (4 μg/ml; Santa Cruz Biotechnology, Dallas, TX). At 48 h after transduction, puromycin (0.5 μg/ml; Thermo Fisher Scientific) was added to the target cells. The expression of fusion proteins was further verified using IF and IB. To generate C2C12 (CRL-1772; ATCC) or Saos-2 cells stably expressing GFP-VRK2A, cells were transfected with GFP-VRK2A construct via Lipofectamine 2000 and selected under Zeocin (100 mg/ml) 48 h after transfection. Upon colony formation, subclones were isolated and screened by IF and IB. To generate HeLa cells stable for both GFP-BAF and tdTomato-VRK2A WT or -VRK2A K169E, HeLa cells stably expressing GFP-BAF were transfected with tdTomato-VRK2A WT or -VRK2A K169E using Lipofectamine 2000, selected by G418 (600 μg/ml), and subcloned. The stable cell lines were maintained in 5.0% CO2 at 37°C in DMEM (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (FBS). A-type lamin–deficient (Lmna-/-) MEFs, which were generously provided by Yixian Zheng (Carnegie Institution for Science), along with WT MEFs were derived from Lmna-knockout mice (Kim and Zheng, 2013) using methods described previously (Guo et al., 2014). Emerin-deficient (Emd-/-) MEFs were generously provided by Colin Stewart (Institute of Medical Biology, Singapore; Sullivan et al., 1999; Lammerding et al., 2005). All cells were tested monthly for mycoplasma contamination.
Immunofluorescence
Cells grown on glass coverslips were fixed in 3% (wt/vol) paraformaldehyde/phosphate-buffered saline (PBS) for 10 min and permeabilized by 0.4% (wt/vol) Triton X-100/PBS for 15 min. For labeling purposes, the following primary antibodies were used: chicken anti-BioID2 using purified glutathione S-transferase–BioID2 (1:5000; Kim et al., 2016), mouse anti-LaA/C (1:100; XB10; Covance), rabbit anti-LaA (1:100; SC20680; Santa Cruz Biotechnology), mouse anti-hemagglutinin (HA; 1:200; 12CA5; Covance), rabbit polyclonal anti-VRK2 (1:200; HPA047503; Sigma-Aldrich, St. Louis, MO), rabbit polyclonal anti-BAF (1:100; ab129184; Abcam), rabbit polyclonal anti-pBAF (specific for phosphorylated-BAF; 1:200; a generous gift from Robert Craigie, National Institutes of Health), mouse monoclonal anti-Nup153 (SA1; Bodoor et al., 1999), and rabbit anti-emerin (1:200; a generous gift from G. Morris, Robert Jones and Agnes Hunt Orthopaedic Hospital, United Kingdom; Liu et al., 2010). Primary antibodies were detected using Alexa Fluor 568–conjugated goat anti-chicken (1:1000; A11041; Thermo Fisher Scientific), goat anti-rabbit (1:1000; A11036; Thermo Fisher Scientific), Alexa Fluor 488–conjugated goat anti-mouse (1:1000; A11029; Thermo Fisher Scientific), and Alexa Fluor 647–conjugated goat anti-mouse (1:1000; A21235; Thermo Fisher Scientific). Alexa Fluor 488–conjugated streptavidin (S32354; Thermo Fisher Scientific) was used to detect biotinylated proteins. DNA was detected with Hoechst dye 33342. Coverslips were mounted using 10% (wt/vol) Mowiol 4-88 (Polysciences). Confocal images were obtained using a Nikon A1 confocal microscope (60 ×/1.49 oil APO TIRF Nikon objective) with a charge-coupled device camera (CoolSnap HQ; Photometrics) linked to a workstation running NIS-Elements software (Nikon, Melville, NY). Epifluorescence images were captured using a Nikon Eclipse NiE (40 ×/0.75 Plan Apo Nikon objective) microscope.
Immunoblot and immunoprecipitation
To analyze total cell lysates by immunoblot, 1.2 × 106 cells were lysed in SDS–PAGE sample buffer, boiled for 5 min, and sonicated to shear DNA. Proteins were separated on 4–20% gradient gels (Mini-PROTEAN TGX; Bio-Rad, Hercules, CA) and transferred to nitrocellulose membrane (Bio-Rad). After blocking with 10% (vol/vol) adult bovine serum and 0.2% Triton X-100 in PBS for 30 min, the membrane was incubated with appropriate primary antibodies: rabbit anti-LaA (1:1000; SC20680; Santa Cruz Biotechnology), mouse monoclonal anti-LaA/C (1:1000; 4777s; Cell Signaling Technology), rabbit polyclonal anti-VRK2 (1:1000; HPA047503; Sigma-Aldrich), rabbit polyclonal anti-VRK1 (1:1000; HPA000660; Sigma-Aldrich), rabbit polyclonal anti-BAF (1:1000; ab129184; Abcam), rabbit polyclonal anti-pBAF (1:1000), and mouse monoclonal anti-tubulin as a loading control (1:2000; T9026; Sigma-Aldrich). The primary antibodies were detected using horseradish peroxidase (HRP)–conjugated anti-rabbit (1:40,000; G21234; Thermo Fisher Scientific) or anti-mouse (1:40,000; F21453; Thermo Fisher Scientific) antibodies. The signals from antibodies were detected using enhanced chemiluminescence via a UVP BioImaging System (UVP, Upland, CA). To detect biotinylated proteins, after blocking with 1% (wt/vol) bovine serum albumin (BSA) and 0.2% Triton X-100 in PBS for 30 min, the membrane was incubated with HRP-conjugated streptavidin (1:40,000; ab7403; Abcam) for 45 min. After detection of biotinylated proteins, HRP activity on the membrane was quenched by incubating the membrane in 30% (vol/vol) H2O2 for 15 min. The blot was blocked with 10% (vol/vol) adult bovine serum and 0.2% Triton X-100 in PBS for 30 min and incubated with chicken anti-BioID2 (1:40,000) antibodies. The primary antibodies were detected using HRP-conjugated anti-chicken (1:40,000; A9046; Sigma-Aldrich).
For coIP analyses, transiently transfected HeLa cells or Saos-2 cells stably expressing GFP-VRK2A (2.4 × 106) were lysed in 1 ml of IP lysis buffer (50 mM Tris, pH 7.5; 150 mM NaCl, 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 1% Triton X-100, and 1× proteinase inhibitor [1861278; Thermo Fisher Scientific]). Lysates were passed through a 21-gauge needle 20 times and centrifuged at 16,500 × g for 10 min at 4°C. The supernatants were rotated overnight at 4°C with 20 µl of protein A–Sepharose beads (20365; Thermo Scientific) and 2 μg of rabbit anti-GFP antibody (1:1000; ab290; Abcam). Samples were washed thoroughly three times with the IP lysis buffer and twice with wash buffer (50 mM Tris, pH 7.5, and 50 mM NaCl) at 4°C. Proteins were solubilized in 25 μl of SDS–PAGE sample buffer and boiled for 5 min. Proteins were detected by IB as described. The coIP experiment was repeated twice for each cell type.
Fluorescence loss in photobleaching
For FLIP studies in VRK2-depleted HeLa GFP-BAF cells, interphase cells were imaged using a Nikon A1 confocal microscope (60×/1.49 oil APO TIRF Nikon objective) linked to a workstation running NIS-Elements software. Three regions of interest (ROIs) were analyzed. One in the nucleoplasm served as the region for photobleaching, one on the NE was used for measuring fluorescence loss, and one in the nucleoplasm was used to compensate for the loss of signal due to photobleaching. HeLa cells stably expressing GFP-BAF were treated with siRNA oligos against VRK2 for 48 h. For each FLIP experiment from two independent experiments of siRNA-treated and control cells, 22 random cells (44 total cells) were subjected to photobleaching for 1.05 s by 403-nm light from a solid-state laser, immediately followed by image acquisition (one-directional scanning, 512 × 512 resolution) for a total of 39 cycles. Normalized fluorescence intensity was averaged and plotted against time. Data are presented as normalized average intensity ± SEM. For FLIP studies in HeLa cells stably expressing GFP-BAF and tdTomato-VRK2A WT or -VRK2A K169E, the cells were cocultured with HeLa cells stably expressing GFP-BAF and tdTomato-Empty as controls. Photobleaching and image acquisition were performed as described. Normalized fluorescence intensity for 10 cells each from three independent FLIP experiments (30 total cells) was averaged and plotted against time. Data are presented as normalized average intensity ± SEM.
Flow cytometry
For cell cycle analysis by propidium iodide (PI) staining, cells were fixed with 70% ethanol, and washed with fluorescence-activated cell sorting buffer (1× PBS, 1% BSA [A3803; Sigma-Aldrich], 0.1% sodium azide) twice. The cells were stained with PI-RNase A solution (1× PBS, 100 µg/ml RNase A [R4642; Sigma-Aldrich], 40 µg/ml PI [P4170; Sigma-Aldrich]) at room temperature for 10 min in dark. At least 5 × 104 cells were analyzed per sample in a BD Accuri C6 (BD Biosciences, San Jose, CA), and data were analyzed with FlowJo, version 10, software (FlowJo, Ashfield, OR). Three independent experiments were performed for each condition. The p values were calculated by unpaired t test. p ≤ 0.05 was considered statistically significant.
BioID pull down
For large-scale BioID pull down, 4 × 107 cells were incubated with 50 μM biotin for 16 h. After two PBS washes, the cells were lysed in 2.4 ml of lysis buffer containing 8 M urea, 50 mM Tris, pH 7.4, 1 mM DTT, and 1× Complete protease inhibitor (Halt Phosphatase Inhibitor Cocktail; Life Technologies). Triton X-100 was added to 1% final concentration. After two times of sonication each for 1 min at 30% duty cycle and an output level of 4 (Sonifer-250; Branson), an equal volume of lysis buffer was added. After centrifugation at 16,500 × g for 10 min, the supernatant was collected to a 15-ml conical tube and incubated with 300 μg of Dynabeads (MyOne Streptavidin C1; Thermo Fisher Scientific) overnight. Beads were collected using a magnetic stand and washed four times with wash buffer containing 8 M urea, 50 mM Tris, pH 7.4, and 0.5% Triton X-100. Beads were resuspended in wash buffer without Triton X-100. Ten percent of the sample was saved for further analysis. The other 90% of the sample was resuspended in 50 mM NH4HCO3 supplemented with 1 mM biotin for MS analysis.
Mass spectrometry analysis and data analysis
After immunoprecipitation, proteins were digested directly on beads. Briefly, proteins bound to the beads were resuspended with 8 M urea and 50 mM ammonium bicarbonate, and cysteine disulfide bonds were reduced with 10 mM tris(2-carboxyethyl)phosphine at 30°C for 60 min, followed by cysteine alkylation with 30 mM iodoacetamide in the dark at room temperature for 30 min. After alkylation, urea was diluted to 1 M urea using 50 mM ammonium bicarbonate, and proteins were finally subjected to overnight digestion with MS-grade trypsin/Lys-C mix (Promega, Madison, WI). Finally, beads were pulled down and the solution with peptides collected into a new tube. The beads were then washed once with 50 mM ammonium bicarbonate to increase peptide recovery. The digested samples were desalted using a C18 TopTip (PolyLC, Columbia, MD), and the organic solvent was removed in a SpeedVac concentrator before liquid chromatography (LC)–MS/MS analysis. Dried samples were reconstituted in 100 mM ammonium formate, pH ∼10, and analyzed by 2D LC-MS/MS using a 2D nanoACQUITY Ultra Performance Liquid Chromatography system (Waters, Milford, MA) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto the first-dimension column, XBridge BEH130 C18 NanoEase (300 µm × 50 mm, 5 µm) equilibrated with solvent A (20 mM ammonium formate, pH 10, first-dimension pump) at 2 µl/min. The first fraction was eluted from the first-dimension column at 17% of solvent B (100% acetonitrile) for 4 min and transferred to the second-dimension Symmetry C18 trap column of 0.180 × 20 mm (Waters) using a 1:10 dilution with 99.9% second-dimension pump solvent A (0.1% formic acid in water) at 20 µl/min. Peptides were then eluted from the trap column and resolved on an analytical C18 BEH130 PicoChip column of 0.075 × 100 mm, 1.7-µm particles (NewObjective, MA), at low pH by increasing the composition of solvent B (100% acetonitrile) from 2 to 26% over 94 min at 400 nl/min. Subsequent fractions were carried with increasing concentrations of solvent B. The following first-dimension fractions were eluted at 19.5, 22, 26, and 65% solvent B. The mass spectrometer was operated in positive data-dependent acquisition mode. MS1 spectra were measured with a resolution of 70,000, an AGC target of 1e6, and a mass range from 350 to 1700 m/z. Up to 12 MS2 spectra per duty cycle were triggered, fragmented by high-energy collision–induced dissociation, and acquired with a resolution of 17,500 and an automatic gain control (AGC) target of 5e4, an isolation window of 2.0 m/z, and a normalized collision energy of 25. Dynamic exclusion was enabled with duration of 20 s.
All mass spectra were analyzed with MaxQuant software, version 1.5.5.1. MS/MS spectra were searched against the Homo sapiens Uniprot protein sequence database (version July 2016) and Global Proteome Machine common Repository of Adventitious Proteins sequences (commonly known protein contaminants). Precursor mass tolerance was set to 20 and 4.5 ppm for the first search where initial mass recalibration was completed and for the main search, respectively. Product ions were searched with a mass tolerance of 0.5 Da. The maximum precursor ion charge state used for searching was 7. Carbamidomethylation of cysteines was searched as a fixed modification, and oxidation of methionines and acetylation of protein N-terminal were searched as variable modifications. Enzyme was set to trypsin in a specific mode, and a maximum of two missed cleavages was allowed for searching. The target-decoy-based false discovery rate filter for spectrum and protein identification was set to 1%.
Common background proteins, including keratins, tubulins, histones, and ribosomal proteins, were removed (Mellacheruvu et al., 2013). Relative peak intensity values from the technical duplicates were added to obtain a combined intensity value. Any candidate identified by N144-BioID2, BioID2-LaA, or BioID2-Sun2 was excluded if the relative peak intensity was less than three times the level in BioID2-only. To achieve a more stringent list of candidates, an arbitrary relative peak intensity of 5 × 107 was set as a minimum threshold for exclusion criteria. For inclusion as a BioID candidate for N144, LaA, or Sun2, the candidate was considered unique if no other bait detected it or if another bait detected it, but it failed to pass the exclusion criteria. For inclusion as an enriched candidate for a specific bait, the protein must have had at least three times the relative peak intensity found by the comparative bait.
Unique or enriched candidates identified by each of the BioID2 baits were used as input for functional annotation analysis by DAVID (https://david.ncifcrf.gov/). Enriched lists were indexed using their corresponding Official Gene Symbol identifier and designated as “Gene list” for list type. Annotations and background were limited to H. sapiens before analysis by functional annotation clustering. Classification stringency was set to “highest” for quality restriction. Unique or enriched candidates identified by BioID2-Sun2 were exceptions and set to “high,” as no categories were identified at the highest stringency. A representative term from each significant cluster is reported, with p value provided.
Supplementary Material
Acknowledgments
We thank Indra Chandrasekar (Sanford Research, Sioux Falls, SD) for helpful guidance in FLIP studies and Christina Amatya (Sanford Research) for assistance with flow cytometry. This work was supported by Grants R01GM102203 and R01EB014869 to K.J.R. from the National Institutes of Health. The Imaging Core, Protein Biochemistry Core, and Flow Cytometry Core at Sanford Research, which facilitated these studies, are supported by Institutional Development Awards from the National Institute of General Medical Sciences and the National Institutes of Health under Grants P20GM103548 and P20GM103620.
Abbreviations used:
- BAF
barrier to autointegration factor
- ER
endoplasmic reticulum
- FLIP
fluorescence loss in photobleaching
- INM
inner nuclear membrane
- LaA
lamin A
- NE
nuclear envelope
- NETC
nuclear envelope targeting cassette
- NPC
nuclear pore complex
- ONM
outer nuclear membrane
- VRK2
vaccinia-related kinase-2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0138) on June 21, 2017.
REFERENCES
- Acar JF. Which antibiotics for prophylaxis. Scand J Infect Dis Suppl. 1982;36:127–128. [PubMed] [Google Scholar]
- Bermejo R, Kumar A, Foiani M. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends Cell Biol. 2012;22:465–473. doi: 10.1016/j.tcb.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Blanco S, Klimcakova L, Vega FM, Lazo PA. The subcellular localization of vaccinia-related kinase-2 (VRK2) isoforms determines their different effect on p53 stability in tumour cell lines. FEBS J. 2006;273:2487–2504. doi: 10.1111/j.1742-4658.2006.05256.x. [DOI] [PubMed] [Google Scholar]
- Blanco S, Santos C, Lazo PA. Vaccinia-related kinase 2 modulates the stress response to hypoxia mediated by TAK1. Mol Cell Biol. 2007;27:7273–7283. doi: 10.1128/MCB.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev. Mol Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol. 2014;109:1–52. doi: 10.1016/B978-0-12-397920-9.00006-8. [DOI] [PubMed] [Google Scholar]
- Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 2001;20:4399–4407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capanni C, Cenni V, Haraguchi T, Squarzoni S, Schuchner S, Ogris E, Novelli G, Maraldi N, Lattanzi G. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle. 2010;9:2600–2610. doi: 10.4161/cc.9.13.12080. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czapiewski R, Robson MI, Schirmer EC. Anchoring a leviathan: how the nuclear membrane tethers the genome. Front Genet. 2016;7:82. doi: 10.3389/fgene.2016.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P, Enarson M, Bastos R, Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 1998;107:228–236. doi: 10.1007/s004120050301. [DOI] [PubMed] [Google Scholar]
- Fernandez IF, Blanco S, Lozano J, Lazo PA. VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol Cell Biol. 2010;30:4687–4697. doi: 10.1128/MCB.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IF, Perez-Rivas LG, Blanco S, Castillo-Dominguez AA, Lozano J, Lazo PA. VRK2 anchors KSR1-MEK1 to endoplasmic reticulum forming a macromolecular complex that compartmentalizes MAPK signaling. Cell Mol Life Sci. 2012;69:3881–3893. doi: 10.1007/s00018-012-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 2007;26:132–143. doi: 10.1038/sj.emboj.7601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell. 2014;25:1287–1297. doi: 10.1091/mbc.E13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A, Hiraoka Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- Huang Y, Cai M, Clore GM, Craigie R. No interaction of barrier-to-autointegration factor (BAF) with HIV-1 MA, cone-rod homeobox (Crx) or MAN1-C in absence of DNA. PLoS One. 2011;6:e25123. doi: 10.1371/journal.pone.0025123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Roux KJ. Making the LINC: SUN and KASH protein interactions. Biol Chem. 2015a;396:295–310. doi: 10.1515/hsz-2014-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014a;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee D, Lee J, Song H, Kim HJ, Kim KT. Vaccinia-related kinase 2 controls the stability of the eukaryotic chaperonin TRiC/CCT by inhibiting the deubiquitinating enzyme USP25. Mol Cell Biol. 2015b;35:1754–1762. doi: 10.1128/MCB.01325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park DY, Lee D, Kim W, Jeong YH, Lee J, Chung SK, Ha H, Choi BH, Kim KT. Vaccinia-related kinase 2 mediates accumulation of polyglutamine aggregates via negative regulation of the chaperonin TRiC. Mol Cell Biol. 2014b;34:643–652. doi: 10.1128/MCB.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng Y. Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun. 2013;440:8–13. doi: 10.1016/j.bbrc.2013.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster OM, Cullen CF, Ohkura H. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J Cell Biol. 2007;179:817–824. doi: 10.1083/jcb.200706067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kim DI, Syme J, LuValle P, Burke B, Roux KJ. Dynamics of lamin-A processing following precursor accumulation. PLoS One. 2010;5:e10874. doi: 10.1371/journal.pone.0010874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, King MC. The nucleus: keeping it together by keeping it apart. Curr Opin Cell Biol. 2017;44:44–50. doi: 10.1016/j.ceb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- Meier I. LINCing the eukaryotic tree of life—towards a broad evolutionary comparison of nucleocytoplasmic bridging complexes. J Cell Sci. 2016;129:3523–3531. doi: 10.1242/jcs.186700. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor TP, Traktman P. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol Biol Cell. 2014;25:891–903. doi: 10.1091/mbc.E13-10-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve DM, Merced T, Fernandez IF, Blanco S, Vazquez-Cedeira M, Lazo PA. Human VRK2 modulates apoptosis by interaction with Bcl-xL and regulation of BAX gene expression. Cell Death Dis. 2013;4:e513. doi: 10.1038/cddis.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Shoemaker CJ, Gucek M, Cole RN, Wilson KL. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One. 2009;4:e7050. doi: 10.1371/journal.pone.0007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Traktman P. Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J Biol Chem. 2004;279:7934–7946. doi: 10.1074/jbc.M310813200. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N' terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Ryu HG, Kim SH, Lee D, Song H, Kim KT. Presumed pseudokinase VRK3 functions as a BAF kinase. Biochim Biophys Acta. 2015;1853:1738–1748. doi: 10.1016/j.bbamcr.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Powell L, Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Comai L. Recent advances in understanding the role of lamins in health and disease. F1000Research. 2016;5:2536. doi: 10.12688/f1000research.9260.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Wolf K, Lammerding J. Bursting the bubble—nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.02.008. 2017(Mar 9), S0962-8924(17)30032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Koujin T, Segura-Totten M, Wilson KL, Haraguchi T, Hiraoka Y. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, FRET analyses in living HeLa cells. J Struct Biol. 2004;147:31–41. doi: 10.1016/j.jsb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamas JA, Capelson M. Nuclear envelope and genome interactions in cell fate. Front Genet. 2015;6:95. doi: 10.3389/fgene.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Cedeira M, Lazo PA. Human VRK2 (vaccinia-related kinase 2) modulates tumor cell invasion by hyperactivation of NFAT1 and expression of cyclooxygenase-2. J Biol Chem. 2012;287:42739–42750. doi: 10.1074/jbc.M112.404285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B, Lorenz M, Moreno-Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S, Antonin W. Nup153 recruits the Nup107– 160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev Cell. 2015;33:717–728. doi: 10.1016/j.devcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu S, Rivolta C, Li LY, Peng GH, Swain PK, Sung CH, Swaroop A, Berson EL, Dryja TP, Chen S. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe VO, McMurtrie PI, Marr J, Amagase Y, Main S, Mills AD, Laskey RA, Takei Y. MCM3AP is transcribed from a promoter within an intron of the overlapping gene for GANP. J Mol Biol. 2011;406:355–361. doi: 10.1016/j.jmb.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Ostlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci USA. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Semenova E, Maric D, Craigie R. Dephosphorylation of barrier-to-autointegration factor by protein phosphatase 4 and its role in cell mitosis. J Biol Chem. 2014;289:1119–1127. doi: 10.1074/jbc.M113.492777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.