The 3D localization of polyglutamylated tubulin in eukaryotic flagella is identified. Polyglutamylated tubulins are located on the interface between the microtubule and its cross-bridging structure, the N-DRC. Flagellar motility is regulated by electrostatic interaction between negatively charged tubulins and positively charged N-DRC.
Abstract
Tubulins undergo various posttranslational modifications. Among them, polyglutamylation is involved in the motility of eukaryotic flagella and the stability of the axonemal microtubules. However, it remains unclear where polyglutamylated tubulin localizes precisely within the axoneme and how tubulin polyglutamylation affects flagellar motility. In this study, we identified the three-dimensional localization of the polyglutamylated tubulin in Chlamydomonas flagella using antibody labeling and cryo–electron tomography. Polyglutamylated tubulins specifically located in close proximity to a microtubule-cross-bridging structure called the nexin–dynein regulatory complex (N-DRC). Because N-DRC is positively charged, we hypothesized that there is an electrostatic interaction between the polyglutamylated tubulin and the N-DRC, and therefore we mutated the amino acid sequences of DRC4 to modify the charge of the N-DRC. We found that both augmentation and reduction of the positive charge on DRC4 resulted in reduced flagellar motility. Moreover, reduced motility in a mutant with a structurally defective N-DRC was partially restored by increasing the positive charge on DRC4. These results clearly indicate that beating motion of flagella is maintained by the electrostatic cross-bridge formed between the negatively charged polyglutamylated tubulins and the positively charged N-DRC.
INTRODUCTION
The functional diversity of microtubules is achieved by various posttranslational modifications of tubulin, including acetylation, tyrosination, glutamylation, glycylation, and phosphorylation (Janke, 2014; Wloga et al., 2017). Glutamylation, the most abundant tubulin modification (Audebert et al., 1994), adds glutamates to glutamate residues in the carboxyl termini of α- and β-tubulin. In particular, axonemal microtubules undergo polyglutamylation, in which as many as 21 glutamates are added to the tubulin (Schneider et al., 1998). This successive addition of glutamates is performed by the members of tubulin tyrosine ligase-like (TTLL) protein family (Janke, 2014). We previously isolated a Chlamydomonas mutant, tpg1, that lacks TTLL9 and thereby has reduced level of tubulin polyglutamylation (Kubo et al., 2010). The phenotype of tpg1 mutant indicates that tubulin polyglutamylation is involved in the regulation of flagellar motility and stability of axonemal microtubules (Kubo et al., 2010; Kubo et al., 2014). Furthermore, previous studies suggested that tubulin polyglutamylation regulates flagellar motility by controlling specific axonemal dynein and the nexin–dynein regulatory complex (N-DRC; Kubo et al., 2012; Alford et al., 2016). Although conventional immuno–electron microscopy showed that polyglutamylated tubulins are located on the B-tubule surface of the outer doublet microtubule (DMT; Lechtreck and Geimer, 2000; Suryavanshi et al., 2010; Kubo et al., 2010), it is necessary to conduct a three-dimensional (3D) structural analysis to investigate the relationship between the polyglutamylated tubulins and the axonemal dyneins or the N-DRC.
The N-DRC is a microtubule-cross-bridging structure that maintains the circular arrangement of the DMT in the axoneme (Heuser et al., 2009). Mutations in N-DRC components result in motility defects, which cause primary ciliary dyskinesia (Huang et al., 1982; Gardner et al., 1994; Austin-Tse et al., 2013; Wirschell et al., 2013). Our previous study suggested that the N-DRC loosely attaches to the DMT, so that it allows DMT sliding during the flagellar beating (Oda et al., 2015). However, it remains unknown how the N-DRC enables DMT cross-bridging and DMT sliding simultaneously. In this study, we identified the 3D localization of the polyglutamylated tubulin within the axoneme and functionally characterized the interaction between the N-DRC and the polyglutamylated tubulin. Our results suggest an electrostatic force–dependent mechanism of motility regulation in eukaryotic flagella.
RESULTS AND DISCUSSION
Labeling polyglutamylated tubulins in intact axonemes
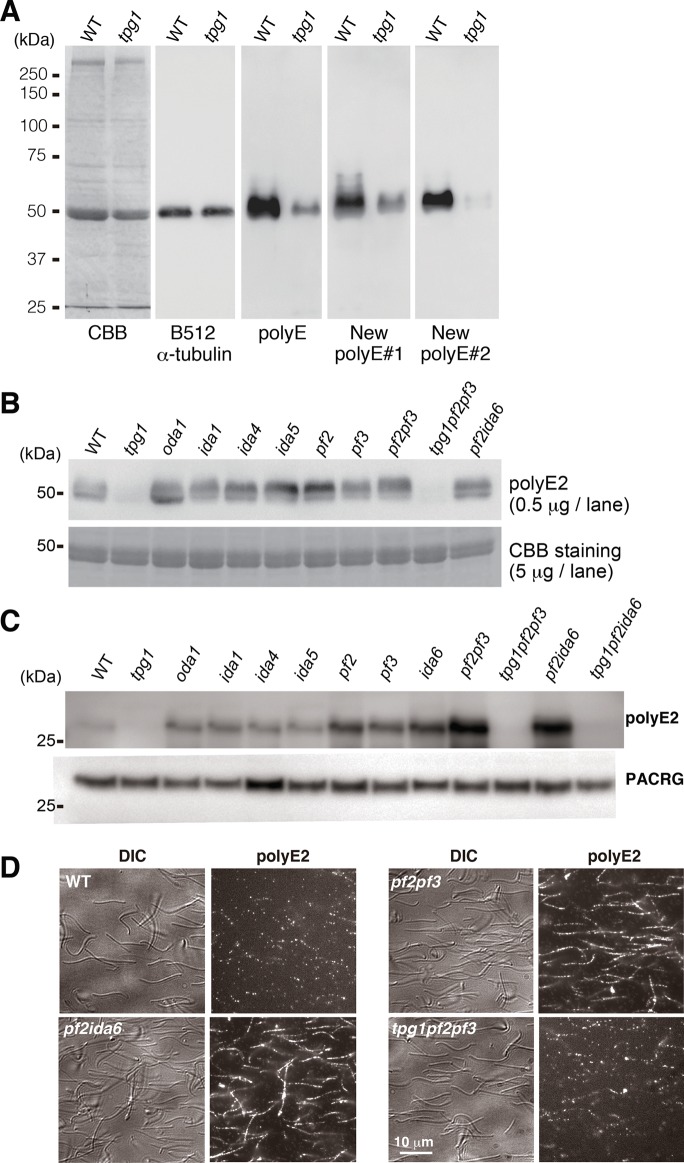
We labeled the polyglutamylated axonemal tubulins in Chlamydomonas flagella by raising polyclonal antibodies against polyglutamate peptide in two rabbits (Shang et al., 2002). The resulting antibodies, polyE#1 and #2, efficiently recognized the denatured axonemal tubulins in immunoblotting (Figure 1, A and B). In the tpg1 mutant, we found that axonemal tubulins could not be labeled with our new polyE antibodies (Figure 1A and B). The polyE#2 antibody showed better specificity than #1, and thus we designated it simply as “polyE2” and used it exclusively in further experiments.
FIGURE 1:
The polyE antibody labeling of the axonemes. (A, B) Immunoblots of denatured axonemal tubulins. (A) Our new polyE antibody (#1 and #2)–labeled polyglutamylated tubulins compared with commercially available polyE antibody (Shang et al., 2002). (B) Probing with the polyE antibodies showed that the polyglutamylation levels were unaffected by deficiency in ODA (oda1), IDA (ida1, ida4, and ida5), and N-DRC (pf2, pf3, pf2pf3, and pf2ida6). (C, D) PolyE2 labeling of the intact axonemes. Axonemes were incubated with the biotinylated Fab fragments of the polyE2 antibody. (C) Labeled axonemes were boiled and separated by electrophoresis. Bound biotinylated Fab fragments were detected using horseradish peroxidase–conjugated streptavidin. Blots with anti-PACRG antibody are shown as a loading control. (D) PolyE2-labeled axonemes were stained with streptavidin–Alexa 546. See Supplemental Figure S1 for fluorescence images of axonemes of other strains.
Next we examined whether polyE2 could label native tubulins within intact axonemes. The immunoglobulin G molecule is too large to penetrate into the axoneme (Oda and Kikkawa, 2013; Oda, 2017), so we digested the polyE2 antibody with papain to generate Fab fragments. We incubated the axonemes with biotinylated Fab fragments of polyE2 antibody and observed binding using both immunoblotting (Figure 1C) and fluorescence microscopy (Figure 1D). In contrast, polyE2 Fab fragments only faintly labeled intact and native wild-type axonemes (Figure 1, C and D). Because polyE antibody can bind to frayed and aldehyde-fixed axonemes (Kubo et al., 2010), we speculated that polyglutamylated tubulins were masked by axonemal structures, such as outer dynein arms (ODAs), inner dynein arms (IDAs), or the N-DRC. Contrary to our expectation, however, polyE2 Fab fragments did not efficiently label ODA-missing (oda1) and IDA-deficient (ida1, ida4, and ida5) axonemes (Figure 1C, Supplemental Figure S1, and Supplemental Table S1; Kamiya, 1988; Takada et al., 2002), whereas N-DRC–deficient (pf2, pf3, and ida6) axonemes were labeled relatively well compared with others, but the staining patterns were sporadic (Figure 1C and Supplemental Figure S1). Biochemically, the N-DRC in pf2 axoneme retains DRC1 and DRC2, whereas DRC3 and DRC4 are missing (Lin et al., 2011), suggesting that the remaining components of the N-DRC in the pf2 axoneme still mask the polyglutamylated tubulins (Oda et al., 2015). Therefore we generated the double mutant pf2pf3, which is deficient in all of the major components (DRC1–7) of the N-DRC (Lin et al., 2011). Surprisingly, we found that polyE2 Fab fragments efficiently labeled the pf2pf3 axonemes (Figure 1, C and D). Similarly, pf2ida6 axonemes were also efficiently labeled with the polyE2 Fab fragments. However, Fab binding was drastically reduced in axonemes carrying the tpg1 background (tpg1pf2pf3 and tpg1pf2ida6; Figure 1, C and D). These results suggest that polyglutamylated tubulins are masked by the N-DRC.
Three-dimensional localization of polyglutamylated tubulins
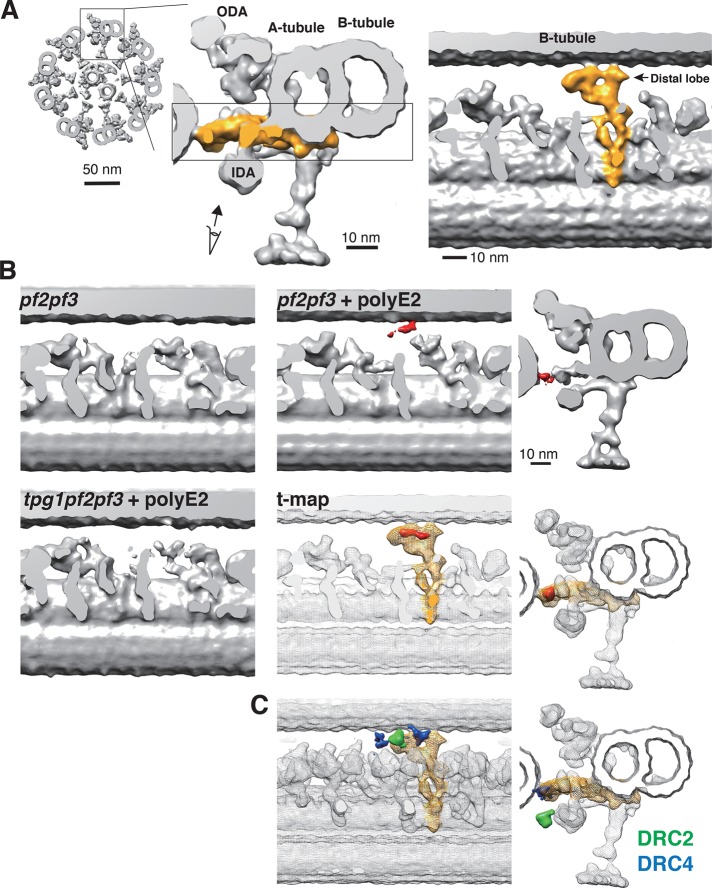
To identify the 3D localization of the polyglutamylated tubulins, we labeled pf2pf3 axonemes with biotinylated polyE2 Fab fragments and then amplified the physical size of the labels using streptavidin and biotinylated cytochrome c (Oda et al., 2014; Oda, 2017). Cryo–electron tomography and subtomogram averaging of the labeled DMTs revealed that the label densities were located on the B-tubule surface, which are masked by the N-DRC in wild-type axonemes (Figure 2, A and B, and Supplemental Figure S2A). In contrast, tpg1pf2pf3 axonemes did not show significant label densities.
FIGURE 2:
Three-dimensional localization of polyglutamylated tubulin. (A) Three-dimensional structure of the N-DRC in the Chlamydomonas axoneme. Right, tip-to-base view of the 9 + 2 structure of the axoneme. Middle, cross-sectional view of the DMT. The N-DRC is shown in yellow. Left, internal slab view of the boxed region. The N-DRC contacts the B-tubule at the distal lobe. (B) DMT structures of unlabeled pf2pf3, polyE2-labeled pf2pf3, and polyE2-labeled tpg1pf2pf3 axonemes. Internal slab (left and middle) and top (right) views. The label densities are in red. A t-value map (t-map, red) was calculated by comparing the labeled and unlabeled pf2pf3 structures. The wild-type DMT structure is superimposed on the t-value map as a transparent mesh isosurface. The N-DRC is shown in yellow. (C) Positions of the amino termini of DRC2 (green) and DRC4 (blue) as reported previously (Oda et al., 2015).
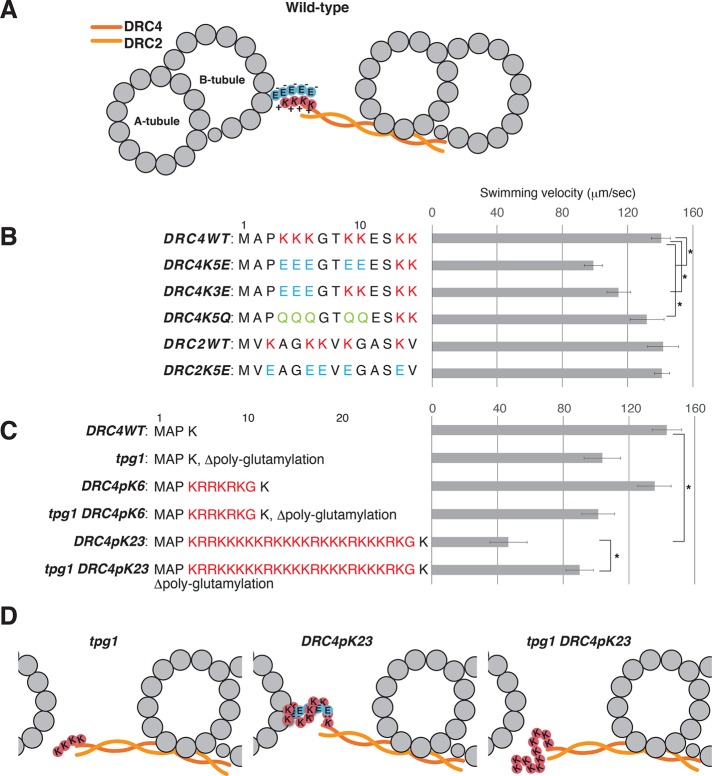
Alford et al. (2016) reported that the motility defect in tpg1 cells is related to the distal lobe of the N-DRC (Figure 2A). In our previous report (Oda et al., 2015), we also showed that the distal lobe of the N-DRC loosely binds to the B-tubule and allows sliding of the DMT. In particular, the amino terminus of DRC4 locates in close proximity to the B-tubule surface (Figure 2C, blue; Oda et al., 2015; Song et al., 2015). The amino terminus of DRC2 also locates in the distal lobe of the N-DRC, but its position is slightly away from the B-tubule surface (Figure 2C, green). Because the amino acid sequence of the amino-terminal domain of DRC4 is rich in lysine residues (Rupp and Porter, 2003), we hypothesized that the N-DRC interacts with the DMT via electrostatically driven “cross-bridges” between the positively charged amino terminus of the DRC4 and the negatively charged polyglutamylated tubulin (Figure 3A).
FIGURE 3:
Effects of replacement and addition of Lys residues on flagellar motility. (A) Schematic diagram of the DMT and the N-DRC, showing the possible electrostatic interaction between the polyglutamate chain (circled E) extending from the B-tubule and the polylysine chain (circled K) on the DRC4. The coiled-coil complex of DRC2 and DRC4 is indicated by orange wavy lines. (B, C) Swimming velocities of DRC4 and DRC2 mutants. Residue replacements and additions are shown next to the strain names. Asterisks indicate statistically significant differences (p < 0.01). The p values were calculated using Student’s t test. Means ± SEM for the mean swimming velocities were calculated from 20 cells. (B) Lys residues on DRC4 and DRC2 were replaced with either Glu or Gln. (C) A total of 6 or 23 residues of Lys and Arg residues were inserted after the Pro-3 of DRC4. (D) Schematic diagrams of the interaction between DRC4 and the B-tubule. In tpg1, the interaction between the poly-Lys on DRC4 and the B-tubule is abolished. In DRC4 pK23, the hyper-poly-Lys peptide on DRC4 forms an excessively strong cross-bridge with polyglutamylated tubulin on the B-tubule.
Net charge of DRC4 affects flagellar motility
To test our “electrostatic interaction hypothesis,” we changed the amino acid composition of the DRC4 amino terminus (Figure 3, B and C, and Supplemental Figure S3A). First, we replaced the lysine residues with glutamate or glutamine and expressed the modified DRC4 in pf2 cells. The transformed cells showed a charge inversion–dependent decrease in swimming velocity (Figure 3B), indicating that the interaction between positively charged DRC4 and polyglutamylated tubulin is required for normal flagellar motility. However, replacement of the lysine residues in DRC2 with glutamate had little effect on the swimming velocity.
To modify the electrostatic cross-bridge, we next added lysine and arginine residues to the amino terminus of DRC4 and expressed the modified protein in pf2 and tpg1pf2 cells. Of interest, the addition of 23 positively charged residues to DRC4 (DRC4pK23) caused a decrease in the swimming velocity, suggesting that excessively strong electrostatic interaction between the N-DRC and the B-tubule impedes microtubule sliding (Figure 3D). This possibility is supported by the observation that the tpg1 DRC4pK23 cells swam faster than DRC4pK23 cells. Finally, we expressed this hyperpositively charged DRC4 in pf2ida6 and tpg1pf2ida6 cells (Figure 4A). Based on the previous reports (Lin et al., 2011; Oda et al., 2015), the N-DRC in ida6 (DRC2-deficient) axonemes is expected to retain the microtubule-cross-bridging capacity via the remaining DRC4. Expression of DRC4pK23 protein partially rescued the motility defect of ida6 cells, suggesting that the augmented positive charges on DRC4 could partially complement the weakened interaction between the defective N-DRC and the B-tubule in ida6 (Figure 4B). In accordance with this model, expression of DRC4pK23 protein did not restore the motility of tpg1pf2ida6 cells.
FIGURE 4:
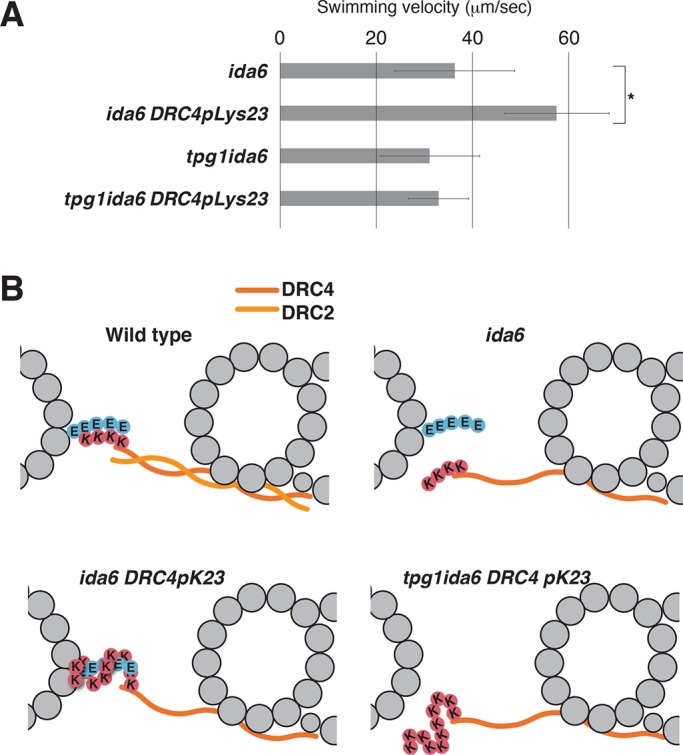
Effect of poly-Lys addition on the motility defect of ida6. (A) The motility defect in ida6 (DRC2-deficient) cells was partially rescued by the addition of hyper-poly-Lys on DRC4. No restoration of the flagellar motility was observed in tpg1ida6. Asterisks indicate statistically significant differences (p < 0.01). The p values were calculated using Student’s t test. Means ± SEM for the mean swimming velocities were calculated from 20 cells. See Supplemental Figure S3B for the swimming velocities of other strains. (B) Schematic diagrams of the cross-bridge between the hyper-poly-Lys peptide on DRC4 and the polyglutamate chain of the B-tubule. In ida6, the absence of DRC2 causes structural defects in the N-DRC and weakens its interaction with the B-tubule. The hyper-poly-Lys peptide on DRC4 reinforces the interaction with the polyglutamylated tubulins to partially restore N-DRC function.
Relationship between IDA e and polyglutamylated tubulin
We previously showed (Kubo et al., 2012) that there is a genetic interaction between IDA e and tubulin polyglutamylation, based on the observation that tpg1ida4 cells were essentially nonmotile, whereas tpg1ida5 cells showed clear motility, although slightly slower than that of ida5 cells. Both ida4 and ida5 mutants lack the IDA subspecies a, c, and d, but only ida5 lacks IDA e (Kamiya et al., 1991; Kagami and Kamiya, 1992; Kato et al., 1993). These results suggested that IDA e can interact with the polyglutamylated tubulins and that IDA e has a negative effect on the flagellar motility in the absence of tubulin polyglutamylation. The present results revealed that the polyglutamylated tubulins are located on the protofilament facing the N-DRC (Figure 2). Lin et al. (2014) reported that the microtubule-binding domains of IDAs and the distal lobe of the N-DRC attach to different protofilaments of the B-tubule. Thus it is unlikely that IDA e interacts with the polyglutamylated tubulins on the B-tubule. The difference in flagellar motility between tpg1ida4 and tpg1ida5 cells may result from the possible functional interaction between the N-DRC and IDA e. N-DRC-defective mutants such as pf2, pf3, and ida6 lack or have reduced amounts of IDA e (Kamiya et al., 1991; Bower et al., 2013; Oda et al., 2013). In contrast, IDA e–deficient ida5 has an intact N-DRC (Bui et al., 2012), suggesting that assembly of IDA e structurally depends on the N-DRC. We previously showed that mechanical signals are transmitted from the central pair apparatus to the ODAs through the radial spokes (Oda et al., 2014). These mechanical signals are likely to travel along the outer–inner dynein linker between the N-DRC and the ODA (Oda et al., 2013, 2015, 2016). Considering the structural interaction between the N-DRC and the IDA e, it is possible that the N-DRC regulates the IDA e by mechanosignal inputs, which can be altered in the absence of tubulin polyglutamylation. Therefore we reasoned that the difference in flagellar motility between tpg1ida4 and tpg1ida5 mutants may result from an indirect effect of the tpg1 background on the IDA e through the aberrant regulatory inputs from the N-DRC. Testing this hypothesis will require functional analyses of the IDA e using a mutant lacking only IDA e. Similarly, the N-DRC is structurally connected to IDA g through DRC3 (Awata et al., 2015). It is of interest to investigate whether the activity of IDA g is also affected by tubulin polyglutamylation.
Determinants of the polyglutamylation sites
In summary, our results suggest that the positively charged surface of the distal lobe of the N-DRC interacts with the negatively charged surface of the B-tubule. The strength of the electrostatic force between the N-DRC and the B-tubule appears to be tightly regulated because both an increase and a reduction of the positive charge on the N-DRC reduced flagellar motility. Of interest, the function of structurally defective N-DRC was partially restored by increasing the positive charge on the N-DRC, suggesting that the N-DRC regulates flagellar motility via a chemically simple (and artificially manipulatable) mechanism.
Our results show that the polyglutamylation sites localize on the interface between the N-DRC and the B-tubule. However, there remains a possibility that the averaged subtomogram fails to visualize other polyglutamylated tubulins, whose localizations do not conform to the 96-nm repeat of the DMT or are structurally inaccessible to polyE2 Fab fragments. To address this question, we roughly estimate the amount of polyglutamylated tubulin based on the previous biochemical data. Two-dimensional gel electrophoresis of wild-type and tpg1 axonemes shows that ∼1.5% of total axonemal tubulin is polyglutamylated (Figure 1D of Kubo et al., 2010). This estimation agrees with the mass spectrometric result on sea urchin sperm axonemes that only a trace amount of tubulin is polyglutamylated (Multigner et al., 1996). The label density of polyE2 Fab on pf2pf3 axoneme spans ∼15 nm (Figure 2B), corresponding roughly to two tubulin dimers. Given that there are 276 tubulin dimers in one 96-nm repeat of the DMT (96/8 nm × 23 protofilaments; Nicastro et al., 2011; Maheshwari et al., 2015), the labeled tubulins in our electron density map represent a good amount but not all of the polyglutamylated tubulins in the axoneme. The localization and function of these unvisualized tubulins remain to be investigated.
Another remaining question is how TTLL9 glutamylates specifically the N-DRC–facing part of the B-tubule. One possibility is that the N-DRC recruits TTLL9 through direct binding. However, the unchanged polyglutamylation levels in pf2, pf3, pf2pf3, and pf2ida6 mutants (Figure 1B) suggest that the N-DRC itself is dispensable for the activity and regulation of TTLL9. It has been reported that TTLL9 forms a complex with an adaptor protein FAP234, which is essential for the axonemal localization of TTLL9 (Kubo et al., 2014). It is possible that FAP234 is also involved in the localized activation of TTLL9. Investigation of the molecular mechanisms for the spatial regulation of polyglutamylation will require identifying the localizations of the TTLL9–FAP234 complex and other glutamylases and deglutamylases.
MATERIALS AND METHODS
Strains and reagents
Chlamydomonas reinhardtii wild-type strain CC-125 cells were grown in Tris-acetate-phosphate (TAP) medium. To screen transformants, cells were grown on TAP agar supplemented with hygromycin B (20 μg/ml; Nacalai Tesque, Kyoto, Japan). The C. reinhardtii strains used in this study are listed in Supplemental Table S1. Expression plasmids for DRC4 were as described previously (Oda et al., 2015). Residue addition and replacement were carried out using an In-Fusion HD Cloning Kit (Clontech, Palo Alto, CA). B512 mouse monoclonal anti–α-tubulin antibody and polyE anti-polyglutamate antibody were purchased from Sigma-Aldrich (St. Louis, MO) and Adipogen Life Sciences (San Diego, CA), respectively. Anti-DRC2 (Oda et al., 2013), DRC4 (Oda et al., 2013), IC1 (Oda et al., 2016), and PACRG (Ikeda et al., 2007) antibodies were described previously.
Preparation of axonemes
Chlamydomonas cells were deflagellated with dibucaine-HCl (Wako Pure Chemical Industries, Tokyo, Japan), and axonemes were collected by centrifugation (Piperno et al., 1977). Flagella were demembranated with 1% Nonidet P-40 in HMDENa buffer or HMDEK buffer composed of 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–NaOH pH 7.2, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ethylene glycol tetraacetic acid, 50 mM NaCl (HMDENa) or 50 mM CH3COOK (HMDEK), and 1× protease inhibitor cocktail (Nacalai Tesque).
Generation of biotinylated Fab fragments of polyE2 antibody
Anti-polyglutamate polyclonal rabbit antisera (polyE#1 and #2) were raised commercially (Hokkaido System Science, Sapporo, Japan; Evebio Science, Wakayama, Japan) against Cys(Glu)9 peptide coupled to keyhole limpet hemocyanin (Shang et al., 2002). The new polyE#2 antibody (referred to as polyE2) was purified by ammonium sulfate precipitation (Grodzki and Berenstein, 2010) and digested at 37°C for 6 h with papain (Nacalai Tesque, Kyoto, Japan) after dialysis against digestion buffer (20 mM HEPES-NaOH, pH 7.0, 10 mM EDTA, 20 mM cysteine HCl). Fab fragments were then cross-linked with a 40-fold molar excess of biotin-OSu (Dojindo Molecular Technologies, Kumamoto, Japan). Unreacted biotin was quenched with 0.1 M glycine and removed by three rounds of desalting using PD Mini-Trap G-25 columns (GE Healthcare, Pittsburgh, PA).
Electrophoresis and immunoblotting
Axonemal proteins were resolved by SDS–PAGE on 5–15% polyacrylamide gradient gels (Nacalai Tesque) and then blotted onto polyvinylidene difluoride membranes. Blots were probed with streptavidin conjugated with horseradish peroxidase (Thermo Scientific, Rockford, IL) or the indicated primary antibodies.
Fab labeling and fluorescence microscopy detection of axonemes
For antibody labeling, demembranated axonemes were blocked with 1 mg/ml bovine serum albumin (BSA) in HMDENa buffer for 1 h at 4°C and then incubated with 10 μg/ml biotinylated Fab fragments of the polyE2 antibody for 1 h at 4°C. Labeled axonemes were washed twice with HMDENa buffer and then attached to glass slides or boiled for the electrophoresis. For fluorescence staining using streptavidin, axonemes and glass slides were blocked with 1 mg/ml BSA in HMDENa buffer and then incubated with 1 μg/ml Alexa Fluor 546–conjugated streptavidin (Invitrogen, Carlsbad, CA) for 1 min. Axonemes were then washed three times with HMDENa buffer and observed using a fluorescence microscope (BX53; Olympus, Tokyo, Japan). Images were recorded using a charge-coupled device camera (ORCA-Flash4.0 V3; Hamamatsu Photonics, Hamamatsu, Japan).
Sample preparation for cryo–electron tomography
Streptavidin–cytochrome c labeling of polyE2 Fab-labeled axonemes was carried out as described previously (Oda et al., 2014) with minor modifications. Demembranated axonemes were incubated with 0.05 mg/ml biotinylated Fab fragments for 1 h at 4°C in HMDENa buffer in the presence of 1 mg/ml BSA. Axonemes were then washed with HMDENa buffer and incubated with 0.05 mg/ml streptavidin for 15 min at 4°C. Next axonemes were washed and again incubated with 0.05 mg/ml biotinylated cytochrome c. This sequence was repeated, so that the biotinylated Fab fragments were labeled with three rounds of streptavidin and two rounds of cytochrome c. Labeled or unlabeled axonemes were resuspended in HMDEK buffer at a concentration of 0.02 mg/ml and mixed with cationic 15-nm colloidal gold (BBI Solutions, Cardiff, United Kingdom). Homemade holey carbon grids were glow discharged for 20 s. Suspended axonemes plus colloidal gold (5 μl) was loaded onto the grids and plunge-frozen in liquid ethane at −180°C with a Vitrobot Mark IV automated plunge-freezing device (FEI, Hillsboro, OR).
Image acquisition
Grids were transferred to a JEM-3100FEF transmission electron microscope (JEOL, Tokyo, Japan) with a Gatan 914 high-tilt liquid nitrogen cryotransfer holder (Gatan, Pleasanton, CA). Tilt series images were recorded at −180°C using a TemCam-F416 complementary metal-oxide semiconductor camera (TVIPS, Gauting, Germany), and automated acquisition was performed using the EM-TOOLs program (TVIPS). The angular range of the tilt series was from –60 to 60° at 2.0° increments. The total electron dose was limited to ∼100 e–/Å2. Images were recorded at 300 keV with 6- to 9-μm defocus at a magnification of 22,300× and a pixel size of 7 Å. An in-column omega energy filter was used to enhance image contrast in the zero-loss mode with a slit width of 20 eV.
Image processing
Image processing for subtomogram averaging of DMT structures was carried out as described previously (Oda and Kikkawa, 2013; Oda et al., 2014). Tilt series images were aligned and backprojected to reconstruct 3D tomograms using the IMOD software package (Kremer et al., 1996). Tomograms of intact axonemes with a high signal-to-noise ratio were selected and used for subtomogram averaging of the 96-nm repeats of DMTs. Alignment and averaging of subtomograms were conducted using custom Ruby-Helix scripts (Metlagel et al., 2007) and the PEET software suite (Nicastro et al., 2006). The number of DMT subtomograms averaged was as follows: 766 for wild type, 963 for pf2pf3, 846 for pf2pf3 plus polyE2, and 952 for tpg1pf2pf3 plus polyE2. The effective resolutions determined by Fourier shell correlation with a cutoff value of 0.5 were ∼5.0 nm (Supplemental Figure S2B). Surface renderings were generated using UCSF Chimera (Pettersen et al., 2004). The electron microscopic maps of averaged DMTs are available at the EM Data Bank (www.emdatabank.org) under the accession numbers EMD-6753–6756.
Statistical analysis
We identified statistically significant differences by applying Student’s t test to compare unlabeled pf2pf3 and polyE2-labeled pf2pf3 axonemes as described previously (Oda and Kikkawa, 2013; Oda et al., 2014). First, unlabeled and labeled subtomograms were randomly divided into three data sets. Subtomograms for each data set were aligned and averaged, and a six averaged subtomograms were created. We calculated the t value for each voxel and present it as a single t-value map. The isosurface threshold values for the data sets were t > 7.17, with a one-tailed probability of <0.1%.
Measurement of swimming velocity
The swimming velocity of Chlamydomonas cells was recorded using an inverted CK40 microscope (Olympus) at a total magnification of 100×. A red filter with a cutoff wavelength of 630 nm was inserted before the condenser lens to suppress the cellular response to light.
Supplementary Material
Acknowledgments
We thank Masahide Kikkawa for his invaluable advice and use of the electron microscope. This work was supported by the Kazato Research Foundation (to T.O.), the Takeda Science Foundation (to T.O.), Japan Society for the Promotion of Science KAKENHI Grants 15H01202 and 17H05057 (to T.O.) and 17K15115 (to T.K.), the Naito Foundation (to T.O.), the Uehara Memorial Foundation (to T.O.), the Senri Life Science Foundation (to T.O.), the Institute for Fermentation, Osaka (to T.O.), and the Ichiro Kanehara Foundation (to T.K.).
Abbreviations used:
- DMT
outer doublet microtubule
- IDA
inner dynein arm
- N-DRC
nexin–dynein regulatory complex
- ODA
outer dynein arm.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-05-0285) on June 21, 2017.
REFERENCES
- Alford LM, Stoddard D, Li JH, Hunter EL, Tritschler D, Bower R, Nicastro D, Porter ME, Sale WS. The nexin link and B-tubule glutamylation maintain the alignment of outer doublets in the ciliary axoneme. Cytoskeleton (Hoboken) 2016;73:331–340. doi: 10.1002/cm.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Edde B. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J Cell Sci. 1994;107:2313–2322. doi: 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 Defects as causing primary ciliary dyskinesia. Am J Hum Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata J, Song K, Lin J, King SM, Sanderson MJ, Nicastro D, Witman GB. DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol Biol Cell. 2015;26:2788–2800. doi: 10.1091/mbc.E15-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell. 2013;24:1134–1152. doi: 10.1091/mbc.E12-11-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol. 1994;127:1311–1325. doi: 10.1083/jcb.127.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzki AC, Berenstein E. Antibody purification: ammonium sulfate fractionation or gel filtration. Methods Mol Biol. 2010;588:15–26. doi: 10.1007/978-1-59745-324-0_3. [DOI] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ikeda T, Morikawa K, Kamiya R. Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil Cytoskeleton. 2007;64:814–821. doi: 10.1002/cm.20225. [DOI] [PubMed] [Google Scholar]
- Janke C. The tubulin code: molecular components, readout mechanisms, functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci. 1992;103:653–664. [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Kagami O, Yagi T, Kamiya R. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct. 1993;18:371–377. doi: 10.1247/csf.18.371. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kubo T, Yagi T, Kamiya R. Tubulin polyglutamylation regulates flagellar motility by controlling a specific inner-arm dynein that interacts with the dynein regulatory complex. Cytoskeleton (Hoboken) 2012;69:1059–1068. doi: 10.1002/cm.21075. [DOI] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Liu Z, Shibuya R, Hirono M, Kamiya R. A conserved flagella-associated protein in Chlamydomonas, FAP234, is essential for axonemal localization of tubulin polyglutamylase TTLL9. Mol Biol Cell. 2014;25:107–117. doi: 10.1091/mbc.E13-07-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Geimer S. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton. 2000;47:219–235. doi: 10.1002/1097-0169(200011)47:3<219::AID-CM5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. Structural mechanism of the dynein power stroke. Nat Cell Biol. 2014;16:479–485. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–29191. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T. alpha- and beta-Tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo-electron microscopy and tomography. Structure. 2015:1584–1595. doi: 10.1016/j.str.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Metlagel Z, Kikkawa YS, Kikkawa M. Ruby-Helix: an implementation of helical image processing based on object-oriented scripting language. J Struct Biol. 2007;157:95–105. doi: 10.1016/j.jsb.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Multigner L, Pignot-Paintrand I, Saoudi Y, Job D, Plessmann U, Rudiger M, Weber K. The A and B tubules of the outer doublets of sea urchin sperm axonemes are composed of different tubulin variants. Biochemistry. 1996;35:10862–10871. doi: 10.1021/bi961057u. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci USA. 2011;108:E845–E853. doi: 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Oda T. Three-dimensional structural labeling microscopy of cilia and flagella. Microscopy (Oxf) 2017 doi: 10.1093/jmicro/dfx018. [DOI] [PubMed] [Google Scholar]
- Oda T, Abe T, Yanagisawa H, Kikkawa M. Structure and function of outer dynein arm intermediate and light chain complex. Mol Biol Cell. 2016;27:1051–1059. doi: 10.1091/mbc.E15-10-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Kikkawa M. Novel structural labeling method using cryo-electron tomography and biotin-streptavidin system. J Struct Biol. 2013;183:305–311. doi: 10.1016/j.jsb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Oda T, Yagi T, Yanagisawa H, Kikkawa M. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr Biol. 2013;23:656–664. doi: 10.1016/j.cub.2013.03.028. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kikkawa M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol Biol Cell. 2015;26:294–304. doi: 10.1091/mbc.E14-09-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol. 2014;204:807–819. doi: 10.1083/jcb.201312014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJ. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, Porter ME. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol. 2003;162:47–57. doi: 10.1083/jcb.200303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Plessmann U, Felleisen R, Weber K. Posttranslational modifications of trichomonad tubulins; identification of multiple glutamylation sites. FEBS Lett. 1998;429:399–402. doi: 10.1016/s0014-5793(98)00644-9. [DOI] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Awata J, Tritschler D, Bower R, Witman GB, Porter ME, Nicastro D. In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J Biol Chem. 2015;290:5341–5353. doi: 10.1074/jbc.M114.626556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell. 2002;13:1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Joachimiak E, Louka P, Gaertig J. Posttranslational modifications of tubulin and cilia. Cold Spring Harb Perspect Biol. 2017;9, pii: a028159 doi: 10.1101/cshperspect.a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.