Abstract
Recent identification of the neonatal 2nd coronary vascular population (2nd CVP) suggests that a subset of these vessels form de novo and mature in the inner myocardial wall of the postnatal heart. However, the origin of smooth muscle cells (SMCs) in the postnatal 2nd CVP remains undetermined. Using a tamoxifen-inducible Wt1-CreER driver and a Rosa26-RFP reporter line, we traced the lineage of epicardial cells to determine if they contribute to SMCs of the 2nd CVP. Late embryonic and postnatal induction of Wt1-CreER activity demonstrated that at these stages Wt1-labeled epicardium does not significantly migrate into the myocardium to form SMCs. However, following tamoxifen treatment at an early embryonic stage (E10.5), we detected Wt1 descendants (epicardium-derived cells, or EPDCs) in the outer myocardial wall at E17.5. When the 2nd CVP forms and remodels at postnatal stage, these early labeled EDPCs re-migrate deep into the inner myocardial wall and contribute to 2nd CVP-SMCs in the adult heart. Our findings reveal that SMCs in the postnatal 2nd CVP are pre-specified as EPDCs from the earliest wave of epicardial cell migration. Rather than the re-activation and migration of epicardial cells at later stages, these resident EPDCs mobilize and contribute to smooth muscle of the 2nd CVP during postnatal development.
Keywords: Lineage tracing, Epicardium, Epicardium-derived cells (EPDCs), Smooth muscle, 2nd coronary vascular population (2nd CVP)
1. Introduction
The cellular origin and formation of the coronary arteries represents a fundamental question in the field of developmental and cardiovascular biology. There is debate over the developmental origin for coronary arteries in the compact myocardium of embryonic heart [1, 2]. A comprehensive understanding of coronary vessel specification and morphogenesis could significantly impact future therapeutic strategies targeting angiogenesis in cardiac injury and regeneration [3–7]. In the embryonic heart, the ventricular wall is composed of two layers: the compact myocardium (outer myocardial wall, OMW) and the trabecular myocardium (inner myocardial wall, IMW). The compact myocardium receives oxygen from coronary vessels that are specified during embryonic development, while the trabecular myocardium receives oxygen via direct diffusion from the blood within the interior chambers of the heart. During subsequent development, the trabecular layer becomes solidified, increasing the compact component of the ventricular wall [8]. Compaction of the trabecular myocardium blocks the direct contact between the oxygen-rich blood in the heart chambers and the deeper myocardial layers, therefore the formation of new blood vessels is required for IMW cardiomyocyte survival in the postnatal heart. We have showed that these new blood vessels do not form by angiogenic expansion of pre-existing coronary vessels. Instead, they form de novo by lineage conversion of endocardial cells to coronary vessels within the IMW [9]. Mirroring the behavior of the coronary vessels forming during early embryonic stage (1st coronary vascular population, 1st CVP), endocardium-derived coronary vessels (2nd CVP) form later in the IMW of the heart. The distribution of the 1st and 2nd CVP is almost mutually exclusive in the adult heart [9]. As a substantial portion of the 2nd CVP forms de novo during neonatal development, these vessels mature and recruit smooth muscle cells (SMCs) at later stages. However, the developmental origin of the smooth muscle cells recruited to the newly formed postnatal 2nd CVP is undetermined, which remains as an open question in the cardiovascular developmental field [10]. A better understanding of how these coronary vessels are built during development will provide important insights for therapeutic research in cardiac diseases and regeneration.
2. Materials and methods
2.1 Animals
All Animals were used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. WT1-CreER, Wt1-GFPCre [11], Rosa26-RFP [12], Tbx18-Cre [13] and Apln-CreER [14] mice lines were maintained on a C129/C57BL6/J-mixed background. Tamoxifen (sigma, T5648) was dissolved in corn oil at the concentration of 20 mg/ml, and mice were given Tamoxifen (0.1–0.15 mg/g body weight) by oral gavage to trigger lineage tracing at indicated time as described previously [15].
2.2 Immunostaining
Immunostaining was performed as described previously [16]. Briefly, hearts or embryos were collected and washed in cold PBS, fixed in 4% PFA at 4 °C for 0.5–1 h, depending on the mice age. After washing 3 times in PBS (10 min each time), hearts or embryos were dehydrated in 30% sucrose at 4 °C over night, and then embedded in OCT (Sakura) for frozen sectioning. 8–10 µm thickness slides were collected on positively charged slides, treated with blocking buffer (5% normal donkey serum in PBS with 0.1% triton X-100) for 30 min at room temperature, and then incubated with primary antibodies at optimized dilution over night at 4 °C. The first antibodies were obtained from companies: PECAM (BD Pharmingen, 5,53,370, 1:500), smooth muscle actin (SMA, Sigma, F3777, 1:500), smooth muscle 22 alpha (SM22, Abcam, ab14106, 1:100), smooth muscle myosin heavy chain (SM-MHC, Biomedical technology, BT-562, 1:100), estrogen receptor (ESR, Abcam, ab27,595, ready-to-use), VE-Cadherin (R & D, AF1002, 1:100), RFP (Rockland, 600-401-379, 1:1000), beta-galactosidase (beta-GAL, MP biomedicals, 55,976, 1:5000). Following washing out the primary antibody, signals were developed with Alexa fluorescent second antibodies and nuclei were visualized by 4′6-diamidino-2-phenylindole (DAPI, Vector labs) included in the mounting medium. For weak signals, secondary antibodies conjugated with HRP enzyme were used and tyramide signal amplification (PerkinElmer) method was used to amplify the signal as described previously [17]. Images were taken by Olympus confocal system (FV1200) or Zeiss confocal system (Zeiss 510).
3. Results
The postnatal 2nd CVP forms in the IMW during neonatal development. Throughout the transition from adolescence to adulthood, many of these vessels gradually become mature coronary arteries and recruit stabilizing cells, such as SMCs or pericytes. In the postnatal 4 weeks old (P4w) Apln-CreER; Rosa26-RFP heart, Apln-CreER genetic labeling of the 1st, but not the 2nd, CVP demarcates the outer myocardial wall (OMW) from the inner myocardial wall (IMW, Fig. S1A) [9]. We could clearly detect SMCs of the 2nd CVP in the IMW (Fig. S1B, C2). To reveal the developmental origin of these newly recruited SMCs, we crossed mice harboring an inducible epicardial-specific Cre, Wt1-CreER [11] with Rosa26-lox-stop-lox-RFP reporter line (Rosa26-RFP) [12]. Postnatal day 1 (P1) tamoxifen induction of Wt1-CreER activity leads to robust labeling of the neonatal epicardium and ectopic labeling of endothelial cells (Fig. 1B). Recent reports demonstrated that Wt1 is also expressed in coronary endothelial cells in addition to epicardium [18–20]. Therefore, Wt1-CreER labeling of endothelial cells at this stage likely results from Cre expression driven by Wt1 within this endothelial cell population, rather than trans-differentiation of Wt1-labeled epicardial cells. While worth noting, our study here focuses on the origin of SMCs of the 2nd CVP, and this ectopic endothelial labeling by Wt1-CreER (following induction at P1) did not significantly influence the outcome of this study and this finding was therefore not pursued.
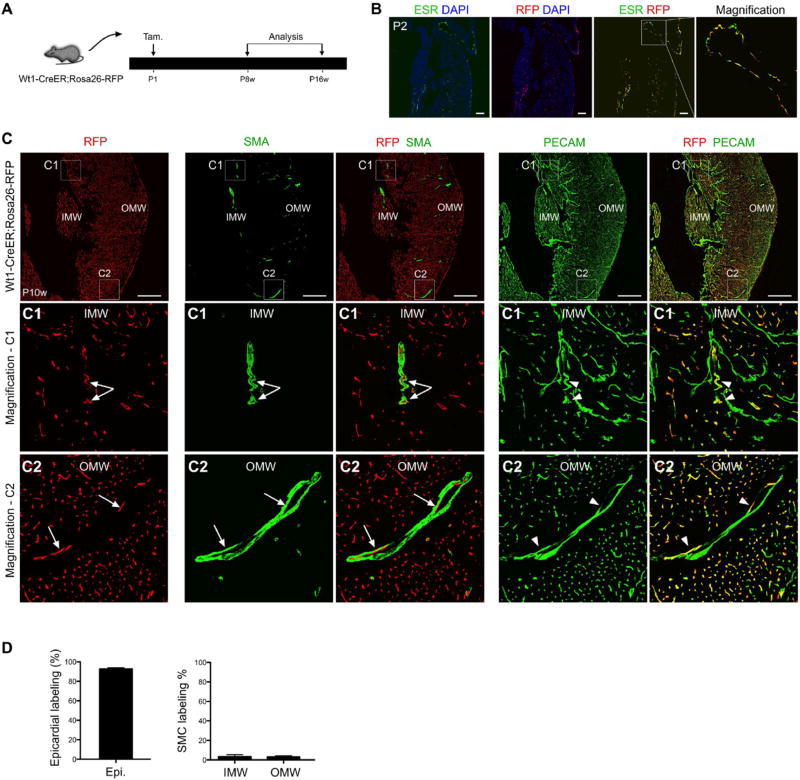
Fig. 1.
Epicardial cells labeled at P1 rarely contribute to SMCs of the 2nd CVP. (A) Strategy showing tamoxifen induction (P1) and analysis of adult Wt1-CreER; Rosa26-RFP mice. (B) Immunostaining of ESR (a surrogate for Wt1), the lineage marker RFP and DAPI on sections of P2 Wt1-CreER; Rosa26-RFP hearts following administration of tamoxifen at P1. Scale bars, 100 µm. (C) Immunostaining for RFP, PECAM, SMA and DAPI on sections of P10w Wt1-CreER; Rosa26-RFP hearts. Wt1-CreER labels coronary endothelial cells (RFP+; PECAM+, arrowheads), but very few vascular SMCs (SMA+; RFP−, arrows) in the IMW or OMW. Scale bars, 0.5 mm. LV, left ventricle; IMW, inner myocardial wall; OMW, outer myocardial wall. Representative figure of 4 individual samples. (D) Quantification of the percentage of epicardial cell labeling or SMCs labeled by Wt1-CreER (RFP). Compared to the robust epicardial cell labeling, few SMCs in the IMW and OMW were labeled in adult hearts following tamoxifen administration at P1. n = 4.
To determine the contribution of the neonatal epicardium within the neonatal OMW to SMCs of the 2nd CVP, we collected P8w-P16w Wt1-CreER; Rosa26-RFP mouse hearts after tamoxifen treatment at P1 (Fig. 1A). While this regimen induced significant epicardial labeling (Fig. 1B), epicardium-derived non-endothelial cells (RFP+;PECAM−) were sparsely detected in either the IMW or OMW (<5%). Similarly, Wt1-CreER labeled SMCs were rarely observed in the IMW and OMW. In the myocardium, the Wt1- lineage (RFP+) was almost exclusively confined to coronary vascular endothelial cells (Fig. 1C). We also used smooth muscle 22 alpha (SM22) and smooth muscle myosin heavy chain (SM-MHC) to verify that labeled postnatal epicardial cells do not contribute to smooth muscle cells (Fig. S2). These data suggested that while the 2nd CVP matures and recruits new SMCs postnatally, postnatal epicardial cells do not contribute significantly to these new SMCs.
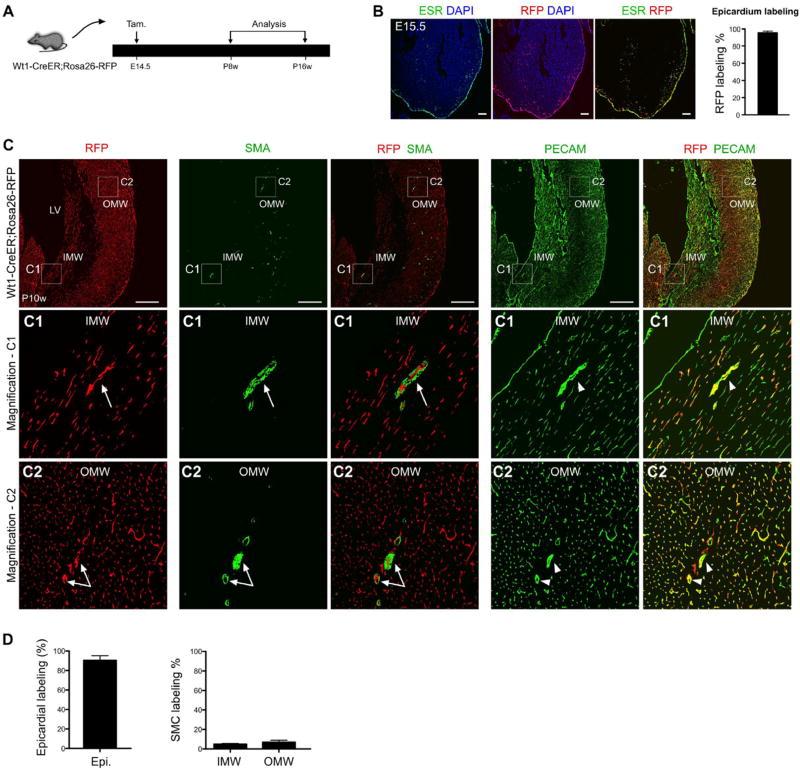
We reasoned that the initiation of 2nd CVP formation or trabecular compaction might trigger a temporal or developmental signal that induces epicardial migration prior to the formation of the neonatal 2nd CVP [9]. Therefore, we induced epicardial labeling at E14.5 before the 2nd CVP forms in the IMW. Induction of Wt1-CreER activity at E14.5 efficiently labels (95.56 ± 2.03%) epicardial cells 24 h later (E15.5, Fig. 2A-B). We observed a subset of coronary endothelial cells that were also RFP+, but this was likely due to the ectopic expression of WT1 in endothelial cells at E15.5 [18–20]. We also collected hearts later at P8w–P16w and found that E14.5 tamoxifen induction failed to efficiently label SMCs in either the IMW or OMW (<5%), yet the epicardium was almost completely RFP+ (Fig. 2C). Furthermore, epicardial cells labeled at late embryonic stage do not contribute to SM22+ or SM-MHC+ cells in the adult heart (Fig. S3). Collectively, these data suggested that the late embryonic stage epicardium does not contribute to SMCs of the postnatal 2nd CVP.
Fig. 2.
Epicardial cells labeled at E14.5 rarely contribute to SMCs of the 2nd CVP. (A) The experimental strategy for pulse-and-chase lineage tracing studies, with tamoxifen induction (the pulse) at E14.5 and analysis (the chase) from postnatal 8 weeks (P8w) to P16w in Wt1-CreER; Rosa26-RFP mice. (B) The active expression domain of WT1, as determined by ESR (estrogen receptor), and the Wt1-lineage, as determined by expression of the RFP reporter, in sections of E15.5 Wt1-CreER; Rosa26-RFP embryonic hearts. Nuclei are stained by DAPI. Tamoxifen was injected at E14.5. Labeling efficiency was calculated as the percentage of RFP + ESR + epicardial cells in total number of ESR + epicardial cells. n = 4. Scale bars, 100 µm. (C) Immunostaining of RFP, SMA, PECAM and DAPI on sections of Wt1-CreER; Rosa26-RFP hearts. Wt1-CreER labels coronary endothelial cells (RFP+; PECAM+, arrow-heads) but very few vascular SMC (RFP+; SMA+, arrows) in the IMW or OMW. Scale bars, 0.5 mm. LV, left ventricle; IMW, inner myocardial wall; OMW, outer myocardial wall. Representative figure of 4 individual samples. (D) Quantification of the percentage of epicardial cell labeling or smooth muscle cell (SMC) labeling by Wt1-CreER mediated lineage tracing (RFP+). Compared with robust epicardial cell labeling, few SMCs were labeled in the IMW and OMW. n = 4.
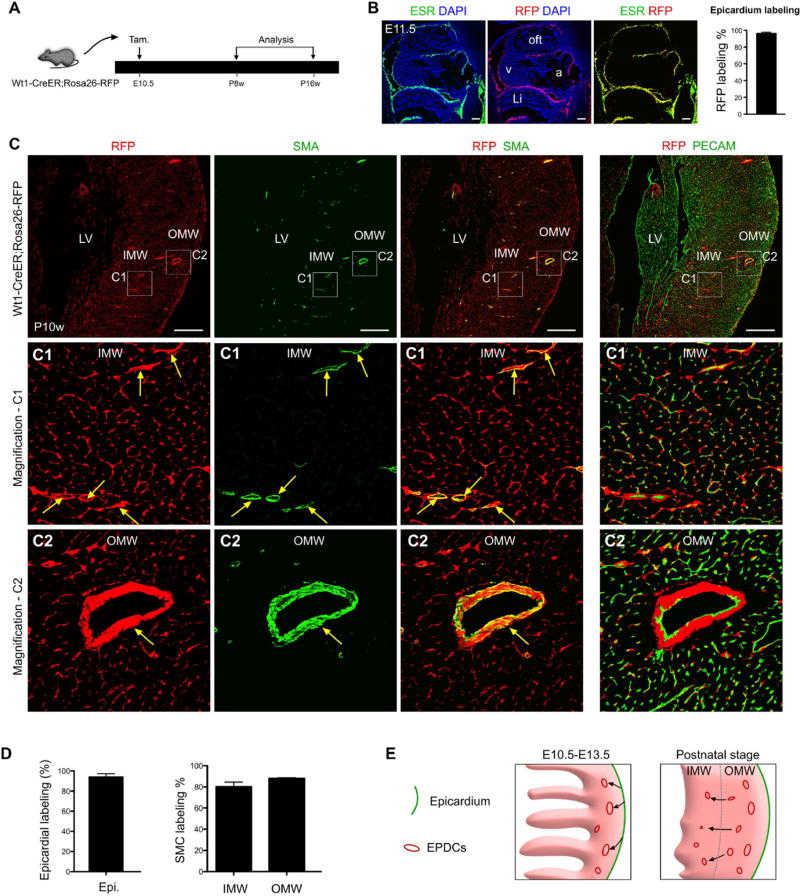
Finally, we traced the contribution of the early epicardium, just as it is forming, to the postnatal 2nd CVP SMCs. At E9.5, the proepicardium migrates onto the surface of the heart to form a single epicardial cell layer [11, 21]. We induced tamoxifen at E10.5 to label this newly formed epicardium (96.11% ± 1.67% labeling), before these cells migrate into the myocardium to form epicardium-derived cells (EPDCs, Fig. 3A-B). Using this strategy, we observed EPDCs have migrated into the compact myocardium at late embryonic stage (E17.5, Fig. S4), without any confounding contribution to the endothelial cells. A subset of these early EPDCs form coronary smooth muscle cells or pericytes that remain close to endothelial cells of 1st CVP, but few adopted an endothelial cell fate (Fig. S4). At this later embryonic stage, these EDPCs remained largely in the compact myocardium (OMW). We also noticed that a few migrated into the trabecular myocardium (IMW) (Fig. S4), which is in consistent with data from chicken model in which a few EPDCs could home to the inner myocardial trabecular layer [22]. When we examined the fate of these early epicardial cells in the adult stage, EPDCs were detected in the IMW and contribute to smooth muscle of 2nd CVP (Fig. 3C), suggesting EPDCs from OMW invade into IMW and contribute to SMCs (Fig. 3E). By immunostaining of SM22 or SM-MHC, we confirmed that these EPDCs contribute to smooth muscle cells of 2nd CVP in adult heart (Fig. S5). Quantification determined that more than 80% of SMCs within the 2nd CVP in the IMW were derived from these EDPCs (RFP+) labeled at E10.5 (Fig. 3D).
Fig. 3.
Early embryonic epicardial cells contribute to SMCs of the 2nd CVP. (A) A schematic figure depicting the experimental time-course shows tamoxifen induction (at E10.5) and endpoint analysis (at postnatal 8 weeks, P8w to P16w) of Wt1-CreER; Rosa26-RFP mice. (B) Following tamoxifen administration at E10.5, Wt1-CreER; Rosa26-RFP embryos were harvested at E11.5. Active WT1 expression was assessed by ESR immunostaining, and epicardial cell genetic labeling was determined by RFP. Nuclei were stained with DAPI. Labeling efficiency was calculated as the percentage of RFP + ESR + epicardial cells divided by the total number of ESR + epicardial cells. Scale bars, 100 µm. Li, liver; a, atrium; v, ventricle; oft, outflow tract. (C) Immunostaining of RFP, SMA, PECAM and DAPI on P10w Wt1-CreER; Rosa26-RFP heart sections. Smooth muscle cells (SMCs) of coronary arteries in both the inner (C1) and outer myocardium wall (C2) were derived from Wt1+ epicardial cells labeled at E10.5 (arrows). Scale bars, 0.5 mm. LV, left ventricle; IMW, inner myocardial wall; OMW, outer myocardial wall. Representative figure of 4 individual samples. (D) Quantification of panels in “C” shows the contribution of the Wt1-CreER lineage to the epicardium and SMCs. n = 4. (E) A hypothetical model illustrating epicardial cell (green) migration (black arrows) and establishment of EPDCs (red) in the compact myocardium at E10.5-E13.5 (left panel). Postnatally, a subset of these resident EPDCs (red) migrate (black arrows) from the OMW into the IMW to form the SMCs of the 2nd CVP.
To study when these EDPCs begin to form SMCs of 2nd CVP, we examined P1, P4, P7 Wt1-CreER; Rosa26-RFP hearts that receive tamoxifen at E10.5. We did not detect any smooth muscle cells in the inner myocardial wall at P1 (Fig. S6A). At P4, we could detect weak expression of SMA, suggesting the differentiation of EDPCs into smooth muscle cells start at P4 (Fig. S6B). At P7, SMA+ cells could also be detected in the EPDCs in the inner myocardial wall (Fig. S6C). These data demonstrated that smooth muscle cells in the inner myocardial wall were differentiated from EPDCs in the neonatal stage.
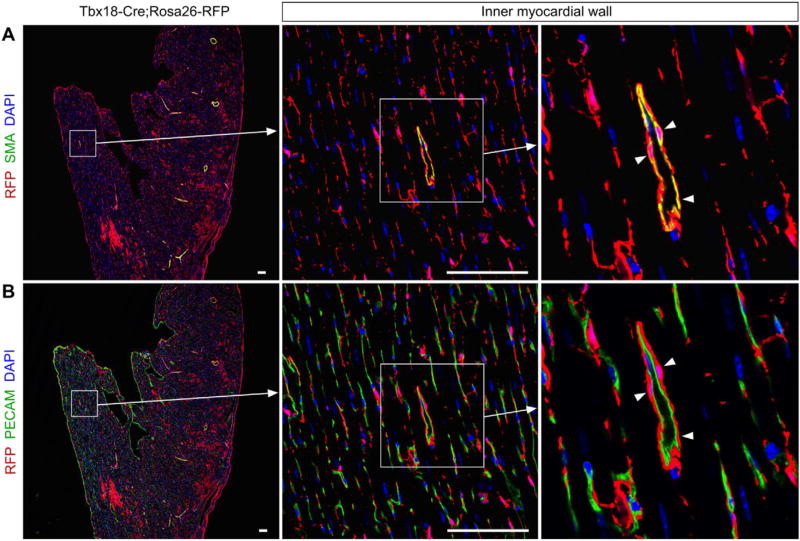
Since proepicardial or epicardial cells are heterogenous population that may have different differentiation potential in developing hearts [23], we therefore used another epicardial marker TBX18 to study if Tbx18+ epicardial cells contribute to the SMCs of 2nd CVP. We crossed Tbx18-Cre [13] with Rosa26-RFP [12] and perform lineage tracing of epicardial cells. In P14 weeks' adult Tbx18-Cre; Rosa26-RFP hearts, Tbx18-derived EPDCs contribute to the most of coronary SMCs in the inner myocardial wall (Fig. 4), confirming that epicardium is the major source for the SMCs of 2nd CVP.
Fig. 4.
Tbx18 + epicardial cells contribute to smooth muscle cells of 2nd CVP. (A, B) Immunostatining for RFP, PECAM and SMA on Tbx18-Cre; Rosa26-RFP heart sections shows Tbx18 + epicardial cells contribute to smooth muscle cells but not endothelial cells of 2nd CVP (arrowheads). Arrows indicate the magnified images of boxed regions. Scale bars, 100 µm.
Previous work showed that PDGFRB is required for EPDCs differentiation into coronary SMCs, as knockout of PDGFRB leads to complete blockade of coronary SMCs formation [24]. We therefore tested if PDGFRB is expressed in EDPCs in the inner myocardial wall before they differentiate into SMCs. Immunostaining of PDGFRB and RFP on P1 Wt1-CreER; Rosa26-RFP heart sections showed that a subset of EPDCs in the inner myocardial wall express PDGFRB before they start to differentiate into mature SMCs at P4 (Fig. S7). Similar to the SMCs in embryonic compaction myocardium [24], PDGFRB signaling may also regulate the differentiation of EDPCs to form SMCs of 2nd CVP in postnatal heart.
4. Discussion
The developmental origin of different components of the coronary arteries remains an unsolved question in the cardiovascular developmental research. The recent identification of the 2nd CVP suggests a new program of coronary artery formation during development [9, 25]. A more detailed understanding of the proper formation and maturation of the 2nd CVP may suggest new mechanistic insights to some forms of congenital heart diseases, such as noncompaction of the ventricular myocardium. While the origin of endothelial component of the 2nd CVP is ventricular endocardium, the cells that contribute to the smooth muscle of the 2nd CVP remains undetermined. Herein, our lineage tracing study reveals that the developmental origin of SMCs of the 2nd CVP in the postnatal heart is derived from EPDCs during the earliest wave of epicardial migration.
The migratory potential of epicardial cells during early embryonic stages of development is well documented [21, 22, 26]. The migration of these epicardial cells and their derivatives EDPCs at proper timing is essential for normal coronary vessel development [27]. EPDCs are known to migrate from the epicardium at E10.5–E13.5, and ultimately reside in the OMW to form smooth muscle cells and pericytes of the 1st CVP during embryogenesis [21, 28, 29]. Later, during postnatal development, these EPDCs residing within the OMW populate the IMW and contribute to the new SMCs in the IMW, where the 2nd CVP locates (Fig. 4E). Epicardial cells gradually lose their migratory potential at later embryonic stages (after E14.5). The temporal window between E10.5 and E14.5 is coincident with the migration of subepicardial coronary vascular endothelial cells into the developing myocardial wall to form the intramyocardial coronary vessels [1, 9, 30]. It remains unknown whether subepicardial endothelial cells and EPDCs share common guidance cues, or is governed by distinct signaling pathways during 1st and 2nd CVP formation. In addition, it is likely that SMCs of the 2nd CVP is derived from the de-differentiation and re-differentiation of the 1st CVP SMCs, which merits further investigation in future studies. In the context of injury to the adult heart, such as myocardial infarction, epicardial cells increase their proliferation and the epicardial layer thickens, but interestingly these cells do not migrate into myocardium [31, 32]. Recent reports showed that administration of thymosin β4 and a modified mRNA encoding VEGF-A could reactivate the epicardial migration following injury [33, 34]. Moreover, heterogeneity in the fetal epicardium is linked to coronary artery integrity, and distortion of the coronaries epicardial origin predisposes to adult onset disease [35]. It is also intriguing to investigate what molecular mechanisms regulate the differential ability of the epicardium to migrate and undergo EMT at early versus late embryonic stage and neonatal stages of development. The mobilization of EPDCs residing in the OMW to enter the IMW and their contribution to the smooth muscle of the 2nd CVP may also involve signals that regulate myocardial compaction and neovascularization.
Supplementary Material
Acknowledgments
We thank Hongkui Zeng and Qiang Shen for providing mice lines.
Sources of funding
This work was supported by National Natural Science Foundation of China (91339104, 31271552, 31222038, 31301188), Ministry of Science and Technology (2012CB945102 and 2013CB945302), Shanghai Basic Research Key Project (14JC1407400) and Zhangjiang Stem Cell Project (ZJ2014-ZD-002), SIBS Postdoc Fund (2013KIP311, 2014KIP314), SIBS President Fund, China Postdoc Fund 2013M541561, Shanghai Yangfan Project, and Scientist Development Grant from the American Heart Association (12SDG12060353) to JDW.
Footnotes
Disclosures
None.
Conflict of interest
The author declare there are no financial conflicts of interest to report.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2016.02.062.
References
- 1.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez R, Markwald R, O'Rourke B, Sharp D, Zheng D, Lenz J, Baldwin HS, Chang C-P, Zhou B. endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley P. Developmental biology: plumbing the heart. Nature. 2010;464:498–499. doi: 10.1038/464498a. [DOI] [PubMed] [Google Scholar]
- 4.del Monte G, Harvey R. An endothelial contribution to coronary vessels. Cell. 2012;151:932–934. doi: 10.1016/j.cell.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Pu WT. Notching up vascular regeneration. Cell Res. 2014;24(7):777–778. doi: 10.1038/cr.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian X, Pu WT, Zhou B. Cellular origin and developmental program of coronary angiogenesis. Circ. Res. 2015;116(3):515–530. doi: 10.1161/CIRCRESAHA.116.305097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF, Lien CL. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat. Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 9.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Yan Y, Yang X, Zhong TP, Pu WT, Zhou B. De novo formation of a distinct coronary vascular population in neonatal heart. Science. 2014;345:90–94. doi: 10.1126/science.1251487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns CG, Burns CE. A crowning achievement for deciphering coronary origins. Science. 2014;345:28–29. doi: 10.1126/science.1256866. [DOI] [PubMed] [Google Scholar]
- 11.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Zhang Z, Zhong TP, Yang X, Yang Z, Yan Y, Baldini A, Sun Y, Lu J, Schwartz RJ, Evans SM, Gittenberger-de Groot AC, Red-Horse K, Zhou B. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013;23:1075–1090. doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B, von Gise A, Ma Q, Hu YW, Pu WT. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev. Biol. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Huang X, Zhang H, Tian X, He L, Yang R, Yan Y, Wang QD, Gillich A, Zhou B. c-kit(+) cells adopt vascular endothelial but not epithelial cell fates during lung maintenance and repair. Nat. Med. 2015;21:866–868. doi: 10.1038/nm.3888. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Hu T, He L, Huang X, Tian X, Zhang H, He L, Pu W, Zhang L, Sun H, Fang J, Yu Y, Duan S, Hu C, Hui L, Zhang H, Quertermous T, Xu Q, Red-Horse K, Wythe JD, Zhou B. Genetic targeting of sprouting angiogenesis using Apln-CreER. Nat. Commun. 2015;6:6020. doi: 10.1038/ncomms7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duim SN, Kurakula K, Goumans MJ, Kruithof BP. Cardiac endothelial cells express Wilms' tumor-1: Wt1 expression in the developing, adult and infarcted heart. J. Mol. Cell. Cardiol. 2015;81:127–135. doi: 10.1016/j.yjmcc.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ. Res. 2012;111(2):165–169. doi: 10.1161/CIRCRESAHA.112.273946. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Pu WT. Genetic Cre-loxP assessment of epicardial cell fate using Wt1-driven Cre alleles. Circ. Res. 2012;111:e276–e280. doi: 10.1161/CIRCRESAHA.112.275784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 22.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 23.Katz T, Singh M, Degenhardt K, Rivera-Feliciano J, Johnson R, Epstein J, Tabin C. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ. Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, Tian X, Zhang H, Wythe JD, Zhou B. Fabp4-CreER lineage tracing reveals two distinctive coronary vascular populations. J. Cell. Mol. Med. 2014;18:2152–2156. doi: 10.1111/jcmm.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter EM, Gittenberger-de Groot AC. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell. Mol. Life Sci. 2007;64:692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eralp I, Lie-Venema H, DeRuiter MC, van den Akker NM, Bogers AJ, Mentink MM, Poelmann RE, Gittenberger-de Groot AC. Coronary artery and orifice development is associated with proper timing of epicardial outgrowth and correlated Fas-ligand-associated apoptosis patterns. Circ. Res. 2005;96:526–534. doi: 10.1161/01.RES.0000158965.34647.4e. [DOI] [PubMed] [Google Scholar]
- 28.Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- 29.Dettman RW, Denetclaw WJ, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 30.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Investig. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B, Honor LB, Ma Q, Oh JH, Lin RZ, Melero-Martin JM, von Gise A, Zhou P, Hu T, He L, Wu KH, Zhang H, Zhang Y, Pu WT. Thymosin beta 4 treatment after myocardial infarction does not reprogram epicardial cells into cardiomyocytes. J. Mol. Cell. Cardiol. 2011;52:43–47. doi: 10.1016/j.yjmcc.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31(10):898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei K, Diaz-Trelles R, Liu Q, Diez-Cunado M, Scimia MC, Cai W, Sawada J, Komatsu M, Boyle JJ, Zhou B, Ruiz-Lozano P, Mercola M. Developmental origin of age-related coronary artery disease. Cardiovasc Res. 2015;162:829–838. doi: 10.1093/cvr/cvv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.