Abstract
Recent identification and isolation of suture stem cells capable of long term self-renewal, clonal expanding and differentiating demonstrate their essential role in calvarial bone development, homeostasis and injury repair. These bona fide stem cells express high level of Axin2 and are able to mediate bone regeneration and repair in a cell autonomous fashion. The importance of Axin2 is further demonstrated by its genetic inactivation in mice causing skeletal deformities resembling craniosynostosis in humans. The fate determination and subsequent differentiation of Axin2+ stem cells are highly orchestrated by a variety of evolutionary conserved signaling pathways including Wnt, FGF and BMP. These signals are often antagonistic of each other and possess differential effects on osteogenic and chondrogenic cell types. However, the mechanisms underlying the interplay of these signaling transductions remain largely elusive. Here we identify Rap1b acting downstream of Axin2 as a signaling interrogator for FGF and BMP. Genetic analysis reveals that Rap1b is essential for development of craniofacial and body skeletons. Axin2 regulates Rap1b through modulation of canonical BMP signaling. The BMP-mediated activation of Rap1b promotes chondrogenic fate and chondrogenesis. Furthermore, by inhibiting MAPK signaling, Rap1b mediates the antagonizing effect of BMP on FGF to repress osteoblast differentiation. Disruption of Rap1b in mice not only enhances osteoblast differentiation but also impairs chondrocyte differentiation during intramembranous and endochondral ossifications, respectively, leading to severe defects in craniofacial and body skeletons. Our findings reveal a dual role of Rap1b in development of the skeletogenic cell types. Rap1b is critical for balancing the signaling effects of BMP and FGF during skeletal development and disease.
Keywords: Craniofacial development, Skeleton, Osteoblast, Chondrocyte, Suture, Skull, Wnt, Bone Morphogenetic Protein, Fibroblast Growth Factor
Introduction
Skeletogenic mesenchyme mediates formation of the craniofacial and body skeletons through two distinct ossification mechanisms (1,2). In the cranium, the skeletogenic mesenchyme, derived from both mesoderm and neural crest, mainly undergoes intramembranous ossification where mesenchymal stem cells directly differentiate into the bone forming cells, osteoblasts (3,4). This bone formation mechanism is also responsible for development of the jaw and collarbones. On the other hand, axial and appendicular skeletons are predominately formed via endochondral ossification for which prior formation of cartilage templates is required. This process requires coordination of two major cell types – osteoblast and chondrocyte – both derived from the skeletogenic mesenchyme. During skeletal development, ossification abnormalities may arise resulting in congenital birth defects. Approximately 5% of children with congenital birth defects have skeletal dysplasia (5). More than 450 different types of skeletal dysplasia have been classified by molecular, biochemical and radiographic criteria (6). Craniosynostosis (7–9), affecting approximately 1 in 2,500, is one of the most common craniofacial deformities (4,10,11). Patients with craniosynostosis develop abnormal skull shape due to excessive intramembranous ossification, leading to aberrant fusion of the cranial suture which serves as a growth center for craniofacial skeletogenesis (4).
The calvarium has provided a convenient and contrasting model for studying the development of multiple mesenchymal lineages (12,13). Although temporal chondrogenesis occurs in craniofacial sutures, most commonly in the sagittal and mid-palatal sutures, it disappears soon after birth (14). Therefore, developmental programming prohibits chondrogenic differentiation of the suture skeletogenic mesenchyme. Axin2, a negative regulator of the canonical Wnt pathway, has previously been shown to regulate skeletogenesis through modulation of β-catenin signaling (15–17). Interplay of Wnt and FGF signaling is critical for fate determination of the mesenchymal stem cell (13). Wnt directly controls expansion of the skeletogenic precursors while indirectly modulating FGF signaling which favors the osteogenic fate. Aberrant FGF signaling promotes osteoblast differentiation by disturbing the balance of these morphogenetic pathways (13,18). In contrast, low levels of FGF signaling alter developmental characteristics of the mesenchymal stem cell to favor the chondrogenic fate, most likely via a de-repressing mechanism (13). BMP signaling is highly elevated implying its crucial involvement in this signaling crosstalk. However, the essential role of BMP signaling and its antagonist effects on FGF signaling in mesenchymal cell fate determination and switching remain to be determined. The mechanism by which the interplay of Wnt, FGF and BMP signaling orchestrates development of the skeletogenic mesenchyme is also largely elusive.
Recent evidence indicates that Axin2-expressing cells contain suture stem cells (SuSCs), which exhibit long-term self-renewing, clonal expanding and differentiating abilities during calvarial development, homeostastic maintenance and injury repair (19). These naïve cells, residing in the suture midline, contribute directly to bone regeneration in a cell autonomous fashion (19). The new findings also demonstrate the true identity of Axin2-expressing cell as bona fide stem cells with innate capacities to replace the damaged skeleton in cell-based therapy (19). Understanding the property and behavior of this stem cell population thus permits further elucidation of the stem cell-mediated craniofacial skeletogenesis, leading to deciphering the complexity of congenital deformities and regenerative medicine.
We report identification of Rap1b as a signaling effector of Axin2 in skeletal development using proteomic analysis. Initially identified as an antagonist of Ras-induced transformation, Rap1 belongs to the Ras-like family of small GTPases with similar effector-binding domains (20). Two highly homologous and evolutionary conserved genes, Rap1a and Rap1b, are found in the mammalian genome. It has been shown that they possess specific as well as redundant functions (21–23). Biochemically, Rap1 acts as a molecular switch by cycling inactive GDP-bound and active GTP-bound forms (24). The transition between these forms is tightly controlled by specific guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Our study reveals that Rap1b is a downstream effector of Axin2, regulating the signaling crosstalk of BMP and FGF in development of skeletogenic cell types. Genetic inactivation of Rap1b in mice causes excessive intramembranous ossification and has negative effects on endochondral ossifications, leading to defects in development of craniofacial and body skeletons, respectively. Rap1b plays a dual role in cell differentiation and developmental programming of the skeletogenic mesenchyme during skeletal development.
Materials and Methods
Animals
The Axin2−/−, FGFR1+/−, Axin2−/−; FGFR1+/−, sβcatAx2, Axin2-rtTA, TRE-Cre, BMPR1ACA/+ and Rap1b−/− mouse strains and genotyping methods were reported previously (13,15,17,19,21,25–28). To generate caBMPR1AAx2 mouse strain, Axin2Cre-Dox males carrying both Axin2-rtTA and TRE-Cre transgenes were first created. They were then crossed with BMPR1ACA/+ mice, permitting the Cre-mediated expression of caBMPR1A, to obtain Axin2Cre-Dox; BMPR1ACA/+ mice. In these mice, Cre induction in the Axin2-expressing cells was controlled by Dox treatment (2 mg/ml plus 50 mg/ml sucrose) for 7 days (13,15,17,25,26) to express a constitutive active form of BMPR1A, thus establishing the caBMPR1AAx2 mouse model. Both male and female mice were used in this study. Mice are maintained in group housing under SPF-free condition and Laboratory Autoclavable Rodent Diet 5010 (LabDiet, St. Louis, MO, cat #0001326). Care and use of experimental animals described in this work comply with guidelines and policies of the University Committee on Animal Resources at the University of Rochester.
Cells and Molecules
Isolation of primary mesenchymal cells from the calvaria was performed basically as described (13,19,29). Both male and female mice were used for cell isolation and subsequent analyses. Specifically, an approximately 1.5 mm width tissue containing sagittal suture and its adjacent parietal bones were harvested. The parietal bones were then separated from the suture to expose the suture mesenchyme, which was subsequently incubated with 0.2% collagenase (Roche, Indianapolis, IN) in PBS at 37°C for 1.5 hours. The dissociated cells were then filtered through a 40 μm strainer, followed by re-suspension in DMEM media for further analyses. For micromass culture, 105 primary cells isolated from E18.5 ribcages were seeded as a 10 μl droplet in a 24 well plate. After one hour of pre-incubation, they were cultured in differentiation media containing 50 μg/ml ascorbic acid (Sigma-Aldrich, St. Louis, MO), and 4 mM β-glycerophosphate (Sigma-Aldrich, St. Louis, MO) with or without GGTI298 (15 μM, Sigma-Aldrich, cat# G5169). C3H10T1/2 cells (ATCC, cat# CCL-226) were cultured in αMEM media containing 10% fetal calf serum. The siRNA from the SMART pool were used for Rap1b knockdown (GE Healthcare, cat #M-062638-01). The addition of U0126 (10 μM, EMD Millipore, Billerica, MA) was used to inhibit Erk1/2. To manipulate gene activity, DNA plasmids, expressing Wnt1, a degradation-deficient β-catenin (ΔNβ-catenin), a fusion chimera of β-catenin and LEF1 (catCLEF), TGFβ receptor II (TβRII), constitutive active BMP receptor 1A (caBMPR1A), myc tagged Axin1, myc tagged Axin2, myc tagged Rap1b and siRNA against Rap1b, were transfected into the cultured cells by lipofectamine (Thermo Fisher Scientific, Grand Island, NY).
Staining, Immunostaining, Immunoblot and in situ hybridization
Skeletal preparation, fixation and embedding for paraffin and frozen sections were performed as described (13,15,17,25,28–31). Samples were subject to hematoxylin/eosin staining for histology, alcian blue staining, von Kossa staining and immunological staining with avidin:biotinlylated enzyme complex as described (13,15,25,28,29,31–37). To quantify the intensify of alcian blue staining, cells were fixed in solution containing 30% ethanol, 0.4% paraformaldehyde and 4% acetic acid for 15 min at room temperature, followed by incubation with 0.05% alcian blue staining solution in 75% ethanol: 0.1M hydrochloride (4:1) overnight at 37°C. The immunological staining was visualized by enzymatic color reaction according to the manufacture’s specification (Vector Laboratories, Burlingame, CA). Immunoblot was performed by isolation of protein extracts from cells or tissues using M-PER (Pierce) in the presence of protease inhibitor cocktail, followed by electrophoresis as described (32,35–39). For expression of the exogenous protein, 1 μg of DNA plasmids were transfected into cultured cells. Cell lysates were collected after 48 hours for immunoblot and subsequent assays as indicated. To measure the GTP bound form of Rap1b, samples were lysed in buffer, containing 50 mM Tris-HCl at pH 7.5, 120 mM NaCl, 1% TritonX-100, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 mM PMSF. Clarified lysates were incubated with 50 μg of GST fusion protein containing the Rap1-binding domain of RalGDS-RBD (GST-RalGDS-RBD). Protein complexes were then precipitated using glutathione-sepharose beads, followed by SDS-PAGE and immunoblot analyses. Rabbit monoclonal antibodies phosphorylated c-Raf (Cell Signaling Technology), phosphorylated Erk1/2 (Cell Signaling Technology), Rap1b (Cell Signaling Technology, Danvers, MA) and polyclonal antibodies FGFR1 (Santa Cruz Biotechnology, Dallas, TX), phosphorylated p38 (Cell Signaling Technology), phosphorylated SAPK/JNK (Cell Signaling Technology), phosphorylated Smad1/5/8 (Cell Signaling Technology), PECAM-1 (Santa Cruz, Santa Cruz, CA, USA), Sox9 (Santa Cruz); mouse monoclonal antibodies Actin (Thermo Fischer Scientific), Rap1 (BD Biosciences, San Jose, CA), Ras (BD Biosciences), type 2 collagen (Thermo Fischer Scientific) were used in these analyses. The in situ hybridization was performed as described (28,30,32,33,36,38,40). In brief, DNA plasmids, containing MMP13, Col1a1 and Col10a1 cDNAs, were linearized for in vitro transcription using T3 or T7 RNA polymerase (Promega, Wisconsin, WI, USA) to generate digoxigenin-labeled RNA probes for in situ hybridization (41–43). Samples were then incubated with the RNA probes, followed by recognition with an alkaline phosphatase conjugated anti-digoxigenin antibody (Roche, Indianapolis, IN, USA). To visualize the bound signals, samples were incubated with BM-purple (Roche) for 4–5 hours. Images were taken using Nikon TS100-F (Nikon, Melville, NY) and Zeiss Axio Observer microscope (Carl Zeiss, Thornwood, NY).
Proteomics
For differential expression analysis, protein extracts isolated from the P3 calvarial bones of three control and mutant male and female mice were subject to two-dimensional (2-D) gel electrophoresis and silver staining analyses. The 2-D gel images of control were first analyzed by PDQuest software to build an archive for standard spot protein (SSP). An experimental condition was set to permit a clear identification of 500–600 SSPs. Next, the 2-D gel images of mutants were compared to the control to identify SSPs displaying a differential expression level between the two samples. In three independent experiments, the SSPs exhibiting differential expression levels were analyzed quantitatively using the PDQuest software and statistical analysis. The identity of the SSPs was then revealed by Mass Spectrometry.
Statistics
R software version 3.2.1 and Microsoft Excel 2010 were used for data analyses. The significance was determined by two-sided Student’s t-tests. Before performing the t-tests, normality of the data distribution was first validated by Shapiro–Wilk normality test. At least 3 independent experiments were performed for statistical analyses of the animal tissues described in figure legends. Immunostaining, in situ hybridization and histology were performed by researchers who are blinded to the animal genotypes. Nine (Figure 1A–G), three (Figure 1H–J, 2C–D, 2F–G, 2I–P, 4A–J, 6A–S, 7A–P) and two (Figure 3A–G) animals of the same genotype were studied for each experimental outcome. Statistical data were presented as mean ± SEM.
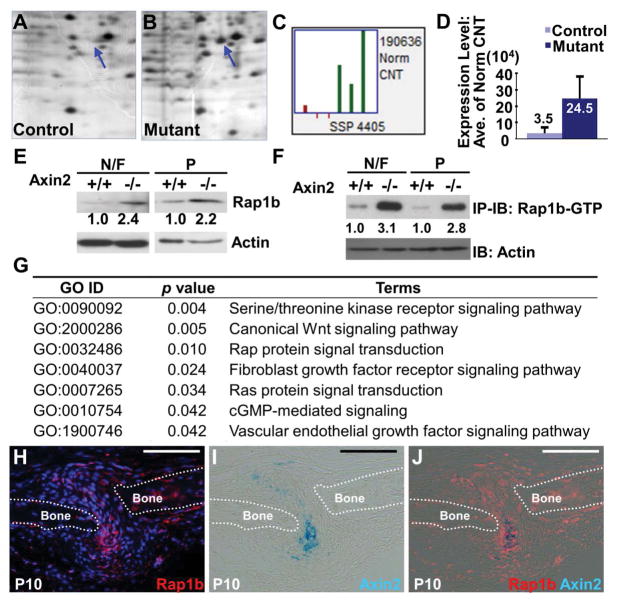
Figure 1.
Proteomic studies identify Rap1b as a potential effector of Axin2 in skeletal development and deformity. Two-dimensional gel electrophoresis analysis of control (A) and Axin2 knockout (B) reveals a spot (SSP 4405) whose protein expression is increased in the newborn nasal and frontal bones. (C) In three independent experiments, SSP 4405 is consistently elevated in the mutant. (D) A graph illustrates the average expression level of SSP 4405 increased 7 fold in the mutant. Mass spectrometry analysis further reveals SSP 4405 containing Rap1b whose altered expression in the mutants is confirmed by immunoblot (E) and immunoprecipitation-immunoblot (F) analyses. Both the total protein and activated form of Rap1b are elevated in the nasal/frontal (N/F) and parietal (P) bones of Axin2−/−, compared to the Axin2+/+. Direct immunoblot analysis of actin is used as protein loading control at 1:20 dilution. (G) Ontology analysis of LC-MS/MS data, consisting of ~2,000 identified proteins by whole proteome study of control and Axin2 knockout skulls, reveals signaling pathways significantly affected by the mutation at P3. Gene ontology ID (GO ID), p value and their terms (terms) of the altered pathways are as indicated in panel G. Sections of the P10 (H–J) calvaria were analyzed for cells expressing Rap1b and Axin2 by immunostaining and β-gal staining, respectively. Images are representatives of three independent experiments. Broken lines highlight the calvarial bone plates (Bone). Scale bars, 100 μm (H–J).
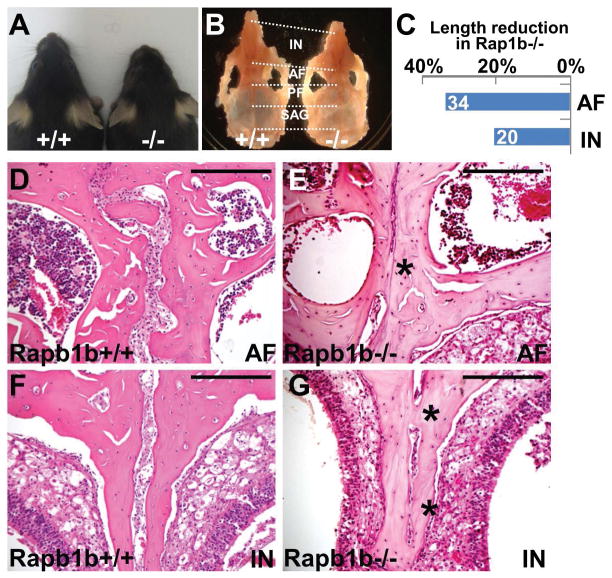
Figure 3.
The disruption of Rap1b in mice causes craniosynostosis. (A) Dorsal view of 2 month-old control (+/+) and Rap1b mutant (−/−) mice shows that genetic inactivation of Rap1b causes shortening of the skull. (B–C) Gross examination reveals the length of anterior frontal (AF) and inter nasal (IN) sutures affected by the Rap1b deletion at 2 months (mean, n=2). (D–G) Hematoxylin and eosin staining of the control and mutant sections evaluates synostosis phenotypes in the AF and IN sutures. Images are representatives of two independent experiments. Scale bars, 200 μm (D–G).
Results
Identification of Rap1b as a downstream target of Axin2 in skeletal development
To further elucidate the mechanism by which Axin2 regulates development of the skeletogenic mesenchyme, we initiated a proteome-wide screening approach to identify potential downstream effectors of Axin2. Protein extracts isolated from the control calvarial bones at postnatal day 3 (P3) were first analyzed by two-dimensional (2-D) gel electrophoresis (Figure 1A), followed by building an archive for standard spot protein (SSP) using PDQuest. An experimental condition was set to permit a clear identification of 500–600 SSPs. Next, the 2-D gel images of the Axin2 mutants were compared to the control to identify SSP displaying differential expression levels between the two samples (Figure 1B). The SSPs exhibiting differential expression levels were examined quantitatively using PDQuest in three independent experiments. Mass Spectrometry analysis was then used to reveal the identity of SSPs showing consistent alterations. Among them, Rap1b is contained in the SSP4405 (Figure 1C) whose expression is consistently elevated and shows an averaged 7-fold increase in the Axin2 mutants (Figure 1D).
Initially identified as antagonists of Ras-induced transformation, Rap1b and its closely related Rap1a belong to the Ras-like family of small GTPase with similar effector-binding domains (20). Rap1 was previously proposed to modulate the Ras-dependent activation of MAPK in a highly cell type-specific manner (44). This notion supported our hypothesis that Rap1b regulates FGF-FGFR signaling through modulation of MAPK, critical for craniofacial bone development and disease (45,46). Immunoblot analysis confirmed that the protein level of Rap1b is elevated in the P3 Axin2−/− calvarial bones (Figure 1E). GTPases are active in their GTP-bound form, and become inactive when GTP is hydrolyzed to GDP (47). The formation of active and inactive forms is promoted by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), respectively. We therefore examined if the active form of Rap1b is also enhanced in the Axin2 mutants. Using protein extracts isolated from the P3 control and Axin2−/− calvaria, Rap1b-GTP pull down assay shows that active Rap1b-GTP is indeed enhanced by the mutation (Figure 1F). The enhancement also appeared to be greater than the total Rap1b (Figure 1E, F).
Furthermore, Rap protein signal transduction is one of pathways significantly altered by the Axin2 deletion based on whole proteome analyses. In this unbiased assay, protein extracts of the control and Axin2−/− calvaria were subject to Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) analysis to identify approximately 2,000 proteins, followed by R package GOstats examination of gene sets differentially expressed in the mutants (48). In addition to several signal transduction pathways, e.g. Wnt, FGF, Ras, Serine/threonine kinase receptor and VEGF, known to modulate skeletogenesis, we found that Rap protein signal transduction and cGMP-mediated signaling are significantly affected by the loss of Axin2 (Figure 1G). Relatively little is known about the role of Rap signaling pathway in skeletal development and disease, providing a new research direction. In addition, Rap1b belongs to the small GTPase family that has been shown to antagonize Ras function. Therefore, we decided to carry out functional studies to further examine the importance of Rap1b in skeletogenesis. To test if Axin2 and Rap1b are simultaneously expressed in cells within the skeletogenic mesenchyme during calvarial bone formation, double labeling was performed (Figure 1H–J). Sections of the P10 calvaria with the expression of lacZ gene under control of the Axin2 promoter were stained by β-gal, followed by immunostaining of Rap1b. During calvarial bone development, Rap1b is widely expressed in the skeletogenic mesenchyme including the Axin2+ SuSCs (Figure 1H–J). These data support potential involvement of Rap1b in the Axin2-mediated signaling effects on suture morphogenesis and craniosynostosis.
Axin2 regulates Rap1b through modulation of BMP signaling
Axin2 is a well-known inhibitor for canonical Wnt signaling. Therefore, we tested if the Axin2-mediated regulation of Rap1b is dependent on transcriptional activity of β-catenin. In mesenchymal C3H10T1/2 cells, expression of Axin2 as well as Axin1 was able to reduce the level of Rap1b (Figure 2A). However, Wnt1, ΔNβ-catenin (a truncated mutant lacking phosphorylation sites for degradation) or catCLEF (a fusion protein of β-catenin and transcriptional activation domain of LEF) showed very little effects on Rap1b expression (Figure 2A). The expression level of transfected genes was examined by immunoblot analyses as well as activation of TOPFlash, a reporter for β-catenin and Lef/Tcf-dependent transcription (Figure 2A, B). These results implied that the Axin2-mediated regulation of Rap1b is independent of any transcriptional activation by β-catenin that may play a role in development of the skeletogenic mesenchyme during calvarial morphogenesis.
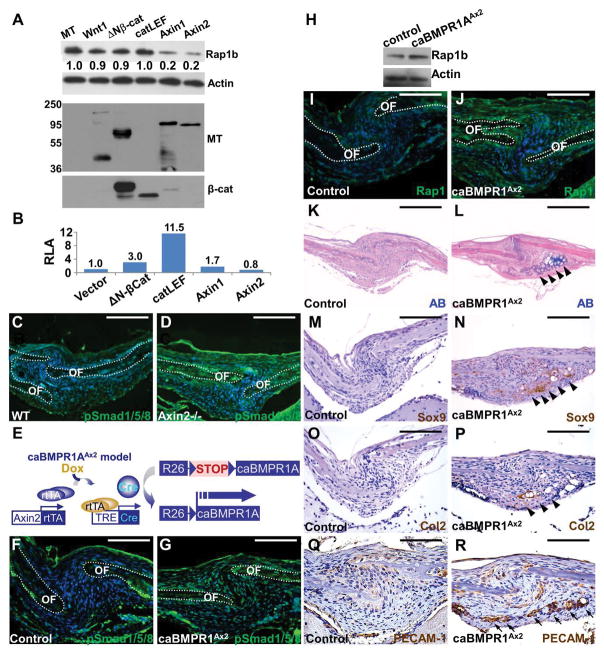
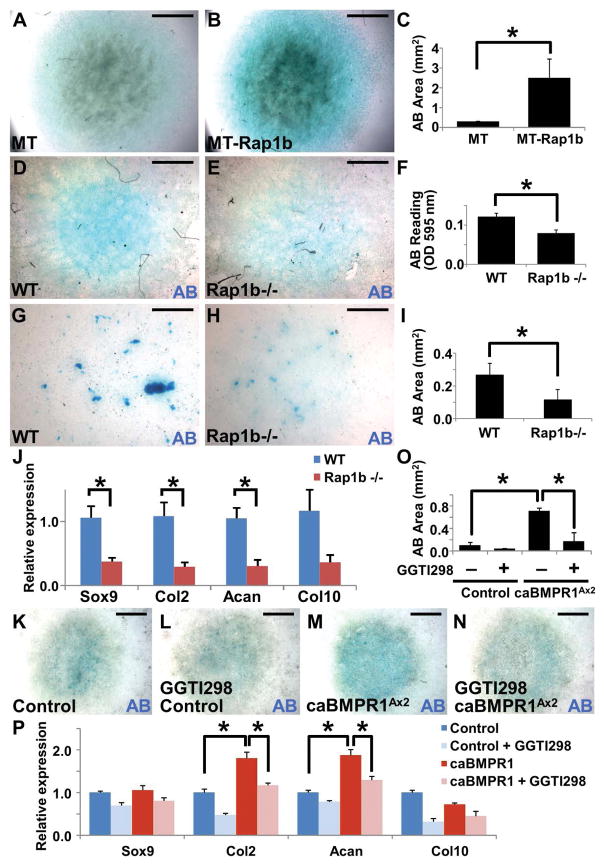
Figure 2.
Rap1b is regulated by Axin2-BMP signaling axis. (A–B) Modulation of Rap1b by Axin2 is independent of β-catenin-mediated transcription. (A) Immunoblot analysis examines the Rap1b level affected by overexpression of myc-tag (MT), Wnt1, ΔNβ-catenin, catCLEF, Axin1 or Axin2, in C3H10T1/2 cells. Actin level is used as a loading control. The expression of transfected genes is shown by immunoblotting of MT and β-catenin. (B) Relative luciferase activity (RLA) examines the β-catenin and Lef/Tcf-dependent transcription of a TOPFlash reporter affected by Wnt signaling effectors. (C–D) Immunostaining of phosphorylated Smad1/5/8 (pSmad1/5/8) shows enhanced canonical BMP signaling in the sagittal suture of Axin2−/−. (E) Diagrams illustrate the creation of caBMPR1AAx2 model where a constitutive active form of BMPR1A is expressed in the Axin2-expressing stem cells within the skeletogenic mesenchyme upon Dox treatment. (F–R) Stimulation of BMP signaling activates Rap1b and promotes chondrogenic fate in the suture mesenchyme at P3. Immunostaining (F–G, I–J) and immunoblotting (H) examines alterations of canonical BMP signaling (F–G) and Rap1b expression (H–J) in the caBMPR1AAx2 sagittal suture. OF, osteogenic front. Actin level is used as a loading control for immunoblot analysis. Alcian blue (AB) staining (K–L), and Sox9, type 2 collagen (Col2) and PECAM-1 immunostaining analyses (M–R) reveal evidence of ectopic chondrogenesis (arrowheads) and vascular invasion positive for endothelial cells (arrows) caused by constitutive activation of BMPR1A in the P3 sagittal suture. Sections are counterstained by DAPI (C–D, F–G, I–J), nuclear fast red (K–L) and hematoxylin (M–R). Images are representatives of three independent experiments. Scale bars, 100 μm (C–D, F–G, I–J, Q–R); 200 μm (K–P).
We previously discovered that the deletion of Axin2 switches the fate of mesenchymal cells to undergo chondrocyte instead of osteoblast differentiation, leading to transient chondrocytes present in the Axin2−/− sagittal suture (13). However, activation of Wnt/β-catenin signaling alone is not sufficient to alter lineage commitment of the suture mesenchyme (17). In the Axin2 nulls, we detected high levels of BMP signaling whose stimulation is also necessary for the SuSC fate switch (13,19). Therefore, we tested whether BMP signaling controls Rap1b because it is regulated by Axin2 via a β-catenin independent mechanism. First, immunostaining of phosphorylated Smad1/5/8 showed an elevation of canonical BMP signaling in the Axin2-null suture mesenchyme at P7, confirming the Axin2-BMP signaling axis (Figure 2C, D). Next, we tested if BMP signaling directly regulates Rap1b during calvarial bone development. We developed a new mouse model (caBMPR1AAx2) in which a constitutive active form of BMP type 1 receptor, caBMPR1A, can be conditionally expressed in the Axin2-expressing SuSCs upon doxycycline administration (Figure 2E). Consistent with the nature of this mutation designed to stimulate canonical BMP pathway, inducible expression of caBMPR1A enhanced phosphorylation of Smad1/5/8 effectors in the sagittal suture at P3 (Figure 2F, G). In these caBMPR1AAx2 calvaria, we detected higher levels of Rap1b using immunoblot and immunostaining analyses (Figure 2H–J; control, 50.1 ± 4.5% and caBMPR1AAx2, 77.6 ± 4.9%, mean ± SEM, p < 0.01, n=3, student’s t-test). Rap1b is stimulated in the suture mesenchyme, osteogenic front and periosteum of the mutant calvaria. BMP signaling may regulate mesenchymal cell fate determination through modulation of Rap1b.
Similar to the Axin2−/− mutants (19), we identified alcian blue staining, suggesting that cell fate switching and ectopic chondrogenesis occur, in the caBMPR1AAx2 suture mesenchyme (Figure 2K, L). Immunostaining analyses further revealed the alcian blued stained area also positive for chondrogenic markers, Sox9 and Col2 (Figure 2M–P). This was accompanied by vascular invasion, a process associated with cartilage resorption during endochondral ossification (Figure 2Q, R). These results demonstrate a causative effect of BMP-BMPR1A signaling on lineage commitment of the skeletogenic mesenchyme to favor chondrogenesis. They also indicate that activation of canonical BMP signaling in SuSCs converts their development from an osteogenic to chondrogenic fate. Therefore, Rap1b is regulated by Axin2-BMP signaling and aberrant stimulation of this regulatory axis is sufficient for mesenchymal cell fate switching. Rap1b is potentially a key regulator in suture morphogenesis orchestrated by the interplay of skeletogenic pathways.
The loss of Rap1b leads to excessive intramembranous ossification and craniosynostosis
To definitively assess the function of Rap1b in craniofacial skeletogenesis, we carried out genetic analysis in mice. The loss of Rap1b resulted in developmental abnormalities of the calvarial bones which are formed via intramembranous ossification (Figure 3). Although a majority of Rap1b homozygotes could not survive to terms due to internal bleeding caused by platelet defects, a small number of them (<5%) were viable (21). However, the survived mutant (<5%) were significantly smaller with abnormal head shape at 2 months (Figure 3A). Similar to the Axin2 nulls, anterior parts of the skull was most severely affected by the Rap1b deletion (Figure 3B). The length of the anterior frontal (AF) and inter-nasal (IN) sutures was significantly shorter than the control (Figure 3C). Histological evaluation revealed that the loss of Rap1b causes aberrant fusion of these sutures (Figure 3D–G).
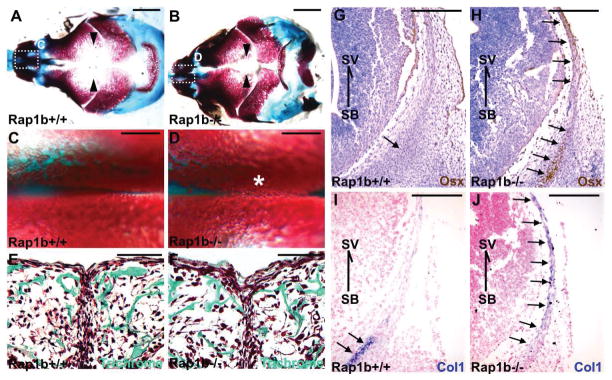
To further elucidate the mechanism underlying craniosynostosis caused by the Rap1b deletion, we examined calvarial bone formation. At E18.5, skeletal staining revealed the non-ossified areas especially those separating the frontal (Figure 4A, B) and nasal (Figure 4C–F) bones were narrower in the Rap1b−/− skulls. This phenotype was often associated with aberrant ossification, suggesting that osteoblast differentiation is enhanced by the Rap1b ablation. The formation of calvarial bone primordia initiates osteogenesis starting at about E12.5. The osteogenic process then extends apically from the skull base to midline of the skull vault within the skeletogenic mesenchyme, characterized by expression of the osteoblast makers, e.g. Osterix and type 1 collagen. At E13.5, the osteogenic process remains confined near the skull base in the control (Figure 4G, I) but has extended apically toward the skull vault in the mutant (Figure 4H, J). The results suggest that the loss of Rap1b induces an early onset of intramembranous ossification, leading to development of craniosynostosis.
Figure 4.
Intramembranous ossification and osteoblast differentiation are enhanced in the Rap1b mutant. Staining of the E18.5 craniofacial skeleton with alcian blue and alizarin red in whole mounts (A–D) reveals skull and suture abnormalities in the Rapb1 nulls. Arrowheads and asterisk indicate the distance between frontal bones and suture deformities caused by excessive ossification, respectively. Enlargements of the insects (A, B) are shown in C and D. Sections of the E18.5 control (+/+) and mutant (−/−) were examined by Goldner’s trichrome staining in green (E, F). Immunostaining of Osterix (Osx; G, H) and in situ hybridization of type 1 collagen (Col1; I, J), marking osteoprogenitors and osteoblasts, respectively, show that osteoblastogenesis is enhanced in the Rap1b knockout frontal bone primordia at E13.5. Arrows indicate area positively stained with Osx and Col1. Orientation is indicated by skull base (SB) and skull vault (SV). Sections were counterstained by hematoxylin (G–H) and nuclear fast red (I–J). Images are representatives of three independent experiments. Scale bars, 2 mm (A, B); 500 μm (C, D); 100 μm (E, F); 400 μm (G–J).
Rap1b antagonizes osteoblast differentiation through modulation of FGF signaling
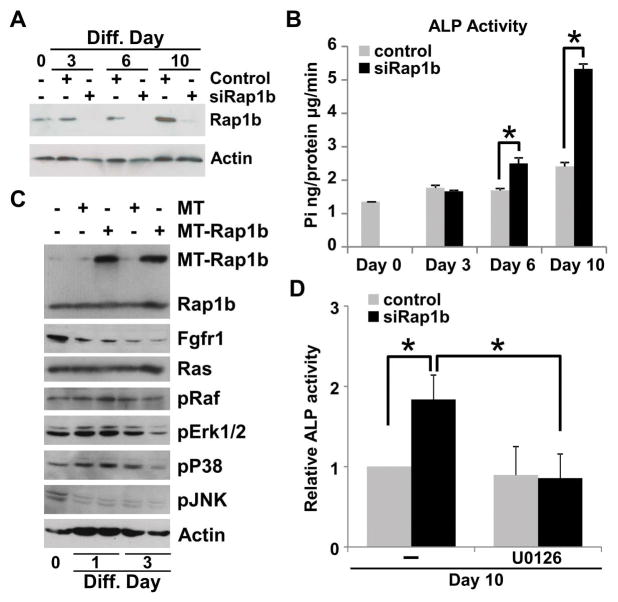
Osteoblast is arguably the most important cell type in intramembranous bone formation, essential for healthy development of the calvarium. It is highly possible that aberrant intramembranous ossification and suture morphogenesis caused by the deletion of Rap1b are attributed to its effects on the osteoblasts. In addition, BMP signaling has been shown to antagonize FGF signaling in order to convert the skeletal stem cell fate from osteoblast to chondrocyte development in the calvarial mesenchyme (13). The pro-osteogenic FGF pathway affected by the pro-chondrogenic BMPR1A-Rap1b regulatory axis might be critical for craniofacial skeletogenesis. Therefore, we tested the ability of Rap1b to influence the osteogenesis of the calvarial mesenchymal cells. First, the knockdown of Rap1b enhanced osteoblast differentiation as determined by alkaline phosphatase activity in C3H10T1/2 cells (Figure 5A, B; mean ± SEM, p < 0.05, n=3). We next examined if Rap1b exerts its effects on osteoblastogenesis through modulation of FGF signaling. High levels of Rap1b expression did not seem to affect Fgfr1, Ras and phosphorylated Raf in C3H10T1/2 cells undergoing osteoblast differentiation (Figure 5C). However, Rap1b prevented the activation of MAP kinases which are known to transduce the FGF pathway and responsible for FGF-mediated synostosis related syndromes (45). Phosphorylated Erk1/2 and p38, but not JNK, were significantly reduced by Rap1b overexpression (Figure 5C), implying that Rap1b possesses negative effects on the MAPK-mediated osteogenesis which is modulated at the level and/or upstream of MAPK. To test the requirement of MAPK signaling in the Rap1b-mediated regulation of osteogenesis we utilized a pharmacogenetic approach. The addition of U0126, an inhibitor for Erk1/2 activation, was able to prohibit the enhanced osteoblast differentiation caused by the Rap1b knockdown (Figure 5D; mean ± SEM, p < 0.05, n=3). The results suggest that Rap1b exerts its effects on osteogenesis through modulation of MAPK signaling. Rap1b negatively regulates osteoblast development mediated by the MAPK pathway. The disruption of Rap1b results in promotion of osteoblastogenesis, leading to excessive intramembranous ossification and craniosynostosis.
Figure 5.
Rap1b negatively regulates osteoblast differentiation through inhibition of MAPK signaling. (A) Immunoblotting of Rap1b and actin shows the knockdown efficiency mediated by siRNA in C3H10T1/2 cells during osteoblast differentiation. (B) Analysis of alkaline phosphatase activity reveals that the Rap1b knockdown enhances osteoblast differentiation (mean ± SEM, p < 0.01, n=3). (C) Immunoblot analysis examines the effect of overexpressing a myc tagged (MT) Rap1b on FGF signaling effectors, including Fgfr1, Ras, phosphorylated Raf (pRaf), phosphorylated Erk1/2 (pErk1/2), phosphorylated P38 (pP38), phosphorylated JNK (pJNK). The level of actin is analyzed as a loading control. (D) Alkaline phosphatase analysis revealed that enhanced osteoblast differentiation caused by the Rap1b knockdown can be alleviated by an inhibitor (10 μM U0126) for Erk1/2 activation (mean ± SEM, p < 0.01, n=3). Imagines are representatives of three independent experiments.
Rap1b promotes chondrocyte differentiation
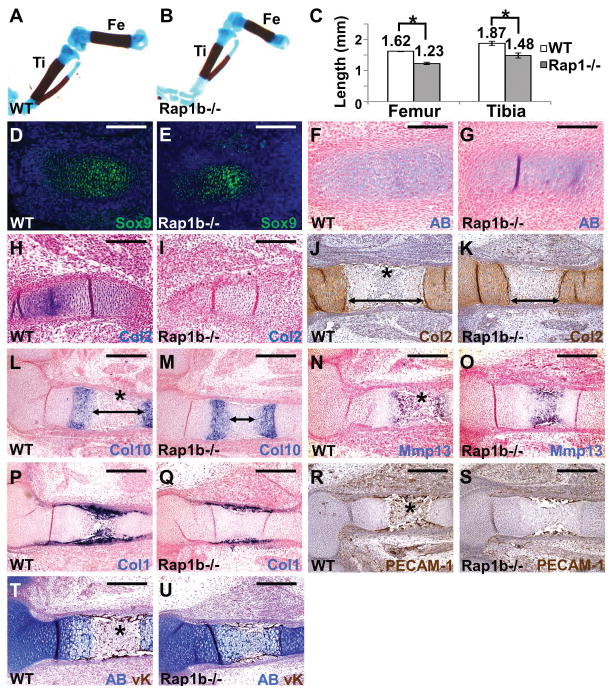
Data obtained from the Axin2−/− and caBMPR1A mutants suggested that Rap1b may exert its effects on the skeletogenic mesenchymal cells through modulation of their differentiation into osteoblasts and chondrocytes. Because of calvarial skeletogenesis independent of chondrogenesis, we tested if development of long bones is affected by the Rap1b ablation. To determine the role of Rap1b in chondrogenesis, we examined formation of the femur and tibia that is mediated through endochondral ossification. All of the Rap1b knockouts appeared to have shorter limbs but were otherwise undistinguishable from their littermates at birth. Alizarin red and alcian blue staining of the skeleton confirmed that development of the femur and tibia has been compromised at E18.5 (Figure 6A–C; mean ± SEM, p < 0.05, n=3). We then examined the endochondral ossification processes initiated by differentiation of condensed mesenchymal cells into chondrocytes. Alcian blue (AB) staining and immunostaining of a chondrogenic progenitor marker, Sox9, showed slight reduction in the mutants at E13.5 (Figure 6D–G). However, immunostaining of a chondrocyte marker, type 2 collagen (Col2) evident in the control but not detectable in the Rap1b nulls (Figure 6H, I), revealed a significant defect in chondrogenesis. The delayed chondrogenesis became evident at E15.5 (Figure 6J–O). Although Col2 and Col10 were expressed, the resorption of mature chondrocytes and formation of marrow cavity were not evident in the E15.5 mutant (Figure 6J–M). The distance between two Col10-expressing zones was also significantly reduced at E15.5 (Figure 6L, M; control, 403 ± 29 μm, Rap1b−/−, 205 ± 51 μm, mean ± SEM, p < 0.01, n=3, student’s t-test). The delay in chondrogenesis led to abnormalities in subsequent processes, e.g. terminal differentiation of chondrocytes (Figure 6N, O), osteoblast differentiation (Figure 6P, Q) and vascular invasion (Figure 6R, S). The marrow cavity is normally evident but not yet developed in the mutant at this developmental stage (Figure 6T, U). To further examine if chondrocytes are indeed affected by the Rap1b deletion, we performed an in vitro study. First, overexpression of Rap1b promotes differentiation of ATDC5 chondrogenic cells in micromass culture analysis (Figure 7A–C; mean ± SEM, p < 0.05, n=3). Next, we isolated primary cells from the E18.5 wild type and Rap1b knockout calvarial bones and ribcages, and assessed their ability to differentiate into chondrocytes (Figure 7D–E, G–H). The loss of Rap1b significantly reduced chondrocyte differentiation (Figure 7F, I; mean ± SEM, p < 0.05, n=3). In addition, qRT-PCR analysis revealed the expression of chondrogenic markers, e.g. Sox9, Col2, and Aggrecan (Acan), significantly reduced in the Rap1b−/− chondrocytes (Figure 7J; mean ± SEM, p < 0.05, n=3). The data thus suggested an important role of Rap1b in chondrocyte differentiation. To further determine the requirement of Rap1b for chondrogenesis stimulated by canonical BMP signaling, primary caBMPR1AAx2 cells were isolated and differentiated into chondrocytes with or without a Rap1 inhibitor (Figure 7K–N). The enhanced differentiation of caBMPR1AAx2 could be prohibited by Rap1b inhibition, suggesting a BMP-Rap1b signaling axis in chondrogenesis (Figure 7O; p < 0.05, n=3). The Rap1b inhibitor was also able to reduce the stimulation of chondrogenic markers caused by the BMPR1A mutant (Figure 7P; p < 0.05, n=3). Furthermore, chondrogenesis essential for endochondral ossification was significantly impaired by the loss of Rap1b during limb development (Figure 6). Together with the osteogenic effects of Rap1b, our findings suggest its dual and determinant role in differentiation of the skeletogenic cell types.
Figure 6.
The Rap1b knockout mice exhibit chondrogenic defects during endochondral ossification. Skeletal staining of E18.5 wild type (WT, A) and Rap1b−/− (B) embryos reveals the development of the body skeleton affected by the mutation. The length of femur and tibia is significantly shorter in the mutant (C; mean ± S.E.M., p < 0.05, n=3, student’s t-test). The E13.5 (D–I) and E15.5 (J–U) WT and Rap1b mutants were analyzed by AB staining (F, G), immunostaining of Sox9 (D, E), Col2 (J, K) and PECAM-1 (R, S), double labeling of AB and von Kossa (T, U), and in situ hybridization of Col2 (H, I), Col10 (L, M), Mmp13 (N, O) and Col1 (P, Q). Arrows and asterisks indicate the distance between two Col2/Col10-expressing zones and marrow cavity, respectively. Sections are counterstained by nuclear fast red (F–I, L–Q) or hematoxylin (J, K, R, S). Images are representatives of three independent experiments. Scale bars, 200 μm (D–I); 400 μm (J–U).
Figure 7.
Rap1b regulates chondrocyte differentiation. AB staining of the in vitro micromass cultures shows differentiation of ATDC5 cells (A, B) or cells isolated from the E18.5 WT (D, G, K), Rap1b−/− (E, H) and caBMPR1AAx2 (K–N) calvaria (D, E) and ribcages (G, H, K–N), into chondrocytes. Chondrogenesis is promoted by overexpression of Rap1b (A–C) while its deficiency causes significant inhibition (D–I), determined by AB stained area and OD595 reading (C, F, I; mean ± S.E.M., p value < 0.05, n=3, student’s t-test) and qRT-PCR analysis of chondrogenic markers, Sox9, Col2, Acan and Col10 (J; mean ± S.E.M., p value < 0.05, n=3, student’s t-test). Enhanced chondrogenesis by the caBMPR1A mutant can be prohibited by Rap1b inhibitor GGTI298, determined by AB stained area (K–O, mean ± S.E.M., p value<0.05, n=3, student’s t-test) and qRT-PCR analysis of chondrogenic markers, Sox9, Col2, Acan and Col10 (P; mean ± S.E.M., p value < 0.05, n=3, student’s t-test). Asterisks indicate statistically significant alterations. Images are representatives of three independent experiments. Scale bars, 2 mm (A, B, D, E and K–N); 1 mm (G, H).
Discussion
This study identifies Rap1b, which acts downstream of Axin2 and is essential for skeletal development. Although Rap1b has been implicated in angiogenesis, lens epithelial maintenance and immune cell functions (21,22,49–53), its role in skeletogenesis has yet to be reported. In humans, multiple omics studies have recently identified Rap1b as one of the genes associated with bone mineral density and male osteoporosis (54). It has also shown to be important for osteoclast function and bone remodeling in mice (55). Most recently, we have demonstrated that Axin2-expressing cells are bona fide stem cells residing in the suture mesenchyme essential for development, homeostasis and injury repair of the calvarial bone (19). The function of the Axin2 gene, which promotes β-catenin degradation, is also required for proper development of the craniofacial skeleton (29). The Axin2-mediated Wnt signaling interplays with two well-known skeletogenic pathways, FGF and BMP, to dictate commitment of the mesenchymal lineages (13). Here the new study further supports our initial hypothesis and provides compelling evidence that Rap1b is a key signaling interrogator critical for developmental programming of the skeletogenic mesenchyme. We propose that Rap1b plays an integral role in signaling crosstalk of Wnt, FGF and BMP during development of the craniofacial and body skeletons. Rap1b expression and activation are regulated by Axin2 but are independent of β-catenin-mediated transcription. Instead, Axin2 controls Rap1b through modulation of the BMP pathway. Stimulation of canonical BMP signaling is necessary and sufficient to switch the fate of suture mesenchymal cells, which are developmentally programed to become osteoblasts, and promote their differentiation into chondrocytes. Consistent with new genetic results reported here, a recent cell transplantation study also supports an inductive role of BMP in chondrocyte fate determination and differentiation (19).
The BMPR1A-Rap1b signaling axis inhibits the Ras-dependent activation of MAPKs that could be highly cell type-specific (44). By modulating the MAPK activities, Rap1b negatively controls osteoblast development. Rap1b also mediates the effects of BMP signaling to enhance chondrogenesis. Therefore, Rap1b has a dual role in proper development of skeletogenic mesenchyme that is manifested in a mouse model for Rap1b deficiency. Skull and limb defects caused by the loss of Rap1b demonstrate its importance in intramembranous and endochondral ossifications, respectively. Genetic analysis has linked several members of FGF receptor (FGFR) family to synostosis-related syndromes and dwarfism (1,18). A common feature of these syndromic diseases is the highly stimulated FGF-MAPK signaling, which enhances osteoblast proliferation and differentiation, leading to excessive intramembranous ossification and premature suture closure (12,56). In addition to the osteoblast, craniosynostosis has been associated with abnormalities in the skeletogenic stem cell (13). A switch of mesenchymal cell fate from osteoblast to chondrocyte, resulting in ectopic chondrogenesis and endochondral ossification is another pathogenic mechanism (13), providing an explanation for peculiar chondrocytes present in the synostosis-related syndromes of human and mouse (57–62). Proper coordination and balance of skeletal stem cell development into chondrocytes and osteoblasts appears to play a crucial role in formation of craniofacial and body skeletons and pathogenesis of skeletal deformities. Our findings linking the FGF/MAPK-mediated skeletal development and deformity to Rap1b also imply its potential involvement in certain cases of dwarfism with apparent synostosis-related syndromes (63,64). Furthermore, our in vivo and in vitro studies reveal novel functions of Rap1b in differentiation of mesenchymal cells into osteoblasts and chondrocytes. The Axin2-BMPR1A-Rap1b signaling axis is critical for skeletal development and disease.
Axin2 is known to control skeletogenesis through modulation of canonical Wnt signaling (29). However, we have detected distinct skeletal defects between the loss of Axin2 and activation of β-catenin-dependent transcription in the suture mesenchyme (17). The loss of Axin2 promotes osteoblastogenesis and bone formation, leading to craniosynostosis (16,29). However, stimulation of β-catenin-dependent transcription in the Axin2-expressing cells does not cause suture abnormality although excessive intramembranous ossification is evident (17). In addition, transient ectopic chondrocytes are present in the Axin2 null suture but absent in the dominant mutant of β-catenin (13). These are two clear indications that Axin2 controls suture morphogenesis via the non-canonical Wnt pathway. Current study further supports our initial hypothesize that Axin2 regulates mesenchymal cell fate determination independent of Wnt/β-catenin signaling. Rap1b is an important target of Axin2 that mediates its downstream effects independent of β-catenin transcription. Future studies focusing on delineating the non-canonical signaling cascade promise new insight into skeletal development and disease.
Supplementary Material
Acknowledgments
We thank Fan Wang at Duke University for providing BMPR1ACA/+ mouse strain. T.M., M.J., A.A., H.I.Y., Q.H. and W.H. conceived, designed and performed the experiments, and analyzed the data. T.M., A.A. and W.H. wrote the paper. E.I.C. performed Mass Spec analyses. M.C-W. provided Rap1b mutant mouse strain. This research work is supported by National Institutes of Health (DE15654) and NYSTEM (C029558) to W.H. T.M. is a recipient of postdoctoral fellowships supported by National Institutes of Health (DE21958) and NYSTEM (C026877).
Footnotes
Disclosure
The authors declare no conflict of interest.
References
- 1.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes & development. 2002 Jun 15;16(12):1446–65. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 2.Erlebacher A, Filvaroff EH, Gitelman SE, Derynck R. Toward a molecular understanding of skeletal development. Cell. 1995 Feb 10;80(3):371–8. doi: 10.1016/0092-8674(95)90487-5. [DOI] [PubMed] [Google Scholar]
- 3.Hall BK. Bone. Caldwell, N.J: Telford Press; 1990. p. v. [Google Scholar]
- 4.Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev Dyn. 2000 Dec;219(4):472–85. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 5.Krakow D. Skeletal dysplasias. Clinics in perinatology. 2015 Jun;42(2):301–19. viii. doi: 10.1016/j.clp.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011 May;155A(5):943–68. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Accogli A, Pacetti M, Fiaschi P, Pavanello M, Piatelli G, Nuzzi D, et al. Association of achondroplasia with sagittal synostosis and scaphocephaly in two patients, an underestimated condition? Am J Med Genet A. 2015 Mar;167A(3):646–52. doi: 10.1002/ajmg.a.36933. [DOI] [PubMed] [Google Scholar]
- 8.Cohen MM., Jr Short-limb skeletal dysplasias and craniosynostosis: what do they have in common? Pediatric radiology. 1997 May;27(5):442–6. doi: 10.1007/s002470050165. [DOI] [PubMed] [Google Scholar]
- 9.De Moerlooze L, Dickson C. Skeletal disorders associated with fibroblast growth factor receptor mutations. Current opinion in genetics & development. 1997 Jun;7(3):378–85. doi: 10.1016/s0959-437x(97)80152-9. [DOI] [PubMed] [Google Scholar]
- 10.Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2(6):458–68. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- 11.Cohen MM, MacLean RE. Craniosynostosis : diagnosis, evaluation, and management. 2. xx. New York: Oxford University Press; 2000. p. 454. [Google Scholar]
- 12.Miraoui H, Marie PJ. Fibroblast growth factor receptor signaling crosstalk in skeletogenesis. Sci Signal. 2010;3(146):re9. doi: 10.1126/scisignal.3146re9. Epub 2010/11/04. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama T, Mirando AJ, Deng CX, Hsu W. The balance of WNT and FGF signaling influences mesenchymal stem cell fate during skeletal development. Sci Signal. 2010;3(123):ra40. doi: 10.1126/scisignal.2000727. Epub 2010/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard JJ, Scott JH, Girgis FG. The structure and development of cranial and facial sutures. Journal of anatomy. 1956 Jan;90(1):73–86. [PMC free article] [PubMed] [Google Scholar]
- 15.Yu HM, Liu B, Chiu SY, Costantini F, Hsu W. Development of a unique system for spatiotemporal and lineage-specific gene expression in mice. Proceedings of the National Academy of Sciences of the United States of America. 2005 Jun 14;102(24):8615–20. doi: 10.1073/pnas.0500124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Yu HM, Hsu W. Craniosynostosis caused by Axin2 deficiency is mediated through distinct functions of beta-catenin in proliferation and differentiation. Developmental biology. 2007 Jan 1;301(1):298–308. doi: 10.1016/j.physletb.2003.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirando AJ, Maruyama T, Fu J, Yu HM, Hsu W. Beta-catenin/cyclin D1 mediated development of suture mesenchyme in calvarial morphogenesis. BMC Dev Biol. 2010 Nov 26;10(1):116. doi: 10.1186/1471-213X-10-116. Epub 2010/11/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coumoul X, Deng CX. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res C Embryo Today. 2003 Nov;69(4):286–304. doi: 10.1002/bdrc.10025. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama T, Jeong J, Sheu TJ, Hsu W. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nature communications. 2016;7:10526. doi: 10.1038/ncomms10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. The Journal of biological chemistry. 2009 Apr 24;284(17):10995–9. doi: 10.1074/jbc.R800061200. Epub 2008/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC., 2nd Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005 Mar;115(3):680–7. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddala R, Nagendran T, Lang RA, Morozov A, Rao PV. Rap1 GTPase is required for mouse lens epithelial maintenance and morphogenesis. Developmental biology. 2015 Oct 1;406(1):74–91. doi: 10.1016/j.ydbio.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Yan J, De P, Chang HC, Yamauchi A, Christopherson KW, 2nd, et al. Rap1a null mice have altered myeloid cell functions suggesting distinct roles for the closely related Rap1a and 1b proteins. J Immunol. 2007 Dec 15;179(12):8322–31. doi: 10.4049/jimmunol.179.12.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. Journal of cell science. 2003 Feb 1;116(Pt 3):435–40. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- 25.Yu HM, Liu B, Costantini F, Hsu W. Impaired neural development caused by inducible expression of Axin in transgenic mice. Mechanisms of development. 2007 Feb;124(2):146–56. doi: 10.1016/j.mod.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu W, Mirando AJ, Yu HM. Manipulating gene activity in Wnt1-expressing precursors of neural epithelial and neural crest cells. Dev Dyn. 2010 Jan;239(1):338–45. doi: 10.1002/dvdy.22044. Epub 2009/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J. BMP signaling in the development of the mouse esophagus and forestomach. Development (Cambridge, England) 2010 Dec;137(24):4171–6. doi: 10.1242/dev.056077. Epub 2010/11/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maruyama T, Jiang M, Hsu W. Gpr177, a novel locus for bone mineral density and osteoporosis, regulates osteogenesis and chondrogenesis in skeletal development. J Bone Miner Res. 2013 May;28(5):1150–9. doi: 10.1002/jbmr.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu HM, Jerchow B, Sheu TJ, Liu B, Costantini F, Puzas JE, et al. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development (Cambridge, England) 2005 Apr;132(8):1995–2005. doi: 10.1242/dev.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu HM, Jin Y, Fu J, Hsu W. Expression of Gpr177, a Wnt trafficking regulator, in mouse embryogenesis. Dev Dyn. 2010 Jun 14;239(7):2102–9. doi: 10.1002/dvdy.22336. Epub 2010/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruyama EO, Yu HM, Jiang M, Fu J, Hsu W. Gpr177 deficiency impairs mammary development and prohibits Wnt-induced tumorigenesis. PLoS ONE. 2013;8(2):e56644. doi: 10.1371/journal.pone.0056644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu SY, Asai N, Costantini F, Hsu W. SUMO-Specific Protease 2 Is Essential for Modulating p53-Mdm2 in Development of Trophoblast Stem Cell Niches and Lineages. PLoS biology. 2008 Dec 16;6(12):e310. doi: 10.1371/journal.pbio.0060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J, Hsu W. Epidermal Wnt controls hair follicle induction by orchestrating dynamic signaling crosstalk between the epidermis and dermis. J Invest Dermatol. 2013 Apr;133(4):890–8. doi: 10.1038/jid.2012.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell HK., Jr A modification of Movat’s pentachrome stain. Archives of pathology. 1972 Aug;94(2):187–91. [PubMed] [Google Scholar]
- 35.Jiang M, Chiu SY, Hsu W. SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ. 2011 Jun;18(6):1005–15. doi: 10.1038/cdd.2010.168. Epub 2010/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu J, Ivy Yu HM, Maruyama T, Mirando AJ, Hsu W. Gpr177/mouse Wntless is essential for Wnt-mediated craniofacial and brain development. Dev Dyn. 2011 Feb;240(2):365–71. doi: 10.1002/dvdy.22541. Epub 2011/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruyama EO, Lin H, Chiu SY, Yu HM, Porter GA, Hsu W. Extraembryonic but not embryonic SUMO-specific protease 2 is required for heart development. Scientific reports. 2016;6:20999. doi: 10.1038/srep20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W. Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proceedings of the National Academy of Sciences of the United States of America. 2009 Nov 3;106(44):18598–603. doi: 10.1073/pnas.0904894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Yu HM, Huang J, Hsu W. Co-opted JNK/SAPK signaling in Wnt/beta-catenin-induced tumorigenesis. Neoplasia (New York, NY. 2008 Sep;10(9):1004–13. doi: 10.1593/neo.08548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu J, Yu HM, Chiu SY, Mirando AJ, Maruyama EO, Cheng JG, et al. Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration. PLoS genetics. 2014 Oct;10(10):e1004579. doi: 10.1371/journal.pgen.1004579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provot S, Zinyk D, Gunes Y, Kathri R, Le Q, Kronenberg HM, et al. Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development. The Journal of cell biology. 2007 May 7;177(3):451–64. doi: 10.1083/jcb.200612023. Epub 2007/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada M, Greer PA, McMahon AP, Bouxsein ML, Schipani E. In vivo targeted deletion of calpain small subunit, Capn4, in cells of the osteoblast lineage impairs cell proliferation, differentiation, and bone formation. The Journal of biological chemistry. 2008 Jul 25;283(30):21002–10. doi: 10.1074/jbc.M710354200. Epub 2008/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development (Cambridge, England) 2005 Jan;132(1):49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt JM, Stork PJ. PKA phosphorylation of Src mediates cAMP’s inhibition of cell growth via Rap1. Molecular cell. 2002 Jan;9(1):85–94. doi: 10.1016/s1097-2765(01)00432-4. [DOI] [PubMed] [Google Scholar]
- 45.Shukla V, Coumoul X, Wang RH, Kim HS, Deng CX. RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nature genetics. 2007 Sep;39(9):1145–50. doi: 10.1038/ng2096. [DOI] [PubMed] [Google Scholar]
- 46.Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010 Jul;120(7):2457–73. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nature reviews. 2012 Jan;13(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007 Jan 15;23(2):257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 49.Lakshmikanthan S, Sobczak M, Chun C, Henschel A, Dargatz J, Ramchandran R, et al. Rap1 promotes VEGFR2 activation and angiogenesis by a mechanism involving integrin alphavbeta(3) Blood. 2011 Aug 18;118(7):2015–26. doi: 10.1182/blood-2011-04-349282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chrzanowska-Wodnicka M, Kraus AE, Gale D, White GC, 2nd, Vansluys J. Defective angiogenesis, endothelial migration, proliferation, and MAPK signaling in Rap1b-deficient mice. Blood. 2008 Mar 1;111(5):2647–56. doi: 10.1182/blood-2007-08-109710. Epub 2007/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caron E, Self AJ, Hall A. The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr Biol. 2000 Aug 24;10(16):974–8. doi: 10.1016/s0960-9822(00)00641-2. [DOI] [PubMed] [Google Scholar]
- 52.Shimonaka M, Katagiri K, Nakayama T, Fujita N, Tsuruo T, Yoshie O, et al. Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. The Journal of cell biology. 2003 Apr 28;161(2):417–27. doi: 10.1083/jcb.200301133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Xu J, Kumar RS, Lakshmikanthan S, Kapur R, Kofron M, et al. The small GTPase Rap1b negatively regulates neutrophil chemotaxis and transcellular diapedesis by inhibiting Akt activation. The Journal of experimental medicine. 2014 Aug 25;211(9):1741–58. doi: 10.1084/jem.20131706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu W, Shen H, Zhang JG, Zhang L, Zeng Y, Huang HL, et al. Cytosolic proteome profiling of monocytes for male osteoporosis. Osteoporos Int. 2016 Nov 14; doi: 10.1007/s00198-016-3825-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou W, Izawa T, Zhu T, Chappel J, Otero K, Monkley SJ, et al. Talin1 and Rap1 are critical for osteoclast function. Molecular and cellular biology. 2013 Feb;33(4):830–44. doi: 10.1128/MCB.00790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morriss-Kay GM, Wilkie AO. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. Journal of anatomy. 2005 Nov;207(5):637–53. doi: 10.1111/j.1469-7580.2005.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreiborg S, Marsh JL, Cohen MM, Jr, Liversage M, Pedersen H, Skovby F, et al. Comparative three-dimensional analysis of CT-scans of the calvaria and cranial base in Apert and Crouzon syndromes. J Craniomaxillofac Surg. 1993 Jul;21(5):181–8. doi: 10.1016/s1010-5182(05)80478-0. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Xiao R, Yang F, Karim BO, Iacovelli AJ, Cai J, et al. Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development (Cambridge, England) 2005 Aug;132(15):3537–48. doi: 10.1242/dev.01914. [DOI] [PubMed] [Google Scholar]
- 59.Yin L, Du X, Li C, Xu X, Chen Z, Su N, et al. A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone. 2008 Apr;42(4):631–43. doi: 10.1016/j.bone.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Marie PJ, Coffin JD, Hurley MM. FGF and FGFR signaling in chondrodysplasias and craniosynostosis. Journal of cellular biochemistry. 2005 Dec 1;96(5):888–96. doi: 10.1002/jcb.20582. [DOI] [PubMed] [Google Scholar]
- 61.Cohen MM, Jr, Kreiborg S. New indirect method for estimating the birth prevalence of the Apert syndrome. International journal of oral and maxillofacial surgery. 1992 Apr;21(2):107–9. doi: 10.1016/s0901-5027(05)80544-2. [DOI] [PubMed] [Google Scholar]
- 62.Cohen MM, Jr, Kreiborg S. Visceral anomalies in the Apert syndrome. American journal of medical genetics. 1993 Mar 15;45(6):758–60. doi: 10.1002/ajmg.1320450618. [DOI] [PubMed] [Google Scholar]
- 63.el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, et al. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nature genetics. 1997 Jan;15(1):42–6. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 64.White KE, Cabral JM, Davis SI, Fishburn T, Evans WE, Ichikawa S, et al. Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am J Hum Genet. 2005 Feb;76(2):361–7. doi: 10.1086/427956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell-cell junction formation. Journal of cell science. 2007 Jan 1;120(Pt 1):17–22. doi: 10.1242/jcs.03306. Epub 2006/12/22. [DOI] [PubMed] [Google Scholar]
- 66.Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Current opinion in cell biology. 2009 Oct;21(5):684–93. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.