Abstract
Seed priming is a commercially used technique for improving seed performance including germination. However, the treatment sometimes reduces seed longevity as a side effect, limiting the storable period or longevity of the seeds. To overcome this problem, molecular mechanisms involved in the loss of seed longevity during priming were analyzed using natural variations of Arabidopsis thaliana. We found that the Est-1 accession retained longevity for longer after priming compared to the reference accession Col-0. QTL analysis using 279 recombinant inbred lines (RILs) derived from the Est-1 × Col-0 detected three QTL regions associated with the loss of seed longevity during priming. Bulked transcriptome analysis (RNA-Seq with bulked RIL populations) revealed that genes related to brassinosteroid (BR) biosynthesis/signaling and cell wall modification were highly expressed in primed seeds with shorter longevity. After priming, BR-deficient mutants cyp85a1/a2 and det2 showed significantly longer longevity than the wild type (WT). Moreover, tetrazolium staining indicated that mutant seed coats were less permeable after priming than those of WT. We suggest that the loss of seed longevity in primed seed is due to increased seed coat permeability, which is positively regulated, at least partly, via BR signaling.
Introduction
Germination is the process during which a quiescent dry seed become a seedling. Germination commences with imbibition of dry seeds, followed by expansion of embryonic cells and rupture of the surrounding layers: endosperm and seed coat (testa). The completion of germination is often defined as radicle emergence from the surrounding layers, whereas dormancy represents a physiological status that enables viable seeds not to germinate even under favorable conditions. Although dormancy is an important trait especially for wild species for their survival in changing environments, extreme dormancy often causes poor germination or non-synchronous germination and the trait is detrimental for crop cultivation. Therefore, less dormant (or rapid germination) traits have been preferentially selected over time through breeding, especially for cereals. However, this can enable pre-harvest sprouting (viviparous germination or precocious germination) which reduces the quality of the products (seeds) not only as foods but also as individuals for the next generation. The precise regulation of these two opposing physiological processes, dormancy and germination, is thus desirable for efficient agricultural production.
Seed priming is a commercially used technique to improve seed performance including germinability, uniformity, vigor and stress tolerance. The treatment involves imbibition of seeds in water under controlled conditions to trigger the metabolic processes that are normally activated during the early phase of germination (pre-germinative metabolism) and subsequent drying of the seeds prior to full germination, or radicle emergence, so that seeds reenter a quiescent state1, 2. Several kinds of priming treatments such as hydropriming, osmopriming, solid matrix priming and biopriming have been developed2. Among them, osmopriming is a widespread pre-sowing procedure that involves treatments with osmotic solutions such as polyethylene glycol or inorganic salts of sodium, potassium and magnesium (most commonly NaCl, NaNO3, MnSO4, MgCl2, K3PO4 and KNO3) at low water potential facilitating controlled water uptake. In contrast, hydropriming is simple and the most traditional type of priming, with seeds soaked in water under optimal temperature conditions (usually in a range from 5 to 20 °C) for a set time period. Imbibition at high or low temperature before sowing promotes germination depending on plant species and this is known as thermopriming. Besides the benefits of priming to enhance seed germination, the treatments applied sometimes reduce seed longevity (total time span during which seeds remain viable) or storability3. Primed seeds with high performances in terms of germination are thought to have a more advanced physiological status in the germination process compared with seeds without priming, and are thus more susceptible to deterioration1. This loss of seed longevity is a disadvantage of priming with regard to commercial distribution.
The molecular aspects of seed longevity have been reviewed4–6. During dry storage of mature seeds, reduction of longevity is mainly caused by oxidation of cellular macromolecules such as nucleic acids, proteins and lipids. To circumvent the effects, seeds possess protective mechanisms such as the formation of a glassy cytoplasm to reduce cellular metabolic activities and the production of antioxidants, as well as repair systems that remove damage accumulated in DNA, RNA and proteins upon seed imbibition through enzymes such as DNA glycosylase and methionine sulfoxide reductase. In addition, the seed coat represents the interface through which the embryo interacts with the external environment and thus its composition and structure are critical factors for seed longevity. Several factors that determine seed longevity during dry storage have been identified so far, however, little is known about the molecular mechanisms involved in the loss of seed longevity during priming treatments.
Arabidopsis is not only a model plant but also a member of one of the largest families of flowering plants, the Brassicaceae, which includes several crop species such as mustard, radish, cauliflower, broccoli and cabbage. Natural variations (also referred to as ecotypes or accessions) of Arabidopsis have tremendous genetic and phenotypic diversity, and QTL mapping has been successfully employed to identify genes that are responsible for the variations in a range of physiological and developmental traits7. Among several mapping population that can be used for QTL analysis, recombinant inbred lines (RILs) offer unique advantages: homozygous lines that can be propagated without genetic changes8 allow us to conduct replicate experiments to evaluate their phenotypes.
In this study, we compared seed longevity of 230 Arabidopsis natural variations before and after priming and identified an accession, Est-1, which can retain a relatively high level of seed longevity after priming when compared with the reference accession, Col-0. Mapping with a RIL population derived from a cross between Col-0 and Est-1 detected three major QTLs associated with the loss of seed longevity during priming. Furthermore we performed transcriptome analysis with the RILs to predict genes that might be involved in the regulation of seed longevity. The relationship between seed coat permeability as modulated by the plant hormone brassinosteroid (BR) and the loss of seed longevity during priming will be discussed.
Results
Effects of priming on seed germination and longevity
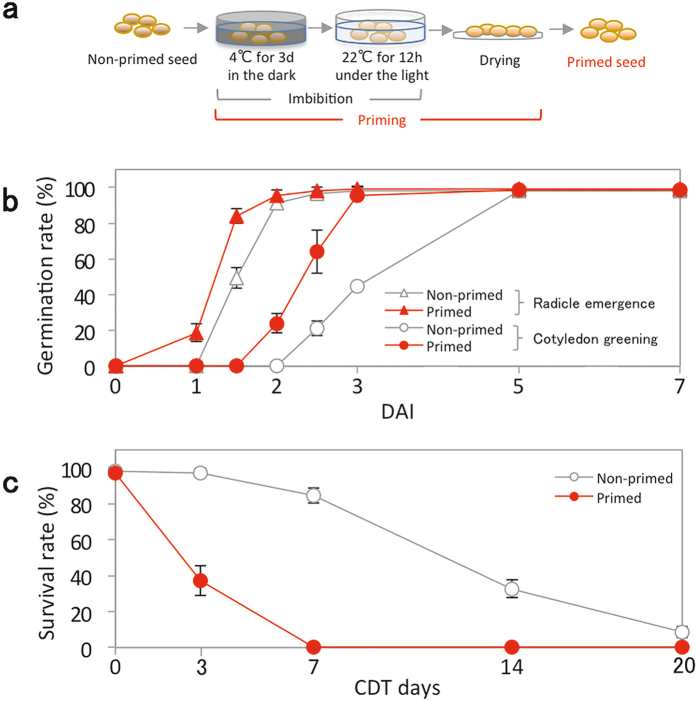
We first set up an experimental system to determine the effects of priming on seed longevity using the Arabidopsis Col-0 reference accession (Fig. 1a). Although Col-0 is known as a weakly dormant accession, germination behavior (e.g. germination speed and final germination percentage) of the seeds varies depending on environmental conditions9. To eliminate the effects caused by variable dormancy status and to synchronize germination, mature dry seeds that had been after-ripened for two to four months were imbibed at 4 °C for 3 days in the dark. This treatment is generally called stratification, but can also be considered as a part of priming (thermopriming). Then the seeds were incubated at 22 °C in the light for 12 h to partially initiate germination events; this treatment is considered as hydropriming. Finally, seeds were dried to a quiescent physiological state. In this manuscript, we refer to the combined series of these treatments as priming. Seeds did not complete germination during these treatments as testa rupture or radicle emergence was not observed. Upon imbibition, radicle protrusion and subsequent establishment of seedlings with green cotyledons was much faster for primed seeds than non-primed seeds (Fig. 1b). Furthermore, the primed seeds were more resistant to high temperature in terms of radicle protrusion compared with non-primed seeds (Supplementary Fig. S1). These observations demonstrated that the priming treatment used in this study effectively enhanced germination performance. Hereafter, we have scored cotyledon greening in germination tests rather than radicle protrusion because this is a better representation of the effect of priming on seedling survival and establishment.
Figure 1.
Effect of priming on germination speed and seed longevity. (a) A simplified scheme for the priming treatment used in this study. (b) Germination speed of primed and non-primed seeds without CDT determined as radicle emergence from the seed coat or cotyledon greening. (c) Survival rates of primed and non-primed seeds after 0, 3, 7, 14 and 20 days of CDT. Values are means ± SD of three replicates for (b) and (c).
To evaluate the effects of priming on seed longevity, we employed a controlled deterioration treatment (CDT); the sensitivity or resistance of seeds to CDT can be used for rapid assessment and prediction of seed longevity during natural aging10, 11. Primed and non-primed seeds were subjected to CDT by incubating the seeds at 37 °C under 80% relative humidity for 0, 3, 7, 14 and 20 days. After CDT, the ability of seeds to establish healthy seedlings with green cotyledons upon imbibition was scored as the survival rate (Fig. 1c). Note that individuals that did not grow into healthy seedlings after radicle protrusion were not considered as survival. Survival rates of the non-primed seeds were not significantly impacted after 3 days of CDT, and more than 80% of non-primed seeds could still establish healthy seedlings after 7 days of CDT. In contrast, survival rates of primed seeds decreased rapidly to 40% within 3 days of CDT, and none of the primed seeds survived after 7 days of CDT. These data indicate that negative impacts of priming on seed longevity can be determined by this experimental system.
Effects of dormancy and endogenous abscisic acid (ABA) levels on the longevity of primed seeds
An earlier study had shown that the Arabidopsis ABA deficient mutant aba1-5 in the Columbia genetic background had reduced seed dormancy and longevity compared to the wild type (WT)12. Conversely, QTL analysis using six Arabidopsis RIL populations showed that natural variations for dormancy and longevity were negatively correlated13. The relationship between seed dormancy and seed longevity were investigated in our experimental conditions, by determining the germination speed and seed longevity of ABA deficient and ABA over-accumulating mutants in the Columbia background. The aba2 mutant is defective in an ABA biosynthesis enzyme, whereas cyp707a mutants accumulate high levels of ABA compared with WT due to their defective ABA catabolism14–16. Although all genotypes tested germinated to around 100% within 7 days after imbibition (DAI), faster germination was still observed for non-primed aba2 seeds compared to WT, indicating that aba2 was less dormant than WT (Fig. 2a). In contrast, seeds of cyp707a mutants, especially cyp707a2, germinated more slowly than WT seeds. This is consistent with previous results that showed cyp707a2 seeds are more dormant than those of cyp707a1 and cyp707a3 16. We then examined longevity, or survival rates, of the seeds after CDT. We found that seedling survival for aba2 was higher than WT after 14 days of CDT, whereas cyp707a2 was more sensitive to CDT and exhibited lower survival rates than WT after 3 and 7 days of CDT (Fig. 2b). These data support the hypothesis that dormancy status is negatively correlated with the longevity of mature dry seeds.
Figure 2.
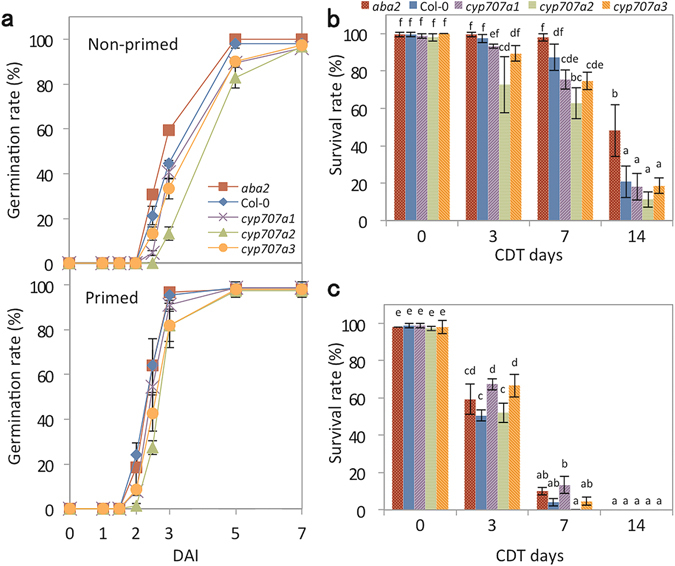
Effect of priming on germination and longevity of ABA-deficient and ABA over-accumulating mutants. (a) Germination speed of wild type Col-0 (Col-0), aba2, cyp707a1, cyp707a2 and cyp707a3 before (non-primed seeds; upper panel) and after (primed; lower panel) priming without CDT. (b) Survival rates of non-primed Col-0, aba2, cyp707a1, cyp707a2 and cyp707a3 seeds after 0, 3, 7 and 14 days of CDT. (c) Survival rates Col-0, aba2, cyp707a1, cyp707a2 and cyp707a3 seeds after priming and 0, 3, 7 and 14 days of CDT. Values presented are means ± SD of three replicates. Different letters in (b) and (c) indicate significant differences (P < 0.05, Tukey-Kramer tests).
The effect of priming on germination speed and longevity was also determined using the same mutants (Fig. 2a). The priming treatment itself increased germination speed for all genotypes. In addition, germination rates became synchronous for all the genotypes suggesting that the priming treatment reduced ABA-mediated seed dormancy. When the primed seeds were subjected to CDT, the survival rates of all genotypes were similarly reduced depending on the duration subjected to CDT and no significant differences were observed between them (Fig. 2c). These results indicate that the dormancy status of seeds prior to priming does not affect the reduction of longevity resulting from priming.
Natural variation in seed longevity after priming
To understand the genetic basis underlying the effect of priming on seed longevity, survival rates of primed and non-primed seeds were compared after CDT for 230 Arabidopsis natural variants (Fig. 3a and Supplementary Table S1). Among the accessions tested, we focused on Est-1 (Rd-0)17 that exhibits a relatively higher resistance to CDT than the reference accession Col-0 after priming (Fig. 3b and c). Survival rates of non-primed Est-1 and Col-0 seeds after 3 days of CDT were comparable, however, non-primed Est-1 seeds exhibited lower survival rates than Col-0 seeds when compared at 7 days of CDT (Fig. 3d). After priming, survival rates of Col-0 seeds were reduced by more than 60% on 3 days of CDT whereas those of Est-1 seeds were reduced by less than 20%. At 7 days of CDT, none of the primed Col-0 seeds were able to establish healthy seedlings in contrast to 33% of Est-1 seeds. These data suggest that Col-0 and Est-1 differ in mechanisms occurring during priming that subsequently impact seed longevity. In non-CDT aged seeds, the germination speed of both accessions either with or without priming was equivalent (Fig. 3e), which indicates that the dormancy status and germination speed of Col-0 and Est-1 were comparable.
Figure 3.
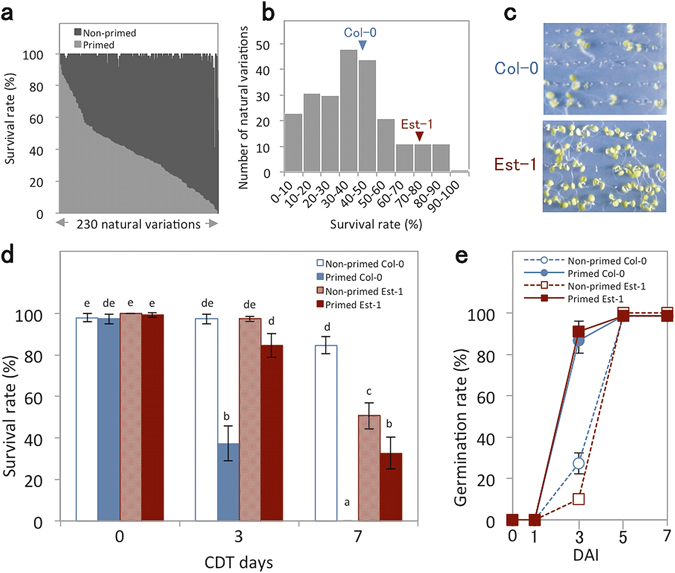
Seed longevity of Arabidopsis natural variants. (a) Survival rates of 230 Arabidopsis natural variations subjected to CDT for 3 days before (non-primed; dark grey bars) and after (primed; light grey bars) priming. (b) Frequency distribution of survival rates among the natural variants. (c) Photographs of Col-0 and Est-1 seedlings developed after priming and subsequent CDT for 3 days. (d) Survival rates of Col-0 and Est-1 before (non-primed) and after (primed) priming and subsequent CDT for 0, 3 and 7 days. (e) Germination speed of Col-0 and Est-1 before (non-primed) and after (primed) priming without CDT determined as the number of seedlings with green cotyledons. For (d) and (e), values presented are means ± SD of three replicates. Different letters in (d) indicate significant differences (P < 0.05, Tukey-Kramer tests).
QTL analysis of seed longevity after priming
To identify loci that account for the variations in seed longevity between the two accessions, QTL analysis was performed using 279 RILs derived from a cross between Col-0 and Est-1, which had been genotyped for 224 molecular markers18. Survival rates for seeds of the RILs following a CDT of 3 days were determined before and after priming (Fig. 4a and Supplementary Table S2). No clear correlation was apparent between seed longevity before and after priming (Supplementary Fig. S2). Based on the survival rate phenotypes, QTLs associated with seed longevity before and after priming were mapped using the interval mapping method (Fig. 4b). Comparison of significant QTLs indicated an overlap of a major QTL detected on chromosome (chr) 3 for non-primed and primed seeds. Other major QTLs were also identified on the lower arm of chr 1 for non-primed seeds and for primed seeds, on the upper arm of chr 1, lower arm of chr 2 and lower arm of chr 5. These suggest that seed longevity of non-primed and primed seeds were determined largely by different genetic factors although longevity at the initial state in mature dry seeds influences seed longevity after priming to some extent. To eliminate the effects of primary longevity in non-primed seeds, we calculated the reduction rates of seed longevity caused by priming in the RILs [(Non-primed) − (primed)/(Non-primed) × 100] (Supplementary Table S2), and mapped QTLs based on this parameter (Fig. 4c). This validated three major QTLs associated with the reduction of seed longevity by priming on chr 1 (LOD 5.17), chr 2 (LOD 3.05) and chr 3 (LOD 4.60). For all these QTLs, Col-0 alleles contributed to the loss of seed longevity in response to priming (Supplementary Fig. S3). To evaluate the explained variance (%), we also analyzed these QTLs with the multiple QTL model and the results are summarized in Supplementary Table S3. The intervals covered by the three QTLs contained a large number of annotated genes (Supplementary Table S4). Identification of causal genes for QTLs by fine mapping generally requires a relatively long time. Thus, in the present study, we rather focused on the characterization of molecular mechanisms involved in the loss of seed longevity during priming than the identification of the causal genes.
Figure 4.
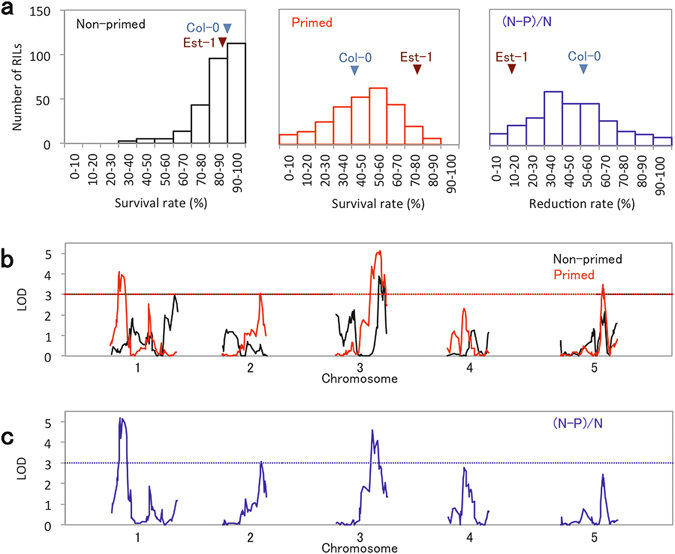
Identification of QTLs for seed longevity. (a) Frequency distributions of survival rates in 279 RILs before (non-primed; left panel) and after (primed; middle panel) priming after 3 days of CDT. Reduction in survival rates caused by priming are also shown [(N − P)/N; right panel]. (b) QTL mapping of seed longevity determined as survival rates after 3 days of CDT before (non-primed) and after (primed) priming. (c) QTL mapping of seed longevity determined as reduction of survival rates by priming [(N − P)/N]. Dotted lines in (b) and (c) indicate threshold (LOD = 3, P < 0.026 from 10,000-time permutation).
Detection of priming responsive genes by RNA-Seq
To better understand the molecular events associated with priming, transcriptome analysis was performed by RNA-Seq using non-primed and primed seeds of Col-0 and Est-1. A positive correlation (R2 = 0.596) was found between the effects of priming on gene expression between Col-0 and Est-1 (Fig. 5a), indicating that many genes were commonly up- or down-regulated in both accessions during priming. These are tentatively named ‘priming responsive genes’ here and were determined using the list of top 1,000 genes whose expression was changed during priming in Col-0 and Est-1 and the TCC package19. From these, 385 genes were commonly up-regulated in Col-0 and Est-1 and termed ‘priming up-regulated genes’ (Fig. 5b and Supplementary Table S5), while 242 genes were down-regulated in both accessions and termed ‘priming down-regulated genes’ (Fig. 5b and Supplementary Table S6). To obtain an overview of the physiological processes in which the priming responsive genes are involved, Gene Ontology (GO) enrichment analysis for biological processes was performed using the PANTHER Classification System20. No significant enrichment of GO terms was observed for priming down-regulated genes, while 17 GO terms including ‘response to oxygen-containing compound’, ‘response to toxic substance’, ‘translation’, ‘response to acid chemical’ and ‘response to karrikin’ were significantly enriched in priming up-regulated genes (Supplementary Table S7).
Figure 5.
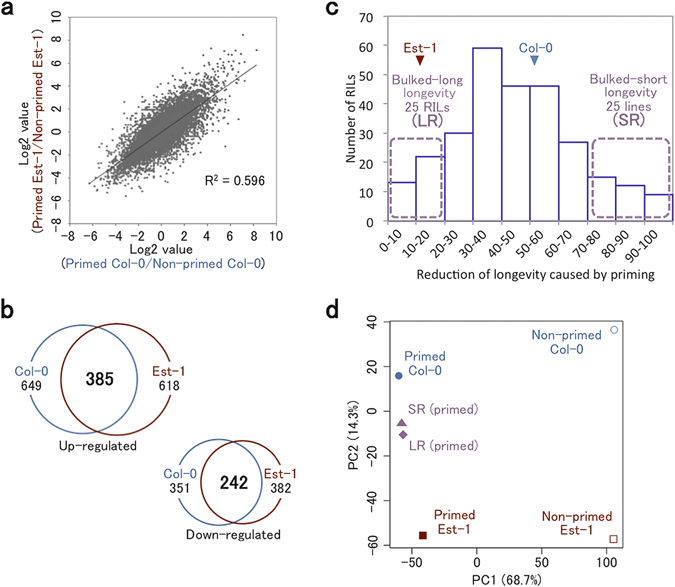
RNA-Seq analysis with seeds having different longevity. (a) Effect of priming on transcriptomes of Col-0 and Est-1 seeds. Fold changes in the gene expression levels by priming were compared between Col-0 and Est-1. (b) Comparison of top 1,000 genes whose expression was up- or down-regulated by priming between Col-0 and Est-1. (c) Frequency distribution of reduction rates of seed longevity after priming for the population of 279 RILs. The top 25 RILs with the greatest reduction were bulked as short RILs (SR). The top 25 RILs with the smallest reduction were bulked as long RILs (LR). (d) PCA of six transcriptome data obtained by RNA-Seq.
Bulked transcriptome analysis using RILs
In addition to the priming responsive genes that were similarly regulated in Col-0 and Est-1, there were many genes whose expression levels differed between the two accessions. The latter might contain key regulatory genes that contribute to the difference in seed longevity between Col-0 and Est-1 under the control of detected QTLs. To efficiently extract possible candidates for these key regulatory genes, we employed ‘bulked transcriptome analysis’21, 22. This approach normalizes the variability among RILs that is not related to longevity by analysis of RNA from bulks of RILs with similar phenotypes. Based on the survival rate phenotypes after priming (Fig. 4a), 25 RILs that were relatively tolerant and 25 that were relatively sensitive to CDT were grouped as bulked-long RILs (LR) and bulked-short RILs (SR), respectively (Fig. 5c), and RNA-Seq were performed on bulks of the primed seeds. Principal component analysis (PCA) was then performed with the 6 transcriptome datasets (non-primed Col-0, non-primed Est-1, primed Col-0, primed Est-1, primed LR and primed SR) (Fig. 5d). The first dimension (PC1) represented variation due to the effects of priming whereas the second dimension (PC2) reflected differences between Col-0 and Est-1, being consistent with the observation that many genes were commonly up- or down- regulated during priming in both Col-0 and Est-1 (Fig. 5a). As expected, only minor differences were observed between the transcriptomes of LR and SR after priming compared to the differences between primed Col-0 and Est-1. In addition, the transcriptomes of LR and SR after priming were intermediate between primed Col-0 and Est-1. Comparison of primed seeds with longer (Est-1 and LR) or shorter longevity (Col-0 and SR) identified 46 genes that are differentially expressed according to following criteria: (i) in the top 100 genes differentially expressed between LR and SR, (ii) more than 1.5-fold differences in normalized read counts between LR and SR, and (iii) more than 1.2-fold differences in normalized read counts between Col-0 and Est-1. Of these, 11 genes were highly expressed in the long life populations (Table 1) and 35 genes highly expressed in the short life populations (Table 2). Comparison of the 46 genes and three major QTL regions for the loss of seed longevity during priming (Fig. 4c) revealed that five (AT1G12650, AT1G12805, AT1G16550, AT1G19540, and AT1G20390), six (AT2G44460, AT2G45400, AT2G45510, AT2G45560, AT2G46450 and AT2G46950) and five (AT3G46000, AT3G46610, AT3G47350, AT3G48580 and AT3G49570) genes were located within the QTL regions on chr 1, 2 and 3, respectively (Tables 1 and 2). In addition, three genes related to brassinosteroid (BR) synthesis or signaling [BEN1 (AT2G45400)23, DWF1 (AT3G19820)24 and EXO (AT4G08950)25] and four cell wall modification-related genes [TRG1/XYL1 (AT1G68560)26, 27, EXPA1 (AT1G69530)28, EXPA2 (AT5G05290)29 and DUF642 (AT5G11420)30] were more highly expressed in the short life populations compared to the long life populations (Table 2). This implies that BR and cell wall-related genes could participate in the regulatory networks determining longevity of primed seeds possibly under the control of the causal genes responsible for the QTLs.
Table 1.
Genes highly expressed in primed seeds of high longevity genotypes (bulked-long RILs and Est-1).
| Gene ID | Gene expression levels (Normalized read counts) | Annotation | Overlapped QTL | |||
|---|---|---|---|---|---|---|
| SRa | LRb | Col-0 | Est-1 | |||
| AT1G12130 | 73.3 | 158.4 | 41.3 | 218.6 | Flavin-binding monooxygenase family protein | —c |
| AT1G16550 | 42.2 | 93.5 | 40.3 | 79.3 | Pseudogene, putative cell division cycle protein | chr 1 |
| AT1G64900 | 582.1 | 880.0 | 218.4 | 1060.7 | CYP89A2 | — |
| AT2G44460 | 23.8 | 66.0 | 10.3 | 16.1 | BGLU28 (beta glucosidase28) | chr 2 |
| AT2G46950 | 187.0 | 354.2 | 21.6 | 500.4 | CYP709B2 | chr 2 |
| AT3G44215 | 145.8 | 430.1 | 0.9 | 606.4 | Copia-like retrotransposon family protein | — |
| AT3G48580 | 131.1 | 253.0 | 100.3 | 422.1 | XTH11 (xyloglucan endotransglucosylase/hydrolase11) | chr 3 |
| AT3G49570 | 229.2 | 387.2 | 159.4 | 350.4 | LSU3 (response to low sulfur3) | chr 3 |
| AT3G60140 | 282.3 | 575.3 | 127.5 | 311.8 | BGLU30 (beta glucosidase30) | — |
| AT4G13180 | 463.8 | 706.2 | 476.3 | 595.7 | NAD(P)-binding Rossman-fold superfamily protein | — |
| AT5G48850 | 132.0 | 218.9 | 46.9 | 63.2 | SDI1 (sulfur deficiency-induced1) | — |
aSR; bulked-short RILs. bLR; bulked-long RILs. cNo overlap with the QTLs.
Table 2.
Genes highly expressed in primed seeds of short longevity genotypes (bulked-short RILs and Col-0).
| Gene ID | Gene expression level (Normalized read counts) | Annotation | Overlapped QTL | |||
|---|---|---|---|---|---|---|
| SRa | LRb | Col-0 | Est-1 | |||
| AT1G01300 | 265.8 | 172.7 | 300.9 | 162.9 | Eukaryotic aspartyl protease family protein | —c |
| AT1G03106 | 797.5 | 513.7 | 927.2 | 455.4 | Unknown protein | — |
| AT1G12650 | 44.0 | 14.3 | 55.3 | 4.3 | Unknown protein | chr 1 |
| AT1G12805 | 240.2 | 101.2 | 496.9 | 77.1 | Nucleotide binding protein | chr 1 |
| AT1G19540 | 235.6 | 93.5 | 309.4 | 76.1 | NmrA-like negative transcriptional regulator family protein | chr 1 |
| AT1G20390 | 62.3 | 9.9 | 73.1 | 0.0 | Gypsy-like retrotransposon | chr 1 |
| AT1G22110 | 138.4 | 73.7 | 302.8 | 114.6 | Structural constituent of ribosome | — |
| AT1G27340 | 99.0 | 46.2 | 74.1 | 61.1 | LCR (leaf curing responsiveness) | — |
| AT1G68560 | 268.6 | 141.9 | 237.2 | 49.3 | TRG1 (thermoinhibition resistant1) | — |
| AT1G69530 | 766.3 | 454.3 | 1154.1 | 173.6 | EXPA1 (expansin A1) | — |
| AT1G80530 | 238.3 | 149.6 | 229.7 | 186.4 | Major facilitator superfamily protein | — |
| AT2G10410 | 286.9 | 156.2 | 410.6 | 0.0 | SADHU1-1 (sadhu non-coding retrotransposon1-1) | — |
| AT2G14878 | 610.5 | 212.3 | 1054.7 | 9.6 | Non-coding RNA | — |
| AT2G14960 | 61.4 | 26.4 | 37.5 | 13.9 | GH3.1 (Protein similar to IAA-amido synthases) | — |
| AT2G16880 | 76.1 | 36.3 | 58.1 | 46.1 | Pentatricopeptide repeat superfamily protein | — |
| AT2G20670 | 33.0 | 8.8 | 17.8 | 8.6 | DUF506 (Protein of unknown function) | — |
| AT2G45400 | 18.3 | 2.2 | 15.9 | 4.3 | BEN1 (BRI1-5 ENHANCED1) | chr 2 |
| AT2G45510 | 256.7 | 166.1 | 239.1 | 159.6 | CYP704A2 | chr 2 |
| AT2G45560 | 246.6 | 134.2 | 298.1 | 48.2 | CYP76C1 | chr 2 |
| AT2G46450 | 55.0 | 22.0 | 101.3 | 2.1 | CNGC12 (cyclic nucleotide-gated channel12) | chr 2 |
| AT3G09210 | 83.4 | 38.5 | 56.3 | 34.3 | PTAC13 (plastid transcriptionally active13) | — |
| AT3G19820 | 74.3 | 27.5 | 58.1 | 48.2 | DWF1 (DWARF1) | — |
| AT3G27770 | 129.3 | 67.1 | 120.9 | 51.4 | Unknown protein | — |
| AT3G42052 | 19.3 | 2.2 | 16.9 | 0.0 | Gypsy-like retrotransposon | — |
| AT3G43580 | 63.3 | 20.9 | 45.9 | 0.0 | Beta-galactosidase related protein | — |
| AT3G43955 | 61.4 | 20.9 | 53.4 | 0.0 | Copia-like retrotransposon | — |
| AT3G44430 | 136.6 | 36.3 | 165.0 | 0.0 | Unknown protein | — |
| AT3G46000 | 221.8 | 138.6 | 190.3 | 122.1 | ADF2 (actin depolymerizing factor2) | chr 3 |
| AT3G46610 | 80.7 | 36.3 | 92.8 | 16.1 | Pentatricopeptide repeat superfamily protein | chr 3 |
| AT3G47350 | 82.5 | 27.5 | 85.3 | 0.0 | HSD2 (hydroxysteroid dehydrogenase2) | chr 3 |
| AT4G08950 | 193.4 | 105.6 | 41.3 | 30.0 | EXO (exordium) | — |
| AT4G10390 | 30.3 | 7.7 | 21.6 | 10.7 | Protein kinase superfamily protein | — |
| AT4G36880 | 1181.6 | 716.1 | 1543.1 | 313.9 | CP1 (cysteine proteinase1) | — |
| AT5G05290 | 102.7 | 35.2 | 134.1 | 8.6 | EXPA2 (expansin A2) | — |
| AT5G11420 | 345.6 | 192.5 | 354.4 | 75.0 | DUF642 (protein of unknown function) | — |
aSR; bulked-short RILs. bLR; bulked-long RILs. cNo overlap with the QTLs.
Effects of BR on seed longevity after priming
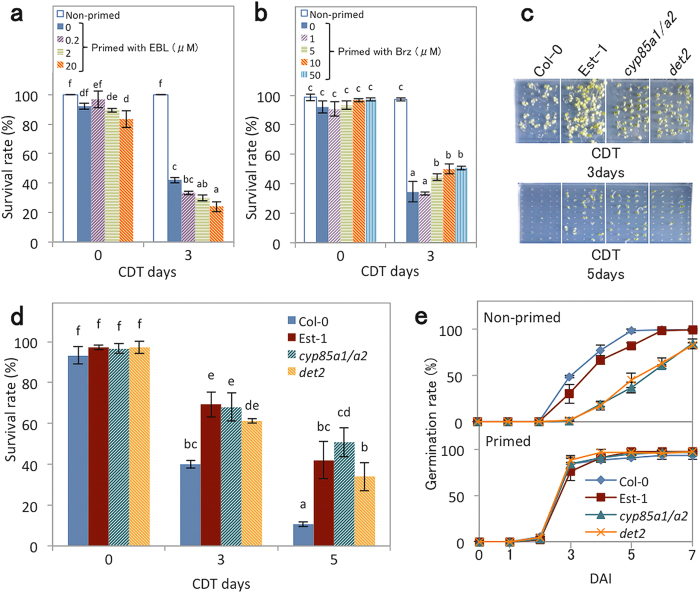
To examine the effect of BR on longevity of primed seeds, Col-0 seeds were subjected to CDT after priming in the presence of a bioactive BR (24-epibrassinolide; EBL) or a BR biosynthesis inhibitor (brassinazole; Brz31) (Fig. 6a and b). EBL promoted the reduction of longevity after priming in a concentration dependent manner (Fig. 6a). Conversely, when Brz was present at concentrations higher than 5 μM during priming, seed longevity was reduced less (Fig. 6b). Furthermore, the reduction of endogenous levels of BR in the BR-deficient mutants cyp85a1 cyp85a2 (cyp85a1/a2)32 and det2 33, in the Columbia, also increased CDT survival rates after priming to levels similar to Est-1 (Fig. 6c and d), although cyp85a1/a2 was significantly more resistant to CDT than other genotypes even before priming (Supplementary Fig. S4). These results suggest that BR promotes the loss of seed longevity during priming. The germination speed of cyp85a1/a2 and det2 before priming was slower than that of Col-0 and Est-1, whereas priming treatment efficiently enhanced the speed of both cyp85a1/a2 and det2, to levels similar to Col-0 and Est-1 (Fig. 6e).
Figure 6.
Effect of BR on seed longevity. (a) Survival rates of wild type Col-0 non-primed seeds or seeds primed with 0, 0.2, 2 or 20 μM EBL after 0 or 3 days of CDT. (b) Survival rates of wild type Col-0 non-primed seeds or seeds primed with 0, 1, 5, 10 or 50 μM Brz after 0 or 3 days of CDT. (c) Germination of wild type Col-0, Est-1, cyp85a1 cyp85a2 (cyp85a1/a2) and det2 after priming and subsequent CDT for 3 or 5 days. Photos were taken after 7 days of imbibition. (d) Survival rates of wild type Col-0, Est-1, cyp85a1/a2 and det2 after priming and subsequent CDT for 0, 3 or 5 days. (e) Germination speed of wild type Col-0, Est-1, cyp85a1/a2 and det2 before (non-primed; upper panel) and after (primed; lower panel) without CDT. For (a), (b), (d) and (e), values presented are means ± SD of three replicates. Wild type Col-0 (Col-0). Different letters in (a), (b) and (d) indicate significant differences (P < 0.05, Tukey-Kramer tests).
Influence of the seed coat on longevity of primed seed
Seed longevity largely relies on viability of the embryos. Nevertheless, although the seed coat that surrounds embryos is formed of maternally derived dead cell layers, mutants that have defects in seed coat properties (e.g. structure and pigmentation) exhibited reduced seed longevity6, 34. This led us to investigate the contribution of the seed coat to the loss of seed longevity during priming. Since it has been reported that seed coat permeability can be evaluated by tetrazolium staining34, we incubated primed and non-primed seeds of Col-0, Est-1, cyp85a1/a2 and det2, with tetrazolium salts for 24 h (Fig. 7a). Non-primed seeds of all genotypes were almost not stained and did not show notable differences in the number of stained seeds (Fig. 7b). In contrast, after priming, significantly more seeds were stained in Col-0, but not in Est-1, suggesting that changes in seed coat properties might be associated with the reduction of seed longevity during priming. In cyp85a1/a2 and det2 the numbers of stained seeds were also increased by priming, however, they were significantly less than for wild type Col-0, implying that seed coat permeability might be at least partially regulated by BR during priming.
Figure 7.
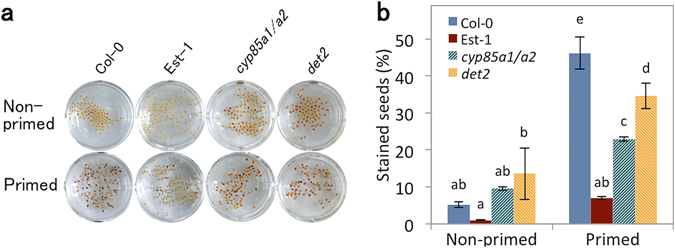
Changes in seed coat permeability after priming. (a) Tetrazolium staining of wild type Col-0 (Col-0), Est-1, cyp85a1 cyp85a2 (cyp85a1/a2) and det2 seeds before (non-primed) and after (primed) priming. (b) Numbers of stained seeds (red colored seeds) before (non-primed) and after (primed) priming for Col-0, Est-1, cyp85a1/a2 and det2. Values presented are means ± SD of three replicates. Different letters indicate significant differences (P < 0.05, Tukey-Kramer tests).
Discussion
Seed priming is a treatment used to enhance seed performance prior to their commercialization. Although the treatment often affects post germination events such as seedling growth (vigor) and resistance to biotic and abiotic stress, here we have focused on the relationship between the promotion of germination and the loss of longevity caused by the treatment. Germination can be considered as the process during which a mature dry seed gradually loose its seed-specific characters and become a seedling, and the loss of seed longevity is a part of the program. Thus, studying the molecular mechanisms involved in the loss of seed longevity during priming is important not only for agricultural applications but also for understanding basic seed physiology. In the present study, hydropriming was used in combination with stratification (or thermoprming) for the assay because it is simple and sufficient to determine the effect of priming on seed germination and longevity (Fig. 1).
QTL analysis using six RIL populations has previously been used to demonstrate that seed dormancy and longevity in mature dry seeds is negatively correlated with DOG QTLs13. Similarly, our present study showed that the dormancy status of Col-0, aba2 and cyp707a2 seeds before priming was negatively correlated with seed longevity, although this is contradictory to the observation that the ABA deficient aba1-5 mutant had shorter longevity than the wild type Columbia background under natural aging. No significant differences were, however, observed in longevity when seeds were subjected to priming, indicating that the reduced survival rate after priming was independent of initial seed dormancy levels.
In this study, we identified an accession, Est-1, which shows longer longevity after priming than the reference accession Col-0 (Fig. 3d). Interestingly, longevity of Est-1 seeds before and after priming did not change drastically, compared to Col-0 seeds, which rapidly lost their longevity upon priming. This suggests that Col-0 and Est-1 have different germination programs. QTL mapping with RILs derived from a cross between Col-0 and Est-1 identified three genomic regions associated with the reduction of longevity during priming. Even though the two accessions showed large differences in this trait, it is generally difficult to extract important gene expression profiles responsible for a unique character by just simply comparing transcriptomes of different accessions. Transcriptomes of germinating seeds are complex, because proteins required for germination are translated not only from mRNAs newly transcribed upon imbibition, but also from mRNA templates that are synthesized during seed maturation and pre-existing in the mature dry seeds (stored mRNA or long-lived mRNA35–37). In addition, transcriptomes in dry seeds of less-dormant Columbia and dormant Cvi accessions resemble each other, suggesting that patterns of stored mRNA accumulation reflect neither the degree of dormancy nor germination potential, but rather the developmental context such as seed maturation38. Here, a bulked transcriptome analysis using RILs, allowed the identification of BR and cell wall-related genes that may contribute to the difference in seed longevity between the two accessions. In effect, BR-deficient mutants exhibited improved longevity after priming compared to WT and priming with a bioactive BR (EBL) and a BR biosynthesis inhibitor (Brz) reduced or prolonged seed longevity, respectively. These data strongly suggest that BR is involved in the loss of longevity during priming and that bulked transcriptome analysis with RILs can be an effective approach for predicting genes involved in traits differing between accessions. Several genes associated with short longevity after priming were found in the QTL regions associated with the loss of seed longevity by priming (Tables 1 and 2). However, a number of single nucleotide polymorphisms (SNPs) were present in the ORF plus 1 kb upstream regions of these genes (Supplementary Table S8). Further fine mapping will be required to identify the causal genes and SNPs responsible for the genetic variations. Characterization of mutants defective in the candidate gens will be also required to determine their involvement in the loss of seed longevity during priming. These studies would contribute to improve seed longevity after priming in crop species.
BR is a plant hormone that regulates a wide range of developmental and physiological processes39, 40. Although little is known about the relationship between BR and seed longevity, it has been shown that BR regulates seed dormancy and/or germination. In Arabidopsis, germination of the BR deficient mutant det2 and the BR-insensitive mutant bri1 is more strongly inhibited by ABA than WT41. The same study also showed that BR treatment induces germination of the severe GA-deficient mutant ga1-3, which normally requires exogenous GA for germination. These data suggest that BR promotes seed germination and are consistence with our observation that BR deficient mutants cyp85a1/a2 and det2 exhibited reduced germination rates compared to WT in the absence of priming (Fig. 6e). We speculate that BR mediates part of the germination process induced during priming, as a result this is only partially initiated in BR-deficient mutants so that seed longevity after priming is less affected compared to WT. Since the effect of BR on seed germination is opposite to that of ABA, one might speculate that BR indirectly affects seed longevity during priming by mediating ABA signaling. However Fig. 2c shows that dormancy levels (or germination speed) determined by ABA do not affect the reduction of longevity during priming. Thus it is likely that the effect BR on the reduction of seed longevity during priming is independent from its effects on ABA signaling.
Cell expansion is dependent on the tension of cell walls of which major components are the cellulose microfibrils and the pectin/hemicellulose matrix. Loosening of the wall allows water influx to drive cell expansion and generate cellular turgor pressure42. Embryonic cell expansion and endosperm weakening during seed germination are controlled by cell wall modification enzymes26. We found that four cell wall modification-related genes TRG1/XYL1 (AT1G68560), EXPA1 (AT1G69530), EXPA2 (AT5G05290) and DUF642 (AT5G11420) were highly expressed in seeds of bulked RILs that had relatively short longevity after priming (Table 2). TRG1/XYL1 encodes an α-xylosidase that is required for xyloglucan maturation and is supposed to be a germination suppressor, because mutants defective in the corresponding gene showed reduced dormancy, thermoinhibition-resistant germination, and resistance to the GA biosynthesis inhibitor paclobutrazol during germination26, 27. The phenotypes of trg1/xyl1 mutant are possibly associated with altered endosperm cell wall composition that impact on its resistance26. In contrast, overexpression of DUF642 increased pectin methylesterase (PME) activity and promoted testa and endosperm rupture in response to matrix based priming30. It has been reported that DUF642 interacts with catalytic domain of AtPME343. This is in accord with the promotion of testa rupture and enhanced testa permeability observed on exogenous treatment of garden cress (Lepidium sativum) seeds with PME during imbibition44. PME controls the methylesterification status of pectin and thereby determines the biophysical properties of cell walls. Interestingly, a relationship between PME action and BR signaling was previously reported. Several growth defects caused by the over-expression of a PME inhibitor, AtPMEI5, were suppressed by a mutation in the BR receptor BRI145. BR has previously been implicated in the activation of a large number of genes encoding cell wall-related enzymes, such as cellulose synthase, pectinesterases, xyloglucosyl transferases, and expansins39. EXP1 was up-regulated by BR39, while mutants defective in EXPA2 showed delayed germination29. These observations suggest that BR-mediated cell wall modification influence the reduction of seed longevity during priming. While TRG1/XYL1 is a negative regulator of germination, DUF642 and EXPA2 promote germination, suggesting that diverse cell wall modifications have different effects on germination and longevity.
Tetrazolium staining demonstrated that the priming treatment increased seed coat permeability (Fig. 7). This is in accord with the high seed coat permeability and reduced seed longevity exhibited by transparent testa (tt) mutants affected in the synthesis of the flavonoids anthocyanins and proanthocyanidins34. Moreover, wrky3 and nfxl1 mutants defective in defense-related transcription factors also have increased seed coat permeability and impaired acquisition of longevity during seed maturation46. These suggest a link between seed coat functions and longevity. The Arabidopsis TT10 laccase is present in the young colorless seed coat, and the oxidation of soluble proanthocyanidins into quinonic compounds by TT10 laccase might increase their capacity to bind to the cell wall during seed desiccation. It has also been pointed out that the formation of antimicrobial quinones and insoluble polymers results in the reinforcement of the seed coat as a barrier to water/oxygen permeation, mechanical damage and biotic/abiotic stresses47. In the present study, we showed that improved seed survival after priming is associated with a better maintenance of seed coat impermeability in Est-1, cyp85a1/a1 and det2 compared with WT Col-0. As mentioned above, BR-mediated cell wall modifications might be involved in the loss of seed longevity during priming. We propose that BR directly or indirectly modulates seed coat permeability during priming.
Methods
Plant materials and growth conditions
The aba2 mutant (aba2-2) used in this study was isolated by Nambara et al. (1998)14. The cyp707a1, cyp707a2 and cyp707a3 mutants (cyp707a1-1, cyp707a2-2 and cyp707a3-1, respectively) were isolated as described previously15, 16. The cyp85a1/a2 mutant (cyp85a1-1 cyp85a2-2) was isolated by Nomura et al.32. To isolate the det2 mutant used in this study, seeds for the T-DNA insertion line CS873725 were obtained from the Arabidopsis Biological Resource Center (ABRC) (http://abrc.osu.edu/). The homozygous mutant was selected from its visual phenotype. T-DNA insertion of the det2 mutant was confirmed by PCR using primers BP (5′-TCAGAAATGGATAAATAGCCTTGCTTC-3′) and RP (5′-CTTGGCAATGTACCACTTGTGACTC-3′) based on their ability to amplify a fragment, and homozygous insertion was confirmed by PCR using primers LP (5′-CCAAGTGGCCAAAACCTAGCTTC-3′) and RP based on their inability to amplify a fragment. Col-0 was used as the wild type control for these mutants. The 230 Arabidopsis natural variations used in this study were obtained from the RIKEN BioResource Center (http://www.brc.riken.jp/). The set of 279 RILs derived from the Est-1 × Col-0 (CS39289) were obtained from Arabidopsis Biological Resource Center.
Seeds were surface-sterilized in a solution containing 5% (v/v) NaClO and 0.05% (v/v) Tween 20, rinsed with water, and sown on Murashige and Skoog media [one-half strength Murashige and Skoog salts, MES (0.5 g/l), pH 6.5] containing 0.8% (w/v) agar. After stratification at 4 °C for 3 days in the dark, plates were incubated at 22 °C under continuous light (approximately 10 W/m2). To propagate seeds, about 1-week-old seedlings were transplanted on pots containing vermiculite and Metro-mix 350 at a 3:1 ratio and grown in growth chambers at 22 °C under continuous white light and supplemented with nutrients (1/1,000 dilution of Hyponex; Hyponex Japan).
Physiological analysis
For priming treatments, surface-sterilized seeds in 1.5 ml tubes were imbibed at 4 °C for 3 days in the dark and then at 22 °C for 12 h under the light. The seeds were dried on filter papers under a laminar flow cabinet for 12 h. For seed priming with EBL or Brz, the stock solutions (200 × concentration) prepared in DMSO were added to the water after sterilization, and water containing 0.5% (v/v) DMSO were used for control experiments.
To evaluate longevity, seeds were subjected to CDT by storing them in a sealed box with an open petri dish containing saturated KCl solution for 0 to 20 days at 37 °C. Relative humidity inside the box was maintained at 80%. Germination assays were performed on Murashige and Skoog media containing 0.8% (w/v) agar with 50 seeds in triplicates. Germination speed was defined as radicle emergence from seed coat or as opening of green cotyledons as described in the figure legends. Seed survival after CDT was defined as establishment of seedlings with green opened cotyledons at 7 DAI.
Seed coat permeability was assessed by staining 50 to 100 seeds with 1% (w/v) 2,3,5-triphenyltetrazolium chloride solution at 30 °C for 24 h. The number of red-colored seeds was counted under a stereomicroscope.
QTL analysis
Survival rates of primed and non-primed seeds after 3 days of CDT were used as phenotype data of 279 RILs. Normalized seed longevity values after priming were calculated as follows: [(Non-primed seed longevity) − (Primed seed longevity)/(Non-primed seed longevity) × 100]. Genotype data of the RILs were obtained from The Arabidopsis Information Resource (ftp://ftp.arabidopsis.org/home/tair/Maps/). QTL analysis was performed using the R-qtl package48 implemented in R (http://www.r-project.org).
Transcriptome analysis
Total RNA was prepared from 20 mg primed or non-primed seeds by acid phenol extraction and lithium chloride precipitation as previously described49. For bulked transcriptome analysis, the top 25 RILs with either longer or shorter longevity were selected for bulks. Total RNA was isolated from primed seeds of individual RILs, and 1 μg of the total RNA from 25 RILs was mixed as LR or SR samples, respectively. Preparation of mRNA-Seq library and sequencing were performed via a commercial service (Eurofins Genomics K.K., Tokyo, Japan). Paired-end libraries were sequenced (2 × 125 bp module) with Illumina Hiseq-2500 version 4 chemistry. Sequenced paired-end reads were mapped onto the Arabidopsis genome (TAIR version 10) obtained from Ensembl Plants (http://plants.ensembl.org/info/website/ftp/index.html) using the Bowtie2 (version 2.2.6)50. Reads mapped onto exon regions were counted using featureCounts (version 1.5.0-p1)51, and summarized in Supplementary Table S9. Normalization of read counts and ranking of differentially expressed genes were performed using the R package TCC19. Based on the normalized read counts, PCA was performed using the prcomp function of R. The GO enrichment analysis was performed using annotation data set ‘GO biological process complete’ (release 20161027) by the PANTHER Classification System (release 20160715)20 with the Bonferroni correction for multiple testing. SNPs information in Supplementary Table S8 was obtained from the 1001 Genomes portal (http://1001genomes.org/).
Statistical analysis
Tukey-Kramer tests were used to determine significant differences in multiple comparisons of seed longevity and seed coat permeability.
Data deposition
RNA-Seq data in this publication have been deposited in DRA (http://trace.ddbj.nig.ac.jp/dra) under accession numbers DRA005725.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This study was supported by RIKEN Special Postdoctoral Researcher Program to N.S. We thank the Arabidopsis Biological Resource Center (ABRC) for providing T-DNA insertion mutants, and RIKEN BioResource Center which is participating in the National Bio-Resource Project of the MEXT, Japan for natural variants. We also thank Ms. Masako Tanaka (RIKEN CSRS) for her assistance with plant growth and seed harvesting.
Author Contributions
N.S. and M.S. designed the experiments. T.N. isolated the det2 mutant. N.S. performed physiological analysis and transcriptome analysis. J.S.K., Y.O., K.M., and M.O. conducted QTL analysis. N.S. and M.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08116-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Varierl A, Vari AK, Dadlani M. The subcellular basis of seed priming. Current Science. 2010;99:450–456. [Google Scholar]
- 2.Paparella S, et al. Seed priming: state of the art and new perspectives. Plant Cell Rep. 2015;34:1281–1293. doi: 10.1007/s00299-015-1784-y. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Bino RJ, van der Burgz WJ, Groot SPC, Hilhorst HWM. Effects of osmotic priming on dormancy and storability of tomato (Lycopersicon esculentum Mill.) seeds. Seed Science Research. 1996;6:49–55. doi: 10.1017/S0960258500003020. [DOI] [Google Scholar]
- 4.Rajjou L, Debeaujon I. Seed longevity: survival and maintenance of high germination ability of dry seeds. C. R. Biol. 2008;331:796–805. doi: 10.1016/j.crvi.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Waterworth W, Bray C, West C. The importance of safeguarding genome integrity in germination and seed longevity. J Exp Bot. 2015;66:3549–3558. doi: 10.1093/jxb/erv080. [DOI] [PubMed] [Google Scholar]
- 6.Sano N, et al. Staying Alive: Molecular Aspects of Seed Longevity. Plant Cell Physiol. 2016;57:660–674. doi: 10.1093/pcp/pcv186. [DOI] [PubMed] [Google Scholar]
- 7.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collard B, Jahufer M, Brouwer J, Pang E. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica. 2005;142:169–196. doi: 10.1007/s10681-005-1681-5. [DOI] [Google Scholar]
- 9.He H, et al. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J Exp Bot. 2014;65:6603–6615. doi: 10.1093/jxb/eru378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajjou L, et al. Proteome-Wide Characterization of Seed Aging in Arabidopsis: A Comparison between Artificial and Natural Aging Protocols. Plant Physiol. 2008;148:620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verma P, et al. Protein L-Isoaspartyl Methyltransferase2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol. 2013;161:1141–1157. doi: 10.1104/pp.112.206243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clerkx E, Vries H, Ruys G, Groot S, Koornneef M. Genetic differences in seed longevity of various Arabidopsis mutants. Physiologia Plantarum. 2004;121:448–461. doi: 10.1111/j.0031-9317.2004.00339.x. [DOI] [Google Scholar]
- 13.Nguyen TPP, Keizer P, van Eeuwijk F, Smeekens S, Bentsink L. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol. 2012;160:2083–2092. doi: 10.1104/pp.112.206649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nambara E, Kawaide H, Kamiya Y, Naito S. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant and Cell Physiol. 1998;39:853–858. doi: 10.1093/oxfordjournals.pcp.a029444. [DOI] [PubMed] [Google Scholar]
- 15.Kushiro T, Okamoto M, Nakabayashi K. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto M, et al. CYP707A1 and CYP707A2, Which Encode Abscisic Acid 8′-Hydroxylases, Are Indispensable for Proper Control of Seed Dormancy and Germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon M, et al. DNA fingerprinting and new tools for fine scale discrimination of Arabidopsis thaliana accessions. Plant J. 2012;69:1094–1101. doi: 10.1111/j.1365-313X.2011.04852.x. [DOI] [PubMed] [Google Scholar]
- 18.Balasubramanian S, et al. QTL Mapping in New Arabidopsis thaliana Advanced Intercross-Recombinant Inbred Lines. PLoS ONE. 2009;4:e4318. doi: 10.1371/journal.pone.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Nishiyama T, Shimizu K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloosterman B, et al. From QTL to candidate gene: genetical genomics of simple and complex traits in potato using a pooling strategy. BMC genomics. 2010;11:158. doi: 10.1186/1471-2164-11-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandit A, et al. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol Genet Genomics. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- 23.Yuan T, Fujioka S, Takatsuto S, Matsumoto S. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 2007;51:220–233. doi: 10.1111/j.1365-313X.2007.03129.x. [DOI] [PubMed] [Google Scholar]
- 24.Fujioka S, Yokota T. Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol. 2003;54:137–164. doi: 10.1146/annurev.arplant.54.031902.134921. [DOI] [PubMed] [Google Scholar]
- 25.Coll-Garcia D, Mazuch J, Altmann T, Müssig C. EXORDIUM regulates brassinosteroid-responsive genes. FEBS letters. 2004;563:82–86. doi: 10.1016/S0014-5793(04)00255-8. [DOI] [PubMed] [Google Scholar]
- 26.Sechet J, et al. Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiol. 2016;170:1367–1380. doi: 10.1104/pp.15.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigeyama T, et al. α-Xylosidase plays essential roles in xyloglucan remodelling, maintenance of cell wall integrity, and seed germination in Arabidopsis thaliana. J. Exp. Bot. 2016;67:5615–5629. doi: 10.1093/jxb/erw321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002;128:854–864. doi: 10.1104/pp.010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan A, et al. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS ONE. 2014;9:e85208. doi: 10.1371/journal.pone.0085208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zúñiga-Sánchez E, Soriano D, Martínez-Barajas E, Orozco-Segovia A, Gamboa-deBuen A. BIIDXI, the At4g32460 DUF642 gene, is involved in pectin methyl esterase regulation during Arabidopsis thaliana seed germination and plant development. BMC Plant Biol. 2014;14:338. doi: 10.1186/s12870-014-0338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asami T, et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura T, et al. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem. 2005;280:17873–17879. doi: 10.1074/jbc.M414592200. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi T, et al. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999;120:833–840. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debeaujon I, Léon-Kloosterziel KM. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajjou L, et al. The Effect of alpha-amanitin on the Arabidopsis Seed Proteome Highlights the Distinct Roles of Stored and Neosynthesized mRNAs during Germination. Plant Physiol. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sano N, et al. Proteomic analysis of embryonic proteins synthesized from long-lived mRNAs during germination of rice seeds. Plant Cell Physiol. 2012;53:687–698. doi: 10.1093/pcp/pcs024. [DOI] [PubMed] [Google Scholar]
- 37.Sano N, et al. Accumulation of long-lived mRNAs associated with germination in embryos during seed development of rice. J. Exp. Bot. 2015;66:4035–4046. doi: 10.1093/jxb/erv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura M, Nambara E. Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol Biol. 2010;73:119–129. doi: 10.1007/s11103-010-9603-x. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung Y, Choe S. The regulation of brassinosteroid biosynthesis in Arabidopsis. Critical reviews in plant sciences. 2013;32:396–410. doi: 10.1080/07352689.2013.797856. [DOI] [Google Scholar]
- 41.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schopfer P. Biomechanics of plant growth. Am J Bot. 2006;93:1415–1425. doi: 10.3732/ajb.93.10.1415. [DOI] [PubMed] [Google Scholar]
- 43.Zúñiga-Sánchez, E. & Buen, GA. The two DUF642 At5g11420 and At4g32460-encoded proteins interact in vitro with the AtPME3 catalytic domain. In Protein Interactions. (ed. Cai, J. & Wang, RE.) 119–142 (InTech, 2012).
- 44.Scheler C, et al. Promotion of Testa Rupture during Garden Cress Germination Involves Seed Compartment-Speciic Expression and Activity of Pectin Methylesterases. Plant Physiol. 2015;167:200–215. doi: 10.1104/pp.114.247429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol. 2012;22:1732–1737. doi: 10.1016/j.cub.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 46.Righetti K, et al. Inference of Longevity-Related Genes from a Robust Coexpression Network of Seed Maturation Identifies Regulators Linking Seed Storability to Biotic Defense-Related Pathways. Plant Cell. 2015;27:2692–2708. doi: 10.1105/tpc.15.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pourcel L, Routaboul JM, Kerhoas L, Caboche M. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broman KW, Wu H, Sen Aunak, Churchill GA. Bioinformatics. 2003. R/qtl: QTL mapping in experimental crosses; pp. 889–890. [DOI] [PubMed] [Google Scholar]
- 49.Oñate-Sánchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.