Abstract
Detection and elimination of virus-infected cells by cytotoxic T lymphocytes depends on recognition of virus-derived peptides presented by MHC class I molecules. A critical step in this process is the translocation of peptides from the cytoplasm into the endoplasmic reticulum by the transporter associated with antigen processing (TAP). Here, we identified the bovine herpesvirus 1-encoded UL49.5 protein as a potent inhibitor of TAP. The expression of UL49.5 results in down-regulation of MHC class I molecules at the cell surface and inhibits detection and lysis of the cells by cytotoxic T lymphocytes. UL49.5 homologs encoded by two other varicelloviruses, pseudorabies-virus and equine herpesvirus 1, also block TAP. Homologs of UL49.5 are widely present in herpesviruses, acting as interaction partners for glycoprotein M, but in several varicelloviruses UL49.5 has uniquely evolved additional functions that mediate its participation in TAP inhibition. Inactivation of TAP by UL49.5 involves two events: inhibition of peptide transport through a conformational arrest of the transporter and degradation of TAP by proteasomes. UL49.5 is degraded along with TAP via a reaction that requires the cytoplasmic tail of UL49.5. Thus, UL49.5 represents a unique immune evasion protein that inactivates TAP through a unique two-tiered process.
Herpesviruses share the conspicuous property that they cause a lifelong infection of their host. Persistence and repeated reactivation of herpesviruses is facilitated by specific evasion of the host immune response. CD8+ cytotoxic T lymphocytes (CTLs) play an important role in the control of viral infections through recognition of viral peptides presented by MHC class I molecules at the surface of infected cells. Consequently, inhibition of MHC class I-restricted antigen presentation represents an attractive strategy for viruses to avoid elimination by the host immune system (1, 2).
Newly synthesized heterodimers of MHC class I heavy chains and β2-microglobulin (β2m) are recruited into the MHC class I peptide-loading complex, a multicomponent assembly of endoplasmic reticulum (ER) proteins that facilitates the loading of high-affinity peptides into class I molecules (3–7). Within this complex, the MHC class I–β2m heterodimers are linked to the transporter associated with antigen processing (TAP) through tapasin. The TAP1/TAP2 heterodimer is a member of the ATP-binding cassette transporter superfamily (8–12). Peptide transport by TAP is a critical step in MHC class I antigen presentation. In the absence of a functional TAP transporter, most class I molecules are not loaded with peptides. They are retained within the ER and ultimately directed for proteasomal degradation (13).
TAP represents an attractive target for viruses that aim to elude CD8+ CTLs. Three different proteins specifically inhibiting peptide translocation by TAP have been described within the herpesvirus family. Herpes simplex virus 1 (HSV-1) and HSV-2 encode a soluble cytoplasmic protein, ICP47, that associates with the peptide binding site formed by the C-terminal cytosolic domains of TAP1 and TAP2, thereby acting as a high-affinity competitor for peptide binding (14–19). Human cytomegalovirus (HCMV) uses an ER-resident type I membrane protein, US6, to inhibit TAP (20–25). This glycoprotein does not affect peptide binding to TAP, but induces conformational changes within the transporter complex that prevent ATP binding and peptide translocation (20, 22, 23). The murine γ-herpesvirus 68-encoded K3 protein (mK3) inhibits antigen presentation by destabilizing MHC class I molecules, TAP, and tapasin (26–28). mK3 carries a cytoplasmic RING finger that catalyzes ubiquitination of substrates, a property essential for targeting MHC class I, TAP, and tapasin for degradation (26–28). Thus, ICP47, US6, and mK3 all inactivate the MHC class I peptide-loading complex, but each acts by a different mechanism.
Here, we identified the viral gene product that is responsible for CTL evasion by several varicelloviruses. Various members of the varicellovirus group down-modulate MHC class I expression (29–34). Among these viruses, bovine herpesvirus 1 (BHV1), pseudorabies virus (PRV), and equine herpesvirus 1 (EHV1) abolish peptide transport by TAP, resulting in down-regulation of MHC class I surface expression. However, neither the viral gene product(s) involved, nor the mechanisms underlying TAP inhibition have been elucidated. None of these viruses encodes a homolog of ICP47, US6, or mK3. In the absence of any genetic or structural characteristics that could support the identification of the viral gene product responsible for inhibition of TAP, we used a biochemical approach to identify the viral proteins involved. Inhibition of TAP by BHV1 relies on a small membrane protein, encoded by the UL49.5 gene. The UL49.5 protein blocks peptide transport by TAP and simultaneously induces degradation of TAP. The homologs of UL49.5 encoded by PRV (glycoprotein N) and EHV1 (the gene 10 product) also inactivate TAP. The present study thus identifies the UL49.5 protein encoded by the varicelloviruses BHV1, PRV, and EHV1 as an immune evasion molecule that uses a two-step reaction to impede TAP-driven peptide import into the ER.
Materials and Methods
Confocal microscopy and fluorescence recovery after photobleaching (FRAP) experiments were performed as described (24). In brief, a circular spot in the ER was bleached at full intensity, and an attenuated laser beam was used to monitor recovery of fluorescence. The half-time for recovery was calculated from each recovery curve after correction for loss of fluorescence caused by imaging (usually <4%). The diffusion coefficient D was determined from at least two independent FRAP experiments (at least eight cells per experiment). The calculated D, assuming 2D diffusion in the ER, is probably an underestimation of the actual D because of a time window between bleaching and the first recovery measurement of 1.2 s and the architecture of the ER.
DNA constructs, retroviral vectors, cell lines, viruses, reagents, and biochemical and immunological assays are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
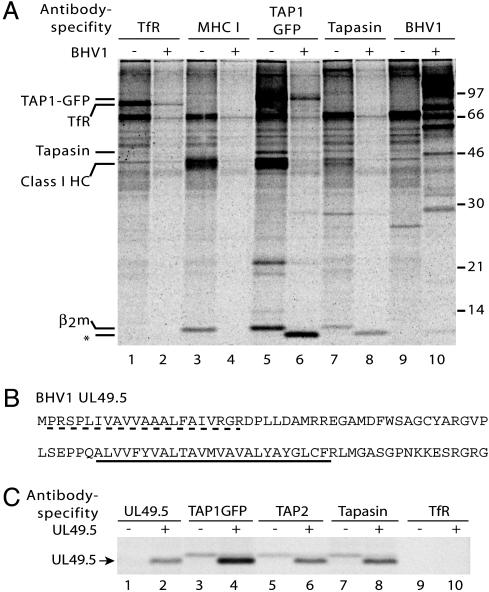
UL49.5 Interacts with the MHC Class I Peptide-Loading Complex. Previously, we have shown that infection of the human melanoma cell line Mel JuSo (MJS) with BHV1 results in down-regulation of MHC class I expression at the cell surface and a complete block of peptide transport by TAP (35). To identify viral proteins interacting with the class I peptide-loading complex, we used MJS cells stably transfected with the human TAP1 subunit tagged with GFP at its cytoplasmic carboxyl terminus (24). BHV1-infected and mock-infected MJS cells were metabolically labeled and solubilized in the presence of digitonin to preserve intermolecular interactions. Isolation of TAP1–GFP from noninfected MJS cells with anti-GFP antibodies yielded the TAP1–GFP fusion protein together with various other proteins, including constituents of the class I peptide-loading complex, such as tapasin, class I heavy chains, and β2m (Fig. 1A, lane 5). In BHV1-infected cells, detection of metabolically labeled host cell proteins is diminished because of a virus host shutoff (vhs) effect (32) (Fig. 1A, lanes 2, 4, 6, and 8). This effect is caused by the BHV1 UL41-encoded vhs protein, which inhibits synthesis of host cell polypeptides (32). As a result, radioactively labeled amino acids were incorporated mostly into newly synthesized viral proteins (Fig. 1A, lane 10).
Fig. 1.
The BHV1-encoded UL49.5 protein interacts with the MHC class I peptide-loading complex. (A) Mock-infected (–) or BHV1-infected (+) MJS TAP1–GFP cells were metabolically labeled, and digitonin lysates were subjected to immunoprecipitation by using antibodies against the transferrin receptor (TfR), MHC class I heavy chain–β2m–peptide complexes (W6/32, labeled MHC I), TAP1–GFP (antibody against the GFP part of the TAP1-GFP fusion protein), tapasin, and BHV1 (polyclonal BHV1 immune serum). Size markers are in kDa. * indicates coprecipitating 9-kDa viral protein. (B) Deduced amino acid sequence of the BHV1 UL49.5 protein. The predicted signal sequence (dashed line) and transmembrane domain (underlined) are indicated. (C) The UL49.5-encoded 9-kDa polypeptide interacts with TAP1, TAP2, and tapasin. MJS TAP1–GFP cells were transduced with control (–) or UL49.5 (+) retrovirus. Cells were metabolically labeled and lysed in the presence of digitonin, and immunoprecipitations were performed with the Abs indicated.
A 9-kDa protein was detected in TAP1–GFP and tapasin immunoprecipitates from infected cells (Fig. 1A, compare lanes 5 and 6 and lanes 7 and 8). This protein did not interact with W6/32-reactive MHC class I heavy chain–β2m–peptide complexes that have dissociated from the class I peptide-loading complex (Fig. 1A, lane 4), nor with the transferrin receptor (Fig. 1A, lane 2). These data indicate that a 9-kDa protein, most likely of viral origin, specifically interacts with the ER-resident MHC class I peptide-loading complex after infection with BHV1.
To characterize the 9-kDa protein, BHV1-infected cells were metabolically labeled with either [35S]methionine or [3H]leucine. The TAP complex was immunoprecipitated from cell lysates, and the 9-kDa polypeptide was isolated from the polyacrylamide gel and subjected to Edman degradation. Radiolabeled methionine residues were identified at positions 8 and 14 of the protein and leucine residues occurred at positions 4 and 5 (Fig. 8, which is published as supporting information on the PNAS web site). Among the 70 proteins encoded by BHV1, only the amino acid sequence of the UL49.5 gene product matched this profile (Fig. 1B). The UL49.5 gene is relatively conserved among varicelloviruses and encodes a type I membrane protein with a cleavable signal sequence (36, 37).
Stable expression of the BHV1 UL49.5 protein in MJS cells using a recombinant retrovirus resulted in synthesis of a 9-kDa polypeptide reacting with a UL49.5-specific antiserum (Fig. 1C, lane 2). The viral protein coprecipitated with TAP1–GFP, TAP2, and tapasin (Fig. 1C, lanes 4, 6, and 8 respectively). Thus, we identified the BHV1-encoded UL49.5 as a protein specifically interacting with the MHC class I peptide-loading complex.
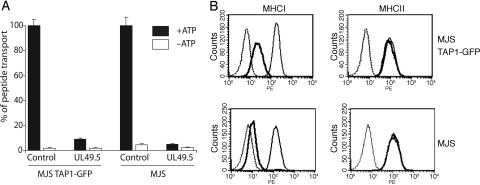
UL49.5 Prohibits CTL Recognition by Blocking TAP. To test whether the UL49.5 protein inhibits TAP, TAP-dependent peptide transport was measured in cells stably expressing UL49.5. A significant reduction of peptide translocation was observed in cells expressing the UL49.5 protein, thereby identifying the viral gene product as an inhibitor of TAP (Fig. 2A). The block in peptide transport coincided with a reduced expression of MHC class I molecules at the cell surface (Fig. 2B). Note that the reduction of class I surface expression was less stringent in MJS TAP1–GFP cells when compared with parental MJS cells.
Fig. 2.
The UL49.5 protein blocks MHC class I-restricted antigen processing and presentation. (A) TAP-dependent peptide transport is inhibited in cells expressing BHV1 UL49.5. Peptide transport is expressed as percentage of translocation, relative to the translocation observed in control cells (set at 100%). (B) Down-regulation of MHC class I surface expression by BHV1 UL49.5. Bold lines indicate UL49.5-expressing cells; thin lines indicate control cells; dashed lines indicate goat-anti-mouse phycoerythrin only.
To assess whether the reduction of MHC class I surface expression caused by BHV1 UL49.5 has functional consequences, we analyzed the capacity of UL49.5-expressing cells to present antigens for recognition by CD8+ T cells (Fig. 9, which is published on supporting information on the PNAS web site). Two well defined CTL clones were used, 5HO11 and 1R35, specific for the minor histocompatibility antigens HA-3 and HY, respectively (38–40). MJS cells and the B-cell derived lymphoblastoid cell line (B-LCL) MoDo express the immunogenic epitopes of the two minor antigens and were lysed by these specific CTL clones (Fig. 9). Upon expression of UL49.5, lysis of MJS cells and the B-LCL by the CTL clones was strongly diminished. UL49.5 thus inhibits TAP and strongly reduces MHC class I expression and antigen presentation to CD8+ cytotoxic T cells.
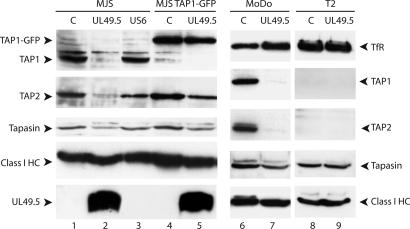
TAP1/TAP2 Protein Levels Are Reduced upon UL49.5 Expression. To investigate the molecular basis for inhibition of TAP-driven peptide transport by UL49.5, we first evaluated the integrity of the peptide-loading complex by immunoblotting. UL49.5 strongly reduced steady-state levels of TAP1 and TAP2 (Fig. 3, compare lanes 1 and 2) and also caused some reduction of tapasin expression (Fig. 3, lane 2). Steady-state levels of class I heavy chains appeared similar in control and UL49.5 cells, which may be related to the relative abundance of these molecules in the cells. For comparison, HCMV US6 did not influence TAP1, TAP2, or tapasin protein levels (Fig. 3, lane 3). Interestingly, the fusion of GFP to TAP1 counteracted the effects of UL49.5 (Fig. 3, lane 5). Note that TAP function was nevertheless inhibited in the UL49.5-expressing TAP1–GFP cells (Fig. 2A). Thus, two UL49.5-associated effects can be distinguished: inhibition of peptide transport by TAP (observed in MJS TAP1–GFP cells) and a reduction of TAP1/TAP2 protein levels (as found in the parental MJS cells). The observation that UL49.5 causes an intermediate down-regulation of MHC class I surface expression in TAP1–GFP cells (Fig. 2B) may be related to the fact that UL49.5 inhibits TAP activity without promoting TAP degradation. This results in a less stringent inhibition of class I expression.
Fig. 3.
Expression of UL49.5 results in reduced TAP1 and TAP2 protein levels. Steady-state protein levels were determined by Western blotting in MJS cells and EBV-transformed B cell lines MoDo and T2. C, control.
Because subunits of the class I peptide-loading complex are expressed at higher levels in B cells, we also determined the effects of UL49.5 in these cells. The dramatic loss of TAP1 and TAP2 expression was also seen in UL49.5-expressing B-LCL MoDo (Fig. 3, lane 7). Again, a moderate reduction of tapasin expression and virtually no effect on class I levels was observed in the presence of UL49.5.
In T2 cells, which lack TAP1 and TAP2, UL49.5 expression had no influence on tapasin levels, showing that this molecule is not directly targeted by the viral protein (Fig. 3, lanes 8 and 9). Moreover, in the absence of TAP, no interaction was observed between UL49.5 and tapasin or class I heavy chains (data not shown). From these experiments we conclude that UL49.5 causes a specific reduction of TAP1 and TAP2 protein levels.
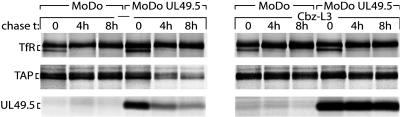
TAP and UL49.5 Are Degraded by the Proteasome. Pulse–chase experiments were performed to explain the observed reduction in TAP1 and TAP2 protein levels. UL49.5-expressing and control B-LCLs (MoDo) were metabolically labeled for 2 h, followed by a chase up to 8 h (Fig. 4). Whereas TAP was stable in control cells, UL49.5 expression reduced the half-life of TAP (Fig. 4 Left). UL49.5 did not affect the half-life of a control protein, the transferrin receptor (Fig. 4 Top). TAP is apparently destabilized by UL49.5 and degraded. Interestingly, UL49.5 itself was degraded with kinetics similar to that observed for TAP, suggesting codegradation. When the pulse–chase experiment was performed in the presence of the proteasome inhibitor Cbz-L3, TAP and UL49.5 were stabilized, indicating that degradation of these proteins was mediated by proteasomes (Fig. 4 Right).
Fig. 4.
UL49.5 targets TAP for degradation by proteasomes. EBV-transformed B cells transduced with retroviruses encoding UL49.5 in the antisense (MoDo) or sense (MoDo UL49.5) orientation were metabolically labeled and chased in the absence or presence of the proteasome inhibitor Cbz-L3 for the times indicated. Transferrin receptor (Tfr), TAP, and UL49.5 were immunoprecipitated from cell lysates and analyzed by SDS/PAGE.
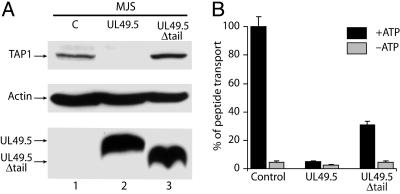
The Cytoplasmic Tail of UL49.5 Is Essential for TAP Degradation. To establish which domains of UL49.5 are involved in the inhibition and degradation of TAP, deletion mutants were constructed lacking the cytoplasmic tail, or the transmembrane domain and cytoplasmic tail. The latter mutant no longer blocked TAP (data not shown). In this respect, UL49.5 differs from US6, the soluble ER-luminal domain of which is sufficient to inhibit TAP function (20, 23, 25). Expression of the UL49.5 mutant lacking only its cytoplasmic tail (UL49.5Δtail) failed to cause degradation of TAP (Fig. 5A), but still inhibited peptide transport (Fig. 5B) and reduced MHC class I surface expression (data not shown), albeit less efficiently than WT UL49.5. These experiments reveal the two effects of UL49.5 on TAP. The cytoplasmic tail of UL49.5 is critical for TAP degradation but not for the inhibition of peptide translocation.
Fig. 5.
The cytoplasmic tail of UL49.5 is essential for degradation of TAP but is not required for inhibition of peptide transport. (A) Detection of TAP1, actin, and UL49.5 by immunoblotting. C, control. (B) Inhibition of peptide transport in MJS cells expressing full-length UL49.5 or UL49.5Δtail.
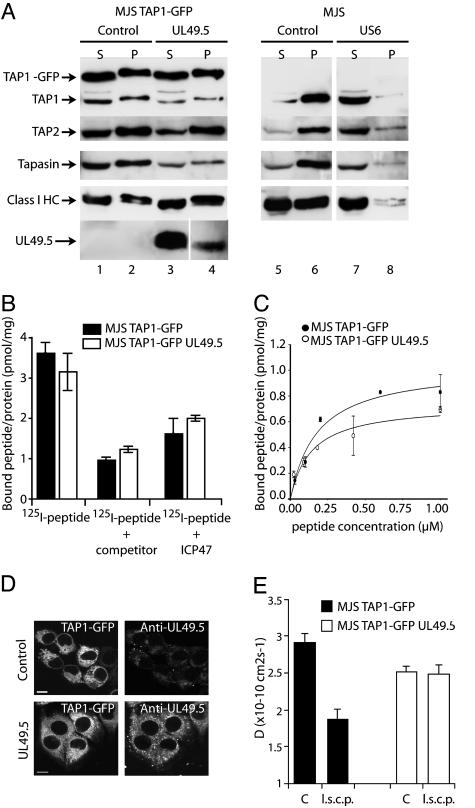
UL49.5 Does Not Block ATP or Peptide Binding by TAP. To investigate the mechanism of UL49.5-mediated inhibition of peptide transport, we evaluated two essential events of the peptide translocation process, binding of ATP and peptides to TAP. To determine the ATP-binding capacity of TAP, cells were lysed and incubated with ATP-agarose (25). Proteins bound to the ATP-agarose (Fig. 6A, pellet P) were eluted with EDTA and displayed next to the proteins in the unbound supernatant fraction (Fig. 6A, S). TAP1 and TAP2 bound to ATP-agarose in lysates from control cells and UL49.5-expressing cells (Fig. 6A, compare lanes 2 and 4), indicating that TAP retains the capacity to bind ATP in the presence of UL49.5. In UL49.5Δtail-transduced MJS cells not expressing TAP1-GFP, similar amounts of TAP1 and TAP2 could be retrieved in the presence and absence of UL49.5Δtail, which confirms the results obtained in TAP1–GFP cells (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 6.
UL49.5 does not block ATP or peptide binding by TAP but arrests the transporter in a translocation-incompetent state. (A) Postnuclear supernatants of digitonin-solubilized cells were incubated with ATP-agarose. Equal cell equivalents of the ATP-agarose bound pellet (P) and unbound supernatant (S) fractions were separated by SDS/PAGE and immunoblotted with Abs against the proteins indicated. (B and C) Peptides can bind to TAP in the presence of UL49.5. (B) Microsomal membranes were incubated with 1 μM 125I-labeled peptide RRYQKSTEL in the absence or presence of 400 μM unlabeled RRYQKSTEL (competitor) or in the presence of 200 μM ICP47. Each data point represents the mean of three measurements. (C) Microsomal membranes were incubated with increasing concentrations of the 125I-labeled peptide RRYQSTEL. Unspecific binding was determined in the presence of 200-fold excess of ICP47. The amount of specifically bound peptide per amount of microsomal protein is plotted against the peptide concentration and fitted with a 1:1 Langmuir equation (44). (D) TAP1–GFP (Left) and UL49.5 distribution (Right) in MJS TAP1–GFP cells. Cells were fixed and stained with anti-UL49.5 antibodies. (Scale bar: 5 μm.) (E) Analysis of TAP activity in vivo as measured by lateral mobility of the TAP complex (D, diffusion coefficient). Cells were microinjected with long side-chain peptides (l.s.c.p.) as indicated. C, control.
Tapasin and class I heavy chains, but also UL49.5, were detected in the ATP-agarose fraction in UL49.5-expressing cells (Fig. 6A, lane 4), indicating that the viral protein associates with peptide-loading complexes capable of binding ATP. UL49.5 blocks TAP through a different mechanism than US6 (Fig. 6A, compare lanes 6 and 8) (23, 25).
The HSV-encoded ICP47 blocks the association of peptides with the cytosolic peptide-binding domain of TAP (14–19). To test whether UL49.5 acts in a similar fashion, peptide binding to TAP was evaluated by using microsomes isolated from UL49.5-expressing and control cells (Fig. 6B). The results show that peptides associate with TAP irrespective of UL49.5 expression (Fig. 6B). When peptide affinity was evaluated at different concentrations, we did not detect a significant change in the Kd value in the presence or absence of UL49.5 (Kd = 160 nM; Fig. 6C). A slight reduction of maximal peptide binding was observed, which may be related to the fact that the level of TAP expression was slightly lower in the presence of UL49.5. Taken together, these results indicate that UL49.5 does not act by affecting the binding of ATP or peptides to TAP.
UL49.5 Arrests TAP in a Translocation-Incompetent State. To define the mechanism underlying the UL49.5-mediated inhibition of TAP, we investigated the mobility of the GFP-tagged TAP complex in the ER membrane by using FRAP. TAP1–GFP-transfected MJS cells display a typical ER staining pattern for TAP1–GFP (Fig. 6D). The translocation cycle is associated with conformational changes within the TAP complex reflected by alterations in the lateral diffusion rate of TAP within the ER membrane. The lateral mobility of TAP molecules is inversely proportional to TAP activity, as peptide-transporting TAP molecules diffuse at a slower rate than inactive, closed TAP complexes (24). In UL49.5-expressing cells, TAP showed a somewhat slower diffusion rate under normal conditions when compared with cells not expressing UL49.5 (Fig. 6E, compare filled and open bars C). This finding suggests that UL49.5 interaction with the TAP complex has an effect on the conformation of the complex, thus reducing its lateral mobility.
To extend our investigations toward the conformational changes of TAP that occur during the translocation of peptides across the ER membrane, MJS TAP1–GFP cells were microinjected with long side-chain peptides. These peptides bind to TAP and stimulate the conformational changes associated with initial translocation steps, but cannot be translocated because of the size of their side chains (41). Accordingly, in control MJS cells the addition of long side-chain peptides markedly affected TAP mobility as the TAP complex was trapped in an “open” conformation (24) (Fig. 6E, compare filled bars). In MJS TAP1–GFP cells expressing UL49.5, the addition of long side-chain peptides did not affect the rate of diffusion (Fig. 6E, compare open bars). Apparently, UL49.5 blocks a conformational transition associated with the translocation of peptides.
Depletion of the peptide pool or ATP resulted in increased mobility of TAP (Fig. 11, which is published as supporting information on the PNAS web site), which supports the conclusion that peptides and ATP can still bind to TAP in the presence of UL49.5 (Fig. 6 A–C). The combined results demonstrate that TAP interacts with ATP and peptides in the presence of UL49.5, but subsequent conformational transitions appear to be prevented by the viral protein, thus inhibiting the cycle of pore opening, peptide translocation, and pore closure.
UL49.5 Is Responsible for Inhibition of TAP in Virus-Infected Cells. Homologs of the BHV1 UL49.5 gene are present in every herpesvirus sequenced to date (36). To examine whether these homologs are able to block TAP, we studied representatives of each herpesvirus group, i.e., HSV-1 and HSV-2, PRV, EHV1 (all alphaherpesviruses), HCMV (a betaherpesvirus), and Epstein–Barr virus (EBV, a gammaherpesvirus). The UL49.5 proteins encoded by HSV-1 and HSV-2, HCMV, and EBV did not influence peptide transport by TAP (Fig. 12, which is published as supporting information on the PNAS web site). In contrast, expression of the UL49.5 homologs encoded by the varicelloviruses PRV and EHV1 resulted in inhibition of peptide transport by 78% and 95%, respectively. The varicella zoster virus-encoded UL49.5 protein did not block peptide transport (Fig. 12), which is in agreement with earlier findings in infected cells (31).
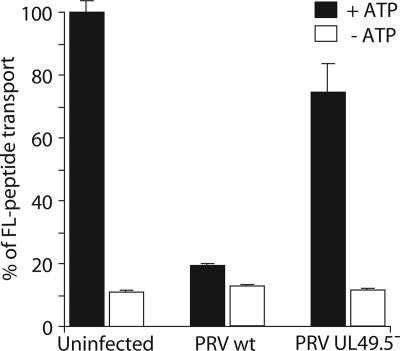
To investigate TAP inhibition by UL49.5 during viral infection, we compared peptide transport activity in cells infected with WT PRV and a mutant virus lacking a functional UL49.5 gene (42). The strong reduction of peptide transport observed in WT PRV-infected cells did not occur in the presence of the PRV mutant lacking the UL49.5 protein (Fig. 7). Therefore, the PRV UL49.5 gene product (glycoprotein N) is necessary and sufficient for the inhibition of peptide translocation during productive PRV infection.
Fig. 7.
UL49.5 is responsible for TAP inhibition during virus infection. TAP-dependent peptide transport in porcine kidney (PK15) cells were infected with either WT PRV or a UL49.5 deletion mutant for 4 h.
Discussion
Here, we describe the identification and characterization of a viral immune evasion molecule, the BHV1 UL49.5 protein. We propose the following model for its action: the ER-luminal domain and the transmembrane domain of UL49.5 interact with the TAP complex and prohibit the conformational transition of TAP that is essential for the peptide-induced ATP-dependent translocation reaction. The cytoplasmic tail of UL49.5 links the complex of UL49.5 and the TAP heterodimer to the ubiquitin–proteasome pathway.
The fact that TAP1-GFP in MJS cells impedes the BHV1 UL49.5-mediated degradation of the TAP complex was critical for the identification of UL49.5. Immunoprecipitation of TAP from parental MJS cells resulted in a weak recovery of UL49.5–TAP complexes because of the degradation of TAP in these cells. The stabilization of TAP by GFP, however, allowed us to distinguish the different events involved in inactivation of TAP by UL49.5. A similar observation has been made in the context of HCMV US11-mediated degradation of MHC class I heavy chains (43, 44). When GFP is linked to the C terminus of the class I heavy chain, the protein is resistant to proteasomal degradation (unpublished observations). Presumably, GFP interferes with the interaction between the substrate and auxiliary proteins essential for degradation.
The observation that TAP1–GFP stabilizes the TAP complex suggests that UL49.5 primarily acts on TAP1. Alternatively, UL49.5 could destabilize TAP indirectly by interfering with the function of tapasin. Tapasin stabilizes TAP and increases its expression (6). Loss of tapasin from the MHC class I peptide-loading complex causes a reduction of class I cell surface expression and destabilization of both TAP subunits (4, 6). However, in T2 cells, which express tapasin in the absence of TAP, coisolation of UL49.5 with tapasin and destabilization of tapasin are not observed, indicating that these events require the presence of TAP. Moreover, in UL49.5-expressing cells, peptide-loading complexes were found to contain both tapasin and UL49.5, indicating that UL49.5 does not act merely by dissociating tapasin from the peptide-loading complex. The observation that also class I heavy chains and TAP are present in complexes containing UL49.5 indicates that the viral protein does not act as a tapasin inhibitor, as do the adenovirus-encoded E19 protein and HCMV US3 (45, 46). Combined, our data indicate that UL49.5 primarily targets TAP.
The BHV1 UL49.5 product is a nonglycosylated type I membrane protein of only 75 amino acid residues that is expressed in virion envelopes and infected cells as a monomer, disulfide-linked homodimer, and disulfide-linked heterodimer with the viral membrane glycoprotein M (gM) (37, 47). The UL49.5 ORF is relatively conserved among herpesviruses, and the interaction with gM is a common property of all UL49.5 homologs (36, 42, 48). So far, we have identified the UL49.5 homologs encoded by BHV1, PRV, and EHV1 as potent inhibitors of TAP. Inhibition of TAP appears to be unique for varicellovirus-encoded UL49.5 proteins, as homologs encoded by simplexviruses (HSV-1 and HSV-2), a betaherpesvirus (HCMV), and a gammaherpesvirus (EBV) had no effect on TAP function. Although the UL49.5 proteins of all herpesviruses are likely to have a common ancestor, their functions apparently diverged during evolution to yield products in certain varicelloviruses that block TAP, in addition to their interaction with gM. Interestingly, the structure of gM is similar to the N-terminal domains of TAP1 and TAP2, with multiple membrane-spanning segments (12, 49). The fact that UL49.5 is capable of interacting with both gM and TAP may be related to these structural similarities.
Preventing TAP-dependent peptide transport by UL49.5 represents an effective immune evasion strategy, as it efficiently blocks cell surface expression of MHC class I molecules and prohibits activation of CTLs. HSV-1, HSV-2, HCMV, and murine γ-herpesvirus 68 also inactivate TAP, but do so by using different strategies. If UL49.5-mediated degradation of the TAP complex is prevented, for example, by GFP tagging of TAP or deletion of the cytoplasmic tail of UL49.5, UL49.5 still blocks peptide transport by TAP. In this situation, UL49.5 does not interfere with the binding of peptides and ATP to TAP, as do ICP47 and US6, respectively (17–20, 22, 23). The FRAP experiments suggest that the conformational rearrangements succeeding ATP and peptide binding are blocked by UL49.5.
The second function of UL49.5, targeting TAP1 and TAP2 for proteasomal degradation, depends on the cytoplasmic tail of UL49.5. UL49.5 is degraded along with TAP1 and TAP2. At this stage it is unclear whether the conformational effect of UL49.5 binding to the TAP complex contributes to TAP degradation. In general, proteins that adopt an abnormal conformation are recognized by ER quality-control mechanisms and are redirected to the cytosol (“dislocated”), where they are degraded by proteasomes (50). The interaction of UL49.5 with TAP could result in an aberrant conformation of the complex, thus triggering its degradation. However, the observation that TAP is not degraded by the UL49.5Δtail mutant (which still interacts with TAP and blocks peptide transport) suggests that UL49.5-induced conformational changes of TAP are insufficient to initiate its degradation.
MK3, a double-pass transmembrane protein with cytoplasmic N and C termini, targets MHC class I, tapasin, and TAP for proteasomal degradation via a reaction that requires its N-terminal RING finger, a structure that is absent in UL49.5 (26–28). Unlike mK3, which targets MHC class I heavy chains for degradation by conjugating ubiquitin to their cytoplasmic tails, UL49.5 does not cause degradation of class I molecules. Thus, UL49.5 and mK3 have a very different structure and act in a different fashion. Neither US6 (Fig. 3) nor ICP47 influence the stability of TAP (16, 18, 20, 21, 25).
In conclusion, we have identified an immune evasion protein, UL49.5, used by several varicelloviruses to specifically block peptide transport by TAP. The UL49.5 protein inactivates TAP through a unique two-stage process, involving inhibition of peptide transport through a conformational arrest of TAP, and degradation of the TAP heterodimer by the proteasome.
Supplementary Material
Acknowledgments
We thank H. L. Ploegh (Harvard Medical School, Boston) for useful discussion and providing reagents; R. Amons for performing Edman degradation; E. Claas (Leiden University Medical Center) for supplying purified viral DNA; E. Goulmy (Leiden University Medical Center) for providing minor histocompatibility antigen-specific CTL clones; E. Spierings, C. Vermeulen, and W. E. Benckhuijsen for advice and technical support; and C. Brown and R. Toes for critical reading of the manuscript. This work was supported by Dutch Cancer Society Grant RUL 1998-1791 (to M.E.R.) and a Federation of European Biochemical Societies Scholarship (to A.D.L.).
Abbreviations: CTL, cytotoxic T lymphocyte; β2m, β2-microglobulin; ER, endoplasmic reticulum; TAP, transporter associated with antigen processing; HSV, herpes simplex virus; HCMV, human cytomegalovirus; mK3, murine γ-herpesvirus 68-encoded K3 protein; BHV1, bovine herpesvirus 1; PRV, pseudorabies virus; EHV1, equine herpesvirus 1; FRAP, fluorescence recovery after photobleaching; MJS, human melanoma cell line Mel JuSo; EBV, Epstein–Barr virus; gM, glycoprotein M; B-LCL, B-cell derived lymphoblastoid cell line.
References
- 1.Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J. & Ploegh, H. L. (2000) Annu. Rev. Immunol. 18, 861–926. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell, J. W. & Hill, A. B. (2002) Nat. Immunol. 3, 1019–1025. [DOI] [PubMed] [Google Scholar]
- 3.Cresswell, P. (2000) Traffic 4, 301–305. [DOI] [PubMed] [Google Scholar]
- 4.Sadasivan, B., Lehner, P. J., Ortmann, B., Spies, T. & Cresswell, P. (1996) Immunity 5, 103–114. [DOI] [PubMed] [Google Scholar]
- 5.Ortmann, B., Copeman, J., Lehner, P. J., Sadasivan, B., Herberg, J. A., Grandea, A. G., Riddell, S. R., Tampe, R., Spies, T., Trowsdale, J. & Cresswell, P. (1997) Science 277, 1306–1309. [DOI] [PubMed] [Google Scholar]
- 6.Lehner, P. J., Surman, M. J. & Cresswell, P. (1998) Immunity 8, 221–231. [DOI] [PubMed] [Google Scholar]
- 7.Garbi, N., Tan, P., Diehl, A. D., Chambers, B. J., Ljunggren, H. G., Momburg, F. & Hammerling, G. J. (2000) Nat. Immunol. 1, 234–238. [DOI] [PubMed] [Google Scholar]
- 8.Androlewicz, M. J., Anderson, K. S. & Cresswell, P. (1993) Proc. Natl. Acad. Sci. USA 90, 9130–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neefjes, J. J., Momburg, F. & Hammerling, G. J. (1993) Science 261, 769–771. [DOI] [PubMed] [Google Scholar]
- 10.van Endert, P. M., Saveanu, L., Hewitt, E. W. & Lehner, P. (2002) Trends Biochem. Sci. 27, 454–461. [DOI] [PubMed] [Google Scholar]
- 11.Abele, R. & Tampe, R. (2004) Physiology 19, 216–224. [DOI] [PubMed] [Google Scholar]
- 12.Koch, J., Guntrum, R., Heintke, S., Kyritsis, C. & Tampe, R. (2004) J. Biol. Chem. 279, 10142–10147. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, E. A., Hammond, C. & Cresswell, P. (1997) Proc. Natl. Acad. Sci. USA 94, 1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, A., Jugovic, P., York, I., Russ, G., Bennink, J., Yewdell, J., Ploegh, H. & Johnson, D. (1995) Nature 375, 411–415. [DOI] [PubMed] [Google Scholar]
- 15.Fruh, K., Ahn, K., Djaballah, H., Sempe, P., van Endert, P. M., Tampe, R., Peterson, P. A. & Yang, Y. (1995) Nature 375, 415–418. [DOI] [PubMed] [Google Scholar]
- 16.Ahn, K., Meyer, T. H., Uebel, S., Sempe, P., Djaballah, H., Yang, Y., Peterson, P. A., Fruh, K. & Tampe, R. (1996) EMBO J. 15, 3247–3255. [PMC free article] [PubMed] [Google Scholar]
- 17.Tomazin, R., Hill, A. B., Jugovic, P., York, I., van Endert, P., Ploegh, H. L., Andrews, D. W. & Johnson, D. C. (1996) EMBO J. 15, 3256–3266. [PMC free article] [PubMed] [Google Scholar]
- 18.Galocha, B., Hill, A., Barnett, B. C., Dolan, A., Raimondi, A., Cook, R. F., Brunner, J., McGeoch, D. J. & Ploegh, H. L. (1997) J. Exp. Med. 185, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann, L., Kraas, W., Uebel, S., Jung, G. & Tampe, R. (1997) J. Mol. Biol. 272, 484–492. [DOI] [PubMed] [Google Scholar]
- 20.Ahn, K., Gruhler, A., Galocha, B., Jones, T. R., Wiertz, E. J., Ploegh, H. L., Peterson, P. A., Yang, Y. & Fruh, K. (1997) Immunity 6, 613–621. [DOI] [PubMed] [Google Scholar]
- 21.Hengel, H., Koopmann, J. O., Flohr, T., Muranyi, W., Goulmy, E., Hammerling, G. J., Koszinowski, U. H. & Momburg, F. (1997) Immunity 6, 623–632. [DOI] [PubMed] [Google Scholar]
- 22.Lehner, P. J., Karttunen, J. T., Wilkinson, G. & Cresswell, P. (1997) Proc. Natl. Acad. Sci. USA 94, 6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyritsis, C., Gorbulev, S., Hutschenreiter, S., Pawlitschko, K., Abele, R. & Tampe, R. (2001) J. Biol. Chem. 276, 48031–48039. [DOI] [PubMed] [Google Scholar]
- 24.Reits, E. A., Vos, J. C., Gromme, M. & Neefjes, J. (2000) Nature 404, 774–778. [DOI] [PubMed] [Google Scholar]
- 25.Hewitt, E. W., Gupta, S. S. & Lehner, P. J. (2001) EMBO J. 20, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boname, J. M. & Stevenson, P. G. (2001) Immunity 15, 627–636. [DOI] [PubMed] [Google Scholar]
- 27.Lybarger, L., Wang, X., Harris, M. R., Virgin, H. W., 4th, & Hansen, T. H. (2003). Immunity 18, 121–130. [DOI] [PubMed] [Google Scholar]
- 28.Boname, J. M., de Lima, B. D., Lehner, P. J. & Stevenson, P. G. (2004) Immunity 20, 305–317. [DOI] [PubMed] [Google Scholar]
- 29.Cohen, J. I. (1998) J. Infect. Dis. 177, 1390–1393. [DOI] [PubMed] [Google Scholar]
- 30.Ambagala, A. P., Hinkley, S. & Srikumaran, S. (2000) J. Immunol. 164, 93–99. [DOI] [PubMed] [Google Scholar]
- 31.Abendroth, A., Lin, I., Slobedman, B., Ploegh, H. & Arvin, A. M. (2001) J. Virol. 75, 4878–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koppers-Lalic, D., Rijsewijk, F. A., Verschuren, S. B., van Gaans-Van den Brink, J. A., Neisig, A., Ressing, M. E., Neefjes, J. & Wiertz, E. J. (2001) J. Gen. Virol. 82, 2071–2081. [DOI] [PubMed] [Google Scholar]
- 33.Rappocciolo, G., Birch, J. & Ellis, S. A. (2003) J. Gen. Virol. 84, 293–300. [DOI] [PubMed] [Google Scholar]
- 34.Ambagala, A. P., Gopinath, R. S. & Srikumaran, S. (2004) J. Gen. Virol. 85, 349–353. [DOI] [PubMed] [Google Scholar]
- 35.Koppers-Lalic, D., Rychlowski, M., van Leeuwen, D., Rijsewijk, F. A., Ressing, M. E., Neefjes, J. J., Bienkowska-Szewczyk, K. & Wiertz, E. J. (2003) Arch. Virol. 148, 2023–2037. [DOI] [PubMed] [Google Scholar]
- 36.Adams, R., Cunningham, C., Davison, M. D., MacLean, C. A. & Davison, A. J. (1998) J. Gen. Virol. 79, 813–823. [DOI] [PubMed] [Google Scholar]
- 37.Liang, X., Chow, B., Raggo, C. & Babiuk, L. A. (1996) J. Virol. 70, 1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spierings, E., Brickner, A. G., Caldwell, J. A., Zegveld, S., Tatsis, N., Blokland, E., Pool, J., Pierce, R. A., Mollah, S., Shabanowitz, J., et al. (2003) Blood 102, 621–629. [DOI] [PubMed] [Google Scholar]
- 39.Meadows, L., Wang, W., den Haan, J. M., Blokland, E., Reinhardus, C., Drijfhout, J. W., Shabanowitz, J., Pierce, R., Agulnik, A. I., Bishop, C. E., et al. (1997). Immunity 6, 273–281. [DOI] [PubMed] [Google Scholar]
- 40.Goulmy, E., Schipper, R., Pool, J., Blokland, E., Falkenburg, J. H., Vossen, J., Gratwohl, A., Vogelsang, G. B., van Houwelingen, H. C. & van Rood, J. J. (1996) N. Engl. J. Med. 334, 281–285. [DOI] [PubMed] [Google Scholar]
- 41.Grommé, M., van der Valk, R., Sliedregt, K., Vernie, L., Liskamp, R., Hammerling, G., Koopmann, J. O., Momburg, F. & Neefjes, J. (1997) Eur. J. Immunol. 27, 898–904. [DOI] [PubMed] [Google Scholar]
- 42.Jons, A., Dijkstra, J. M. & Mettenleiter, T. C. (1998) J. Virol. 72, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiertz, E. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996) Cell 84, 769–779. [DOI] [PubMed] [Google Scholar]
- 44.Wiertz, E. J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T. R., Rapoport, T. A. & Ploegh, H. L. (1996) Nature 384, 432–438. [DOI] [PubMed] [Google Scholar]
- 45.Bennett, E. M., Bennink, J. R., Yewdell, J. W. & Brodsky, F. M. (1999) J. Immunol. 162, 5049–5052. [PubMed] [Google Scholar]
- 46.Park, B., Kim, Y., Shin, J., Lee, S., Cho, K., Fruh, K., Lee, S. & Ahn, K. (2004) Immunity 20, 71–85. [DOI] [PubMed] [Google Scholar]
- 47.Wu, S. X., Zhu, X. P. & Letchworth, G. J. (1998) J. Virol. 72, 3029–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolph, J., Seyboldt, C., Granzow, H. & Osterrieder, N. (2002) J. Virol. 76, 2952–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seyboldt, C., Granzow, H. & Osterrieder, N. (2000) Virology 278, 477–489. [DOI] [PubMed] [Google Scholar]
- 50.Ellgaard, L. & Helenius, A. (2003) Nat. Rev. Mol. Cell. Biol. 4, 181–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.