Abstract
Helicobacter pylori is generally viewed as an extracellular pathogen. We have analyzed the tropism of H. pylori clinical isolates in a gnotobiotic transgenic mouse model of human chronic atrophic gastritis, a preneoplastic condition. These mice lack acid-producing parietal cells and have an amplified population of dividing gastric epithelial progenitors (GEPs) that express NeuAcα2,3Galβ1,4-glycans recognized by H. pylori adhesins. Scanning confocal and transmission electron microscopic studies of stomachs that had been colonized for 1 month or 1 year revealed intracellular bacterial collections (IBCs) in a small subset of multi- and oligopotential epithelial progenitors. Transmission electron microscopic and multilabel immunohistochemical analyses disclosed bacteria with several morphotypes, including spiral-shaped, in the cytoplasm and endosomes. Several stages in IBC evolution were documented, from a few solitary bacteria to consolidated populations in dividing and nondividing GEPs, to microorganisms traversing breaches in the GEP plasma cell membrane. IBC formation was not a unique feature of H. pylori strains isolated from patients with chronic atrophic gastritis. The notion that adult mammalian epithelial progenitors can function as a repository for H. pylori broadens the view of host habitats available to this and perhaps other pathogens.
Keywords: adult mammalian epithelial progenitors, bacterial pathogenesis, intracellular bacterial communities, gnotobiotic mice, chronic atrophic gastritis
The stomachs of more than half of all humans are colonized by Helicobacter pylori. Typically acquired during childhood, this Gram-negative bacterium can persist in the gastric ecosystem throughout the life span of untreated hosts (1). H. pylori is found mainly in the mucous layer of the stomach; at any given moment, only a small fraction appears to adhere to the gastric epithelium (2). H. pylori expresses a number of adhesins that mediate attachment to gastric epithelial glycan receptors, including SabA, which binds to NeuAcα2,3Galβ1,4-containing glycans such as sialyl-Lewisx (3). These glycans are prominently represented in the stomachs of H. pylori-infected individuals who have developed chronic atrophic gastritis (4), a preneoplastic condition characterized by loss of acid-producing parietal cells (5).
H. pylori has long been viewed as an extracellular bacterium, although the ineffectiveness of non-cell-penetrating antibiotics in eradicating infection in a subset of individuals, and the predominance of a T helper 1 adaptive immune response characteristic of invasive pathogens, suggest the existence of an intracellular population (6, 7). Several electron microscopic studies of gastric biopsy samples from infected individuals have reported intact and degraded forms of the bacterium within epithelial cells (8). Additionally, time-lapse microscopic analyses have documented H. pylori invasion of a gastric adenocarcinoma-derived epithelial cell line (9, 10).
Recently, uropathogenic strains of Escherichia coli, which cause acute and recurrent urinary tract infections, were found to spend part of their life cycle as intracellular bacterial communities within bladder urothelial cells (11). These communities progress through distinct developmental stages, characterized by changes in the growth rate, morphology, and motility of its members. Deconstruction occurs as community members exit the host cell and disperse to other urothelial cells. This mechanism allows a reservoir of quiescent bacteria to be established that helps support persistent infection (12). This discovery raises the question of whether other bacterial pathogens form communities within differentiated host epithelial cells and/or epithelial progenitors.
In this current study, we address this issue using a gnotobiotic transgenic mouse model of persistent H. pylori infection in humans with chronic atrophic gastritis. Colonizing germ-free animals with clinical isolates greatly simplifies analysis of bacterial tropism, because mice normally contain a complex gastric microbiota due to their coprophagy. Our transgenic mice have an engineered ablation of parietal cells, achieved by expressing an attenuated diphtheria toxin A fragment (tox176) under the control of a lineage-specific promoter (Atp4b; ref. 13). Parietal cells were selected for ablation because they function as a guardian of the stem cell niche: they are the only major lineal descendant of the multipotent gastric stem cell that completes its differentiation within the niche; their acid impedes colonization of the niche; and their presence affects the proliferative activity of multi- and oligopotential progenitors (ablation stimulates proliferation and produces a progressive amplification of normally rare NeuAcα2,3Galβ1,4 glycan-positive GEPs; ref. 14 and Fig. 1 A and B). The loss of parietal cells and the increased representation of NeuAcα2,3Galβ1,4 glycans are features that tox176 mice share with humans with chronic atrophic gastritis (14).
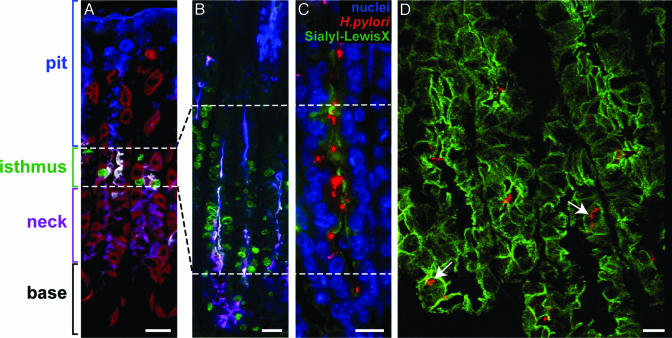
Fig. 1.
Distribution of H. pylori in gnotobiotic tox176 mice. (A and B) Multilabel immunohistochemical study of stomachs from 7-week-old germ-free normal (A) and tox176 (B) mice showing that tox176-mediated parietal cell ablation results in amplification of mitotically active GEPs. Animals were treated with BrdUrd (green) 90 min before death to label cells in S-phase. Dolichos biflorus agglutinin (red) is used to mark parietal cells. Pit cells are tagged with Alexa Fluor 350-conjugated Anguilla anguilla agglutinin (blue), and neck cells are tagged with Alexa Fluor 350- and Alexa Fluor 647-tagged Griffonia simplicifolia II lectin (purple). GEPs produce NeuAcα2,3Galβ1,4 glycans (white after staining with biotinylated MAA and Cy3-labeled streptavidin). (C) Attachment of CAG7:8 (red) to NeuAcα2,3Galβ1,4-positive GEPs (marked green with MAA) in a stomach from a 15-week-old gnotobiotic tox176 mouse killed after 4 weeks of infection. Nuclei are stained blue with bis-benzimide. (D) Frame from a 3D confocal microscopic projection of a focal area of infection in a tox176 stomach showing intracellular bacteria (red, arrows). Cell borders are delineated by using an antibody to E-cadherin (green after treatment with Alexa Fluor 488-tagged donkey anti-rat Ig). To view the 3D projection in its entirety, see Movie 1, which is published as supporting information on the PNAS web site. (Bars, 10 μm.)
Below, we report how scanning confocal microscopy, combined with multilabel immunohistochemistry plus transmission EM (TEM), has revealed that a subset of dividing and nondividing gastric epithelial progenitors (GEPs) provides a milieu that supports formation of intracellular collections of H. pylori strains recovered from patients with or without chronic atrophic gastritis. The development of intracellular bacterial collections (IBCs) in adult mammalian epithelial progenitors provides a previously unappreciated view of how H. pylori may persist in some of its hosts, as well as an opportunity to consider how the biological features of these progenitors, revealed from ongoing functional genomics studies, may not only support but also be influenced by IBCs.
Materials and Methods
Animals. Germ-free FVB/N transgenic mice that express tox176 under the control of nucleotides –1,035 to +24 of Atp4b (noncatalytic β-subunit of mouse H+/K+ ATPase) and their normal littermates were maintained in plastic gnotobiotic isolators (15) under a strict 12-h light cycle (lights on at 0600 hours). Animals were given an autoclaved chow diet (B & K Universal, East Yorkshire, U.K.) ad libitum. All manipulations of mice were performed by using protocols approved by the Washington University Animal Studies Committee.
Monoassociations. H. pylori strains CAG7:8 (16) and Hp1 (17) were cultured under microaerophilic conditions for 3 d at 37°C on selective medium [brain heart infusion agar, supplemented with 10% calf blood/vancomycin (6 μg/ml)/trimethoprim (5 μg/ml)/amphotericin B (8 μg/ml)]. Bacteria were collected, concentrated to 108 colony-forming units (cfu)/ml sterile PBS, and brought into the gnotobiotic isolator (16), and one or the other strain was inoculated (single gavage of 107 cfu) into the stomachs of adult germ-free tox176 and normal littermates. Selected normal and transgenic mice received an i.p. injection of BrdUrd (120 mg/kg) and 5-fluoro-2′-deoxyuridine (12 mg/kg), 1.5 h before they were killed, to label gastric epithelial cells in S phase.
The stomach was removed from each mouse at the time of death, 4, 8, or 56 weeks after inoculation of H. pylori (CAG7:8, 4-week infection, 56 animals; 56-week infection, 30 animals; Hp1, 4-week infection, 13 animals; 8-week infection, 29 animals). All stomachs were cut in half along their cephalocaudal axis. One half was homogenized in 0.5 ml of PBS and plated on selective medium to assay for the presence of viable H. pylori. The other half was immediately frozen in OCT compound (Sakura, Torrance, CA) or fixed overnight at 4°C in Bouin's solution for immunohistochemical analysis (18).
Immunohistochemistry. Cryosections (5–10 μm thick) were cut and postfixed in methanol for 5 min at –20°C. Paraffin-embedded tissue sections (5–10 μm thick) were deparaffinized in xylene and rehydrated in isopropanol. Sections were rinsed in deionized water (5 min at 25°C) and washed in PBS (three cycles, 5 min each at 25°C). Sections were then placed in blocking buffer (1% BSA/0.3% Triton X-100 in PBS) for 1 h at 25°C and subsequently incubated overnight at 4°C with primary antibodies and/or biotinylated lectins. The antibody panel included: (i) rabbit anti-H. pylori (final dilution in blocking buffer, 1:1,000, obtained from Dako), (ii) rat anti-mouse E-cadherin (1:500, Zymed), (iii) mouse anti-β-catenin (1:50, Zymed), (iv) rat anti-mouse Lamp1 (1:100, BD Pharmingen), and (v) goat anti-BrdUrd (1:1,000). The following lectins, obtained from EY Laboratories, were used at a final concentration of 20 μg/ml blocking buffer: (i) Maackia amurensis agglutinin (MAA) (detects GEPs expressing NeuAcα2,3Galβ1,4-containing glycans); (ii) Dolicos biflorus agglutinin (specific for parietal cells in the FVB/N strain); (iii) Anguilla anguilla agglutinin (pit cells); or (iv) Griffonia simplifolica II (neck cells). After three PBS washes (5 min each at 25°C), antigen–antibody complexes and bound biotinylated lectins were detected with Alexa Fluor-(Molecular Probes), cyanine (Cy)3-, Cy5-, or FITC-conjugated secondary antibodies or streptavidin (Jackson ImmunoResearch). In some cases, lectins were directly conjugated to Alexa Fluor before use (Alexa Fluor Protein Labeling Kit, Molecular Probes). Stained sections were examined with a Zeiss LSM 510 inverted confocal microscope.
TEM. Protocols used for TEM analysis of the mouse stomach have been described in detail previously, as have the morphologic criteria used to define the presumptive granule-free multipotential gastric stem and its immediate committed oligopotential daughters that give rise to the pit and neck cell lineages (19).
Results
Identifying Intracellular H. pylori in the Stomachs of tox176 Mice by Scanning Confocal Microscopy. Germ-free 6- to 17-week-old tox176 mice and their age- and gender-matched normal littermates were gavaged once with 107 colony-forming units (cfu) of a H. pylori strain recovered from a patient with chronic atrophic gastritis. This strain, CAG7:8, expresses adhesins that bind to cellular NeuAcα2,3Galβ1,4 glycan receptors (16). The efficiency of infection with CAG7:8 is high in germ-free tox176 mice, even though it had never been adapted to the mouse stomach by serial passage. Four weeks after inoculation, viable organisms (cfu) were recovered from the stomachs of 27/29 tox176 animals (93%) compared with 17/27 normal littermates (63%; P ≤ 0.01; Mann–Whitney U test). This enhanced efficiency was sustained even after 1 year of infection [15/17 (88%) for tox176 vs. 4/13 (31%) for normal littermates (P ≤ 0.001)].
The distribution of CAG7:8 was defined by staining serial sections of stomach with antibodies to H. pylori surface proteins. In normal gnotobiotic mice, the strain was restricted to a narrow band of epithelium positioned at the junction of the distal boundary of the forestomach's squamous epithelium and the proximal boundary of the glandular epithelium that lines the rest of the stomach. This junctional region, also known as the forestomach/zymogenic (FS/Z) transition (Fig. 5, which is published as supporting information on the PNAS web site), is notable for its paucity of parietal cells and its large collection of NeuAcα2,3Galβ1,4-positive mature pit cells. The FS/Z represents the only habitat in the stomach where these sialylated glycans are expressed in pit cells (18).
In gnotobiotic transgenic mice, there was an expanded distribution of CAG7:8 from the FS/Z into the glandular epithelium, where parietal cells had been removed by tox176-mediated lineage ablation [20/29 (69%) of mice surveyed]. Conventional fluorescence microscopy indicated that the majority of the bacteria were located near or on the surface of epithelial cells in the glandular epithelium that express NeuAcα2,3Galβ1,4 glycans (Fig. 1C).
To further define the interaction between CAG7:8 and host epithelial cells, 7- to 10-μm-thick cryosections were prepared from the stomachs of tox176 mice that had been killed after a 4-week infection. Cryosections were stained with antibodies specific for the epithelial plasma membrane marker, E-cadherin, and with antibodies to H. pylori surface proteins. Focal areas of infection were identified where the number of bacteria present in an arbitrarily defined volume [150 μm (x axis) × 150 μm (y axis) × 5–10 μm (z axis)] exceeded an arbitrarily selected threshold (10 organisms). These focal areas (5–10 per mouse; n = 10 animals) were then scanned with a confocal microscope at an optical plane thickness of 0.4–0.6 μm. Serial images (15–25 per focus) were aligned at 7–10° intervals, compiled, and viewed as they were rotated around the x, y, and z axes.
The results confirmed that the majority of bacteria were extracellular (i.e., on, or remote from, surface membranes). However, 1–5% of epithelial cells with associated bacteria appeared to contain an intracellular population of CAG7:8 (Fig. 1D).
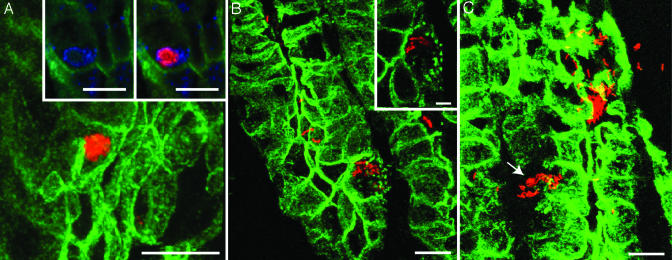
Several Configurations of Internalized Bacteria. The majority (>80%) of epithelial cells harboring bacteria contained fewer than five organisms (Fig. 1D). The remaining contained numerous bacteria, often in the form of a large conglomerate. Fig. 2 presents scanning confocal microscopic views of these IBCs at several stages, i.e., as a consolidated collection where bacteria no longer have their distinct spiral shape and appear rather featureless (Fig. 2 A), as a more dispersed collection of spiral-shaped organisms in epithelial cells that have developed focal areas of dissolution of their surface membranes (Fig. 2B), and as spiral-shaped bacteria that are trafficking across multiple breaches in the plasma membrane (Fig. 2C). The subcellular localization of IBCs was initially confirmed by colocalization of H. pylori and the late endosomal marker Lamp1 (see Fig. 2 A Inset plus the TEM studies described below).
Fig. 2.
Confocal microscopic study of CAG7:8 IBCs. Sixteen-week-old gnotobiotic tox176 mice were killed after a 4-week infection with CAG7:8. Sections were processed exactly as in Fig. 1D.(A) A spherical IBC (red). (Insets) A single section stained with antibodies H. pylori (red) and the late endosomal marker, Lamp1 (blue). The results reveal bacterial cells residing within this endosomal compartment. (B) IBC with a dispersed phenotype in a host cell that has developed punctate disruptions of its plasma membrane (the affected cell is also shown in the Inset). (C) CAG7:8 is seen traversing a breach in its host epithelial cell plasma membrane (arrow). 3D reconstructions of serial confocal scans of the sections presented can be found in Movies 2–4, which are published as supporting information on the PNAS web site. [Bars, 10 μm (5 μm in B Inset).]
Intracellular CAG7:8 were not observed in the FS/Z of tox176 stomachs (n = 29) where, unlike the rest of the glandular epithelium, the dominant NeuAcα2,3Galβ1,4-expressing population is comprised of mature pit cells rather than GEPs, nor were intracellular bacteria detected in any region of the stomachs of infected normal littermates (n = 27), where the census of GEPs is 10-fold lower than in infected tox176 animals (18). Scanning confocal microscopic studies of serial gastric sections, prepared from tox176 stomachs and subjected to multilabel immunohistochemical analysis with antibodies to CD11b, H. pylori surface proteins and E-cadherin established that intracellular CAG7:8 was not associated with macrophages (data not shown).
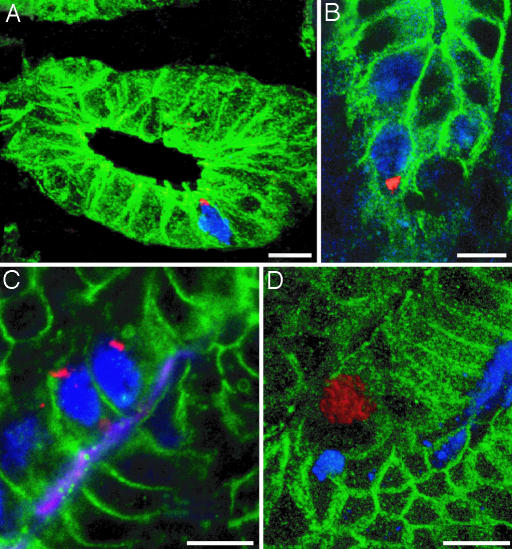
IBCs in Dividing and Nondividing GEPs. Together, these findings suggested a linkage between IBC formation, the loss of parietal cells, and/or amplification of NeuAcα2,3Galβ1,4-positive GEPs. GEPs represent the dominant mitotically active epithelial cell population in tox176 stomachs (20). Scanning confocal microscopy, combined with multilabel immunohistochemistry of cryosections prepared from 4-week-infected mice that had received BrdUrd 1.5 h before death, revealed epithelial cells in S-phase harboring CAG7:8 IBCs (e.g., Fig. 3 A and B). As shown in Fig. 1 A and B, GEP surface-associated NeuAcα2,3Galβ1,4 glycans are detectable with MAA. We observed that dividing and nondividing MAA-positive GEPs in gnotobiotic tox176 mice contain IBCs (Fig. 3 C and D).
Fig. 3.
Intracellular collections of H. pylori form in actively dividing GEPs. Six- to 10-week-old tox176 mice were infected for 4 weeks with CAG7:8 and given an i.p. injection of BrdUrd (blue) 1.5 h before death. Cryosections from the zymogenic region of their glandular epithelium were stained with antibodies to H. pylori (red) and E-cadherin (green) and then serially scanned with a confocal microscope at an optical plane thickness of 0.4–0.6 μm. (A and B) S-phase epithelial cells containing small IBCs. Images were taken from 3D reconstructions of serial confocal scans (see Movies 5 and 6, which are published as supporting information on the PNAS web site). A z axis scan of the IBC portrayed in B can be found in Movie 7, which is published as supporting information on the PNAS web site. (C) Conventional photomicrograph of a section containing BrdUrd-positive (blue) and MAA-positive (magenta) GEPs with intracellular CAG7:8 (red). GEP membrane-associated β-catenin appears green. (D) Confocal microscopic image of a MAA-positive (blue) GEP (membranes appear green after staining with antibodies to β-catenin), harboring H. pylori (red). See Movie 8, which is published as supporting information on the PNAS web site, for the corresponding 3D reconstruction. (Bars, 10 μm.)
The multipotent gastric stem cell gives rise to three principal epithelial lineages: parietal, neck/zymogenic, and pit. Multilabel confocal microscopic studies using two lineage-specific lectins, Anguilla anguilla agglutinin and Griffonia simplicifolia II, indicated that IBCs were not present in mature members of the pit or neck/zymogenic cell lineages (n = 10 tox176 mice studied; data not shown).
TEM Studies of GEPs Harboring H. pylori. We used TEM to examine gastric sections prepared from tox176 mice colonized for 4 weeks to further define (i) the progenitor populations that harbor CAG7:8, (ii) the subcellular location of H. pylori, and (iii) the morphotypes assumed by the bacterium within GEPs.
Previous studies of a gastric adenocarcinoma-derived cell line revealed internalized H. pylori within Lamp1-containing vacuoles, suggesting that cellular entry may occur via zipper-like receptor-mediated endocytosis, as occurs with Yersinia pseudotuberculosis, Yersinia enterocolitica, Neisseria gonorrhoeae and Listeria monocytogenes infections (21). In the latter infections, intracellular bacteria located within vacuolar compartments use several different mechanisms to prevent maturation of host cell phagosomes (i.e., their fusion to lysosomes), including alterations in the composition of the phagosomal membrane and segregation from the endocytic pathway. Alternatively, they escape a degradative fate by liberating themselves from the phagosome and entering the cytoplasm (22). Intriguingly, scanning confocal microscopy indicated that not all IBCs were associated with Lamp1-positive compartments, suggesting there might be intracellular H. pylori in the cytoplasm of GEPs.
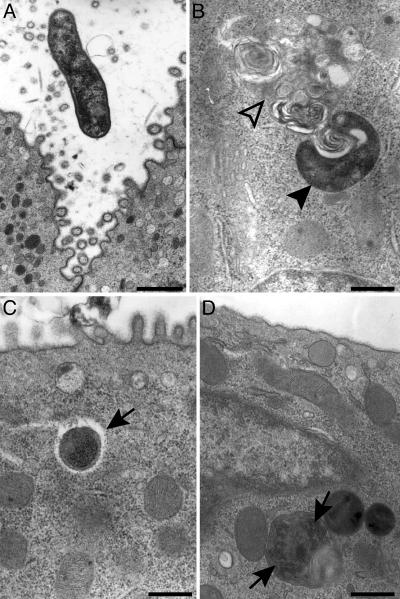
TEM disclosed CAG7:8 residing within granule-free multipotential and oligopotential epithelial lineage progenitors that give rise to the pit and neck cell lineages. Bacteria were observed in both the cytoplasmic and late endosomal compartments. Morphotypes ranged from spiral-shaped, consistent with viable bacteria (23), to organisms in the cytoplasm surrounded by a clear halo, to what appeared to be dividing organisms within endosomes (Fig. 4).
Fig. 4.
TEM study showing H. pylori strain CAG7:8 within GEPs. (A) Extracellular CAG7:8 in the vicinity of an oligopotential precursor to the pit cell lineage. (B) A spiral-shaped bacterium (closed arrowhead) near a late endosome (open arrowhead) in a preneck cell progenitor. (C) Cross-sectional view of CAG7:8 in the cytoplasm of a preneck cell progenitor surrounded by a clear halo (arrow). (D) Two intracellular H. pylori (arrows) enclosed within a late endosome of a preneck cell. [Bars, 800 nm (A) and 500 nm (B–D).]
Long-Term Infections with H. pylori Strains Recovered from Patients With and Without Chronic Atrophic Gastritis. Confocal microscopic analysis of gnotobiotic tox176 mice infected with a single gavage of CAG7:8 at 13 weeks of age and killed 56 weeks later revealed bacteria at all stages (solitary intracellular organisms, IBCs, and traversing the plasma membrane) in the expanded GEP population (n = 5; data not shown). In contrast, intracellular bacteria were not evident in any gastric epithelial cell type present in normal littermates infected for the same period (n = 10). Analysis of serially sectioned stomachs did not reveal gastric adenocarcinomas in any tox176 or normal mouse infected for 56 weeks (n = 30; data not shown).
The intracellular localization of CAG7:8 in tox176 mice was not a unique feature of a strain that had adapted itself to life in a human stomach lacking parietal cells. An identical pattern of IBC formation was observed after a 4- or 8-week infection of tox176 mice with a cagA-strain (Hp1) isolated from a patient with an intact parietal cell mass who had acid peptic disease (n = 5 mice examined by confocal microscopy per time point). Like CAG7:8, Hp1 expresses adhesins that recognize NeuAcα2,3Galβ1,4 glycan receptors (14).
Discussion
Our analysis suggests that the interplay of several factors facilitates formation of intracellular collections of H. pylori in a subset of actively dividing GEPs. Loss of parietal cells results in removal of the acid barrier to colonization of the glandular epithelium. Removal of parietal cells from the stem cell niche in gastric units leads to amplification of GEPs. These amplified GEPs express NeuAcα2,3Galβ1,4 glycans that serve as receptors for H. pylori adhesins. Our failure to detect IBCs in normal gnotobiotic mice is consistent with this formulation: (i) in normal mice, GEPs represent <3% of the steady state population of 200 epithelial cells that populate each of the innumerable gastric units embedded in the proximal two-thirds of the glandular epithelium (24), whereas they comprise ≈20% of the population in 12- to 14-week-old tox176 animals; (ii) IBC formation, when it does occur, is a rare event (<0.5% of GEPs); and (iii) the parietal cell guardian to H. pylori colonization of the stem cell niche is present in normal mice. Our failure to detect IBCs in NeuAcα2,3Galβ1,4-expressing pit cells in the parietal cell-deficient FS/Z transition zone of tox176 (or normal) mice or in differentiated pit and neck cells that are abundantly represented throughout their glandular epithelium suggests that the presence of sialylated glycan receptors for bacterial adhesins is not sufficient for IBC formation and/or that GEPs provide a unique habitat permissive for community formation.
The observation that a pathogenic bacterium is able to invade an adult mammalian epithelial progenitor cell and assemble into IBCs is, to our knowledge, unprecedented. In the case of H. pylori, failure to appreciate the presence of IBCs in human gastric biopsies may reflect a sampling problem. Our findings suggest that searches for IBCs in human stomachs should focus on patients possessing the histologic equivalent of tox176 mouse stomachs, i.e., those with chronic atrophic gastritis.
Although the spiral-shaped bacterial morphotype observed within GEPs is consistent with living organisms, we cannot deem the intracellular collections “communities” until viability is demonstrated directly. Moreover, the impact of H. pylori invasion and IBC formation on progenitor cell biology and bacterial physiology remains unclear. Addressing these questions is very challenging, given the limited number of GEPs harboring IBCs in tox176 mice and the absence of cell lines that phenocopy multi- and/or oligopotential GEPs.
We have embarked on a project to profile gene expression in the expanded population of GEPs present in germ-free tox176 mice. To preserve the effects of the local microenvironment on GEP gene expression, we used laser-capture microdissection to harvest cells from cryosections of stomach and then sequenced cDNA libraries prepared from the microdissected progenitors. Results obtained to date (http://genome.wustl.edu/GSCGAP) provide insights about how H. pylori may be affected by, and/or may affect, GEP biology. For example, ornithine decarboxylase (ODC; EC 4.1.1.17), the first and rate-limiting step in synthesis of polyamines (putrescine, spermidine, and spermine), is expressed in GEPs. ODC activity and high levels of polyamines in epithelium are associated with H. pylori infection and pathology (25). There are no proteins in the sequenced J99 and 26695 strains with homologies to ODC, agmatinase, or agmatine deiminase, which produce putrescine, yet H. pylori has a gene (HP0832) with high homology to spermidine synthase (EC 2.5.1.16), an enzyme that uses putrescine and S-adenosylmethioninamine to generate 5′-methylthioadenosine and spermidine. Polyamines stimulate bacterial growth. Thus, H. pylori may use polyamines generated by GEPs, such as putrescine, to stimulate their intracellular growth. Because polyamines stimulate cell division in a number of mammalian cell lineages (26), regulation of polyamine availability by intracellular H. pylori may also affect the proliferative status of their host GEP.
An intracellular habitat for H. pylori in GEPs provides a theoretical opportunity for the organism to be sheltered within a cell population that has a high capacity for self renewal. The efficiency of transmission of H. pylori among GEPs is unknown. Because of the infrequent and dispersed nature of IBC formation in the gastric epithelium, we have been unable to determine whether the number of GEPs with and without IBCs changes over time. Such determinations await identification of GEP or microbial biomarkers of IBC formation that can be scored in whole stomachs. These biomarkers will also be needed to compare the capacities of different H. pylori isolates or isogenic mutants to form IBCs in our tox176 gnotobiotic model and/or in cultured progenitor cell lines. These types of analyses should provide a framework for exploring IBC formation in human hosts and its possible relationship to gastric neoplasia.
Supplementary Material
Acknowledgments
We thank Maria Karlsson and David O'Donnell for their assistance with rearing and analyzing gnotobiotic mice, Janaki Guruge for culture-based assays, Jaime Dant for help preparing samples for TEM studies, and Sabrina Wagoner for superb technical assistance. This work was supported by grants from the National Institutes of Health (DK58529 and DK63483).
Author contributions: J.D.O. and J.I.G. designed research; J.D.O. and S.M.K. performed research; J.D.O., S.M.K., and J.I.G. analyzed data; and J.D.O. and J.I.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IBC, intracellular bacterial collection; GEP, gastric epithelial progenitor; MAA, Maackia amurensis agglutinin; TEM, transmission EM; FS/Z, forestomach/zymogenic transition.
References
- 1.Mitchell, H. M., Li, Y. Y., Hu, P. J., Liu, Q., Chen, M., Du, G. G., Wang, Z. J., Lee, A. & Hazell, S. L. (1992) J. Infect. Dis. 166, 149–153. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J. & Kirschner, D. (1999) Proc. Natl. Acad. Sci. USA 96, 8359–8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahdavi, J., Sonden, B., Hurtig, M., Olfat, F. O., Forsberg, L., Roche, N., Angstrom, J., Larsson, T., Teneberg, S., Karlsson, K. A., et al. (2002) Science 297, 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farinati, F., Nitti, D., Cardin, F., Di Mario, F., Costa, F., Rossi, C., Marchett, A., Lise, M. & Naccarato, R. (1988) Eur. J. Cancer Clin. Oncol. 24, 923–927. [DOI] [PubMed] [Google Scholar]
- 5.Ye, W., Held, M., Lagergren, J., Engstrand, L., Blot, W. J., McLaughlin, J. K. & Nyren, O. (2004) J. Natl. Cancer Inst. 96, 388–396. [DOI] [PubMed] [Google Scholar]
- 6.Engstrand, L., Graham, D., Scheynius, A., Genta, R. M. & El-Zaatari, F. (1997) Am. J. Clin. Pathol. 108, 504–509. [DOI] [PubMed] [Google Scholar]
- 7.Lindholm, C., Quiding-Jarbrink, M., Lonroth, H., Hamlet, A. & Svennerholm, A. M. (1998) Infect. Immun. 66, 5964–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen, A. M. & Krogfelt, K. A. (2003) FEMS Immunol. Med. Microbiol. 36, 117–126. [DOI] [PubMed] [Google Scholar]
- 9.Bjorkholm, B., Zhukhovitsky, V., Lofman, C., Hulten, K., Enroth, H., Block, M., Rigo, R., Falk, P. & Engstrand, L. (2000) Helicobacter 5, 148–154. [DOI] [PubMed] [Google Scholar]
- 10.Amieva, M. R., Salama, N. R., Tompkins, L. S. & Falkow, S. (2002) Cell. Microbiol. 4, 677–690. [DOI] [PubMed] [Google Scholar]
- 11.Anderson, G. G., Palermo, J. J., Schilling, J. D., Roth, R., Heuser, J. & Hultgren, S. J. (2003) Science 301, 105–107. [DOI] [PubMed] [Google Scholar]
- 12.Justice, S. S., Hung, C., Theriot, J. A., Fletcher, D. A., Anderson, G. G., Footer, M. J. & Hultgren, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Q., Karam, S. M. & Gordon, J. I. (1996) J. Biol. Chem. 271, 3671–3676. [PubMed] [Google Scholar]
- 14.Syder, A. J., Guruge, J. L., Li, Q., Hu, Y., Oleksiewicz, C. M., Lorenz, R. G., Karam, S. M., Falk, P. G. & Gordon, J. I. (1999) Mol. Cell. 3, 263–274. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, L. V., Mills, J. C., Roth, K. A., Stappenbeck, T. S., Wong, M. H. & Gordon, J. I. (2002) in Methods in Microbiology, eds. Sansonetti, P. & Zychlinksy, A. (Academic, San Diego), Vol. 31, pp. 559–589. [Google Scholar]
- 16.Bjorkholm, B. M., Guruge, J. L., Oh, J. D., Syder, A. J., Salama, N., Guillemin, K., Falkow, S., Nilsson, C., Falk, P. G., Engstrand, L., et al. (2002) J. Biol. Chem. 277, 34191–34197. [DOI] [PubMed] [Google Scholar]
- 17.Guruge, J. L., Falk, P. G., Lorenz, R. G., Dans, M., Wirth, H. P., Blaser, M. J., Berg, D. E. & Gordon, J. I. (1998) Proc. Natl. Acad. Sci. USA 95, 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syder, A. J., Oh, J. D., Guruge, J. L., O'Donnell, D., Karlsson, M., Mills, J. C., Bjorkholm, B. M. & Gordon, J. I. (2003) Proc. Natl. Acad. Sci. USA 100, 3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karam, S. M., Li, Q. & Gordon, J. I. (1997) Am. J. Physiol. 272, G1209–1220. [DOI] [PubMed] [Google Scholar]
- 20.Mills, J. C., Andersson, N., Hong, C. V., Stappenbeck, T. S. & Gordon, J. I. (2002) Proc. Natl. Acad. Sci. USA 99, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok, T., Backert, S., Schwarz, H., Berger, J. & Meyer, T. F. (2002) Infect. Immun. 70, 2108–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso, A. & Garcia-del Portillo, F. (2004) Int. Microbiol. 7, 181–191. [PubMed] [Google Scholar]
- 23.Nilsson, H. O., Blom, J., Abu-Al-Soud, W., Ljungh, A. A., Andersen, L. P. & Wadstrom, T. (2002) Appl. Environ. Microbiol. 68, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karam, S. M. & Leblond, C. P. (1992) Anat. Rec. 232, 231–246. [DOI] [PubMed] [Google Scholar]
- 25.Patchett, S. E., Katelaris, P. H., Zhang, Z. W., Alstead, E. M., Domizio, P. & Farthing, M. J. (1996) Gut 39, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace, H. M., Fraser, A. V. & Hughes, A. (2003) Biochem. J. 376, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.