Abstract
This paper reports that the abundances of endogenous cardiolipin and phosphatidylethanolamine halve during elongation of the Gram-positive bacterium Listeria innocua. The lyotropic phase behaviour of model lipid systems that describe these modulations in lipid composition indicate that the average stored curvature elastic stress of the membrane is reduced on elongation of the cell, while the fluidity appears to be maintained. These findings suggest that phospholipid metabolism is linked to the cell cycle and that changes in membrane composition can facilitate passage to the succeding stage of the cell cycle. This therefore suggests a means by which bacteria can manage the physical properties of their membranes through the cell cycle.
Introduction
The bacterial cell cycle is an example of binary fission that is typically both rapid and reliable. It is characterised by periods known as B, C and D1–4. The B period represents the phase between division and the initiation of replication, whereas the C period (sometimes called the replication phase) represents the phase in which the nucleolus is replicated and in which the bacterium reaches its mature size. The D period is that between the replication of the DNA and fission of the cell envelope. The B, C and D periods can therefore considered analogous to the eukaryotic G1, S and G2 phases, respectively. However, despite considerable research, little is understood about how progress from the B to D periods is controlled.
In Eukaryotes it is known that the cell cycle is controlled through the interaction of cyclin and cyclin-dependent kinases5–7. However, to date there is no evidence for the presence of homologues of cyclin in bacteria. Furthermore, studies of Gram-negative species have been unable to produce evidence for regulation of cell division either through changes in gene expression8 or changes in the concentration of the proteins that comprise the divisome9, 10. Existing models for the control of bacterial cell division allow for several possible mechanisms. One is based on regulation of increase in cell size (the size adder theory11–13). Another is that cell size is regulated in advance of division, possibly through dilution of a ‘timekeeper’ protein, though the mechanism underlying this model is not known13, 14 and it is not clear whether such proteins would control, or are rate-limiting, with respect to the cell cycle. There are also hints that cell structure is a determinant in controlling the binary fission in bacteria15. In principle, control of progress through the cell cycle through changes in cell structure could be based on both structural proteins and lipids, but knowledge of the role of lipids in this is lacking.
Recent work on lipids and their metabolism in the Gram-positive bacterium Bacillus subtilis has revealed a link between the synthesis of phospholipids and the divisome. This evidence suggests that de novo synthesised lipids are produced at the septum during fission16. Furthermore, there is evidence for an overall change in the lipid profile of the Gram-negative Escherichia coli through its cell cycle15. The latter work showed that the abundance of curvature-forming cardiolipin (CL) falls whilst that of the bilayer-forming phosphatidylglycerol (PG) increased on elongation of the cell at the C/D boundary. Evidence from biophysical studies conducted over the last half-century shows that modulation of lipid composition can have a considerable impact on the lyotropic phase behaviour of lipid systems17, 18. For example, an increase in the fraction size of phosphatidylethanolamine (PE) increases the propensity of a lipid system to assemble into curved phases rather than bilayer-like ones19, 20. This is known as increasing stored curvature elastic stress, SCES21.
Determining the relationship between lipid composition and phase behaviour complements the interpretation of lipid profiling studies. Data from studies of the phase behaviour of lipids suggest that the increase in abundance of a bilayer-forming lipid and the reduction in abundance of a non-bilayer forming lipid that occurs in E. coli on elongation, reduces SCES22–25. This is consistent with the changes in morphology of the cell during elongation because average curvature is lower in elongated cells and the evidence that the location of the Z-ring may be determined by cell membrane geometry26–28. This led us to the hypothesis that the production and shape of the cell envelope is involved in controlling the cell cycle of bacteria. If correct, this hypothesis implies that the topology of the plasma membrane and thus the physical properties of its lipids play a significant role in progress through the cell cycle in bacteria.
In order to investigate whether modulation of lipid composition through the cell cycle is a general feature of bacteria we measured global changes in the lipid profile of a bacterium that is evolutionarily distinct from E. coli. Listeria innocua NCTC 11288 was chosen because, unlike the commonly studied model Gram-positive B. subtilis, it does not sporulate under stress. L. innocua also has clinical relevance29, due to its biological similarity to the pathogen L. monocytogenes 30, 31.
Cultures of L. innocua in the exponential phase were stopped after elongation but before cell division (around the C/D period boundary) with the bacteriostatic antibiotic rifampicin (RIF) using an adapted15 version of an established method32–34. The lipid fractions (LF) were isolated from freeze-dried, pelleted cultures whose cell walls had been thoroughly digested. The LFs were profiled using a combination of solution phase 31P NMR and HR-MS/MS35. The relationship between changes in lipid composition and membrane morphology observed in L. innocua was investigated using broad line 31P NMR to characterise the phase behaviour of model systems that corresponded to the lipid compositions observed.
Results
Culture growth and synchronisation
Cultures of L. innocua NCTC 11288 were grown in tryptic soya broth at 37 °C and the growth phase synchronised using RIF15. A growth curve was used to identify the appropriate point to administer the arrest drug (determined empirically to be 240 min (Fig. 1).
Figure 1.

Identification of growth phases and image analysis of cultures. Panel A, Growth curve for L. innocua NCTC 11288 grown in TSB at 37 °C (n = 3 independent experimental measurements per point). The lag phase lasted for approximately 150 min (------); exponential phase (untreated cultures) began at about 150 min and by 400 min the cells were in the stationary phase. In cultures treated with RIF, stationary phase was induced immediately although some growth was still detected for ~120 min. Samples in panels B and C (untreated controls) were collected after 240 and 360 min and sample in panel C was collected 240 min after RIF administration (at 480 min). Panels B–D show light and fluorescence micrographs of L. innocua; for fluorescent imaging samples were stained with Nile red (1000 × magnification; scale bar = 1 µm). A graph of the distribution of cell lengths is shown for each sample with Gaussian distribution (red line). Data represents 180–200 cells from at least three images from four independent cultivations. The Box plots show medians, 50% of data in the range (box), non-outliers range (whiskers) and outliers (circles and stars). Sample B is representative of cells in exponential phase growth (240 min after inoculation) with the majority of cells appearing to be in the B period (>80% synchrony; mean = 1.34 µm, ± 0.29 µm). Sample C is representative of cells at the end of the exponential phase (360 min after inoculation) and represents the B period of older cultures (>80% synchrony; mean = 1.22 µm, ±0.26 µm). Sample D is representative of cells arrested at the C/D boundary by RIF (80-90% synchrony; mean = 1.92 µm, ±0.44 µm). Only 6.7% of cells in the C/D boundary samples are as short or shorter than those seen to predominate in the untreated samples (t-test, p = < 0.01).
The experimental steps for preparation of samples and collection of profiling data are shown in Fig. S1. Based on their size distribution the synchrony of the untreated exponentially growing cultures was determined to be >80% (and the cell size analysis suggested that these were predominantly in the B period) (Fig. 1). The cell size was measured using cells stained with Nile Red observed using fluorescence microscopy. These data were used to establish the efficacy of RIF in arresting and holding cells at the C/D boundary and cells from such cultures were found to be significantly longer (mean = 1.92 µm, ± 0.44 µm) than those in the untreated samples (mean = 1.34 µm, ± 0.29), with only 6.7% being as short or shorter than those seen to predominate in the untreated samples (t-test, p = < 0.01). Based on these results the untreated samples were taken to be representative of cells in the B period, and the RIF-treated samples representative of cells at the C/D boundary.
We took several steps in order to avoid chemical and enzymatic degradation of lipids. After centrifugation cells were resuspended in PBS to suppress PI-PLC activity36, and a mixture of lipase inhibitors was added to suppress the activity of these enzymes ex vivo 15, 35. A mixture of mutanolysin37, lysozyme and RNAase was added to digest the cell wall and thus maximise access to the lipid fraction38.
Possible contamination of the cultures was checked for by inspecting the 1H spectrum of the FA fraction but none was found (that of Listeria ssp. 39–42 is distinct from other commonplace bacteria, such as E. coli 43–46. See Preparation of fatty acid isolates in Experimental Methods, and Fig. S2).
Isolation of the lipid fraction
The LFs were extracted using a two-step method that was designed to address the apparent short-comings of traditional methods47 (see Experimental for details). Triethylammonium ions were used to provide a counter-ion that is soluble in organic solvents and thus favours migration of phospholipid species into the organic phase. Dichloromethane was used in place of chloroform in order to minimise chemical degradation of lipid species47. Lastly, in order to maximise the extraction efficacy, the freeze-dried free-flowing powdered cellular matter was washed in a mixture of organic solvents (dichloromethane/methanol/triethylammonium chloride, 3:1:0.0005, v/v/w) before being dispersed in water and methanol, and extracted with dichloromethane. In order to test the effectiveness of this procedure, the aqueous fraction and precipitate from the isolation of the LF were freeze-dried and treated with strong acid (5% H2SO4) in order to hydrolyse any FA ester bonds and thus release any remaining FAs. The amount of FA found is treated as proportional to the mass of the lipid not collected during the isolation. The data from these measurements (1H NMR, see Experimental and Fig. S3) suggest that the procedure used in this study extracted >99% of the LF. This compares favourably with around 85% for the original Bligh & Dyer method48.
Profiling of the lipid fraction
The LFs were profiled using a combination of solution phase 31P NMR and HRMS/MS35. 31P NMR was used for quantification because it allows non-destructive and unambiguous high-resolution spectroscopy of the organically-soluble phosphorus-containing molecular species present. Lysylated lipids have not been profiled using 31P NMR before and so the shifts of lysyl-dioleoyl phosphatidylglycerol (lysyl-DOPG) were determined (Fig. S4). Representative spectra of the lipid profiling and the relative abundance of lipids in the B period and at the C/D boundary are shown in Fig. 2A. A combination of high resolution and tandem mass spectrometry were used to identify both the head groups present and the range of lipid isoforms present (Table S1 shows the lipid abundance and isoform profiles. Representative lipid fragmentation is shown in Fig. S5).
Figure 2.
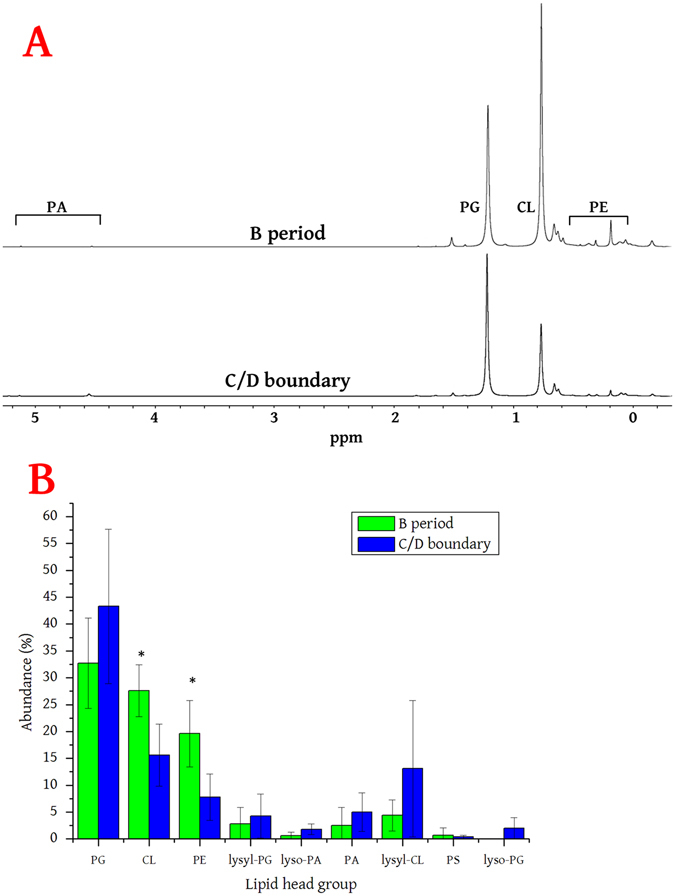
Profiling of the phospholipids in L. innocua NCTC 11288 at different stages of the cell cycle. Panel A shows representative 31P NMR spectra of the lipid fraction collected from B period cultures and cultures at the boundary of the C and D periods. The relative area of the integrations of the appropriate resonance(s) were taken as a fraction of the total integrations for each spectrum and used to generate the fraction size given for each sample. The shift of phosphate mono-ester-containing lipids such as PA, is pH dependent. PE exhibits several resonances due to concentration and pH-dependent solvent interactions15, 78. Panel B shows the abundance of lipids in the B period and at the C/D period boundary from cells collected in the exponential phase. Asterisks mark the head groups (CL, PE) for which the difference in abundance is considerable (when comparing standard deviations, n = 5 independent samples profiled using quantitative 31P NMR). CL, cardiolipin; lyso-PA, lyso-phosphatidic acid; lyso-PG, lyso-phosphatidylglycerol; lysyl-CL, lysyl-cardiolipin; lysyl-PG, lysyl-phosphatidylglycerol; PA, phosphatidic acid, PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine.
These data show that the abundance of both cardiolipin (CL) and phosphatidylethanolamine (PE) fall significantly between the B period and at the C/D period boundary. The fraction size of CL falls by around 40% whereas that of PE falls by about 60%. The absolute value for the abundance of PG varies between cultures, but does not appear to be statistically significant. This is perhaps surprising in view of the significant decreases in both CL and PE, however it is not clear why this should be. It seems unlikely that this is due to lysylation as the abundance of lysyl-PG is also not statistically significant, despite notable shifts in the absolute values. The abundance of major structural lipids CL and PE does not change substantially between cultures at the beginning and end of the exponential phase, while PG changes to some extent (Table S1). Unlike E. coli, the age of cultures of Listeria does not appear to correlate with changes in its lipid profile.
Profiling of the fatty acid residues (FARs) of the total lipid fraction did not indicate a difference in the FA profile between periods, suggesting that FA production is not linked to the cell cycle in this organism (Fig. S6). These data therefore indicate that although the lipid profile changes through the cell cycle of L. innocua it does not change with the age of rapidly-dividing cells. Furthermore, cell-cycle dependent changes are restricted to the head groups of the major structural species. Taken together, these data suggest that synthesis of the principal head groups in particular is linked to the expansion of the membrane during the B and C periods.
These data (Fig. 2B, Table S1) are consistent with the published evidence that the bulk of the Listeria LF comprises PE49 and lysyl-derivatives of PG and CL50, but is at odds with reports in which PE appears not to have been found39, 50. Conversely, phosphatidylinositol (PI) was not detected in any sample in this study despite the presence of several PLC inhibitors, contrasting with reports of the presence of PI in a closely-related strain of Listeria 42, 51. We note that the lipid profile of biological samples can be influenced by myriad factors, including the medium used, how cultures are incubated and handled, how the LF is isolated, handled and profiled, and even the activity of endogenous lipases ex vivo 47.
Lyotropic phase behaviour of model membrane systems
The evidence for modulations in the abundance of CL and PE during cell elongation raises the question of whether membrane properties change with these changes in composition. Studies of the phase behaviour of PG, PE and CL over the last half-century have shown that they have distinct properties. PE’s spontaneous curvature is negative19, 52–54, as is CL’s but only in the presence of a high concentration of divalent cations23, 55, 56. PG’s phase behaviour is dominated by lamellar phases, suggesting that its spontaneous curvature is negligible24, 25, 57–59. Modulation of the abundance of PE and CL (with a low concentration of divalent cations) is therefore a change in the abundance of two major structural lipids with contrasting spontaneous curvature. That these changes in abundance occur through the cell cycle therefore suggests that the stored curvature elastic stress of the membrane also changes through the cell cycle. We investigated this by modelling the behaviour of two-lipid mixtures using broad line 31P NMR.
Model systems comprised the di-oleoyl isoforms of PG and PE (DOPG and DOPE), and tetra-oleoyl isoform of CL (TOCL). Temperature scans of the individual lipids revealed the established behaviour, i.e. systems comprising either TOCL or DOPG showed fluid lamellar (Lα) phase over the temperature range studied (Fig. S7A,B), with DOPE undergoing a transition from Lα to inverse hexagonal (HII) only above 273 K (Fig. S7C).
Mixtures of DOPG with TOCL show a strong preference for Lα, even at relatively low temperatures (273 K, Fig. 3A; Fig. S8A–C), suggesting that both lipids confer fluidity on systems in which they are present. The Lα phase also dominates across most of the concentration range in mixtures of DOPG and DOPE at 273 K (Fig. 3B), but higher temperatures favour curved phases (e.g. 310 K, Fig. 3C; Fig. S9). These data indicate that PE confers stored curvature elastic stress on PG systems. In particular, a reduction in PE from 36% to 18%, similar to that on elongation of L. innocua, reduces the ability of that system to form curved phases, suggesting a significant reduction in SCES. The Lα dominates in mixtures of DOPE with TOCL where the concentration of DOPE is below 50% (273 K, Fig. 3D). This also indicates the preference for lamellar phase(s) by TOCL at low concentrations of divalent cations (Fig. S10A), and curved phases by PE (Fig. S10C). Thus, although the fraction of CL shrinks on elongation of L. innocua, the increase in that of the DOPG ensures that the reduction in the abundance of PE reduces SCES.
Figure 3.
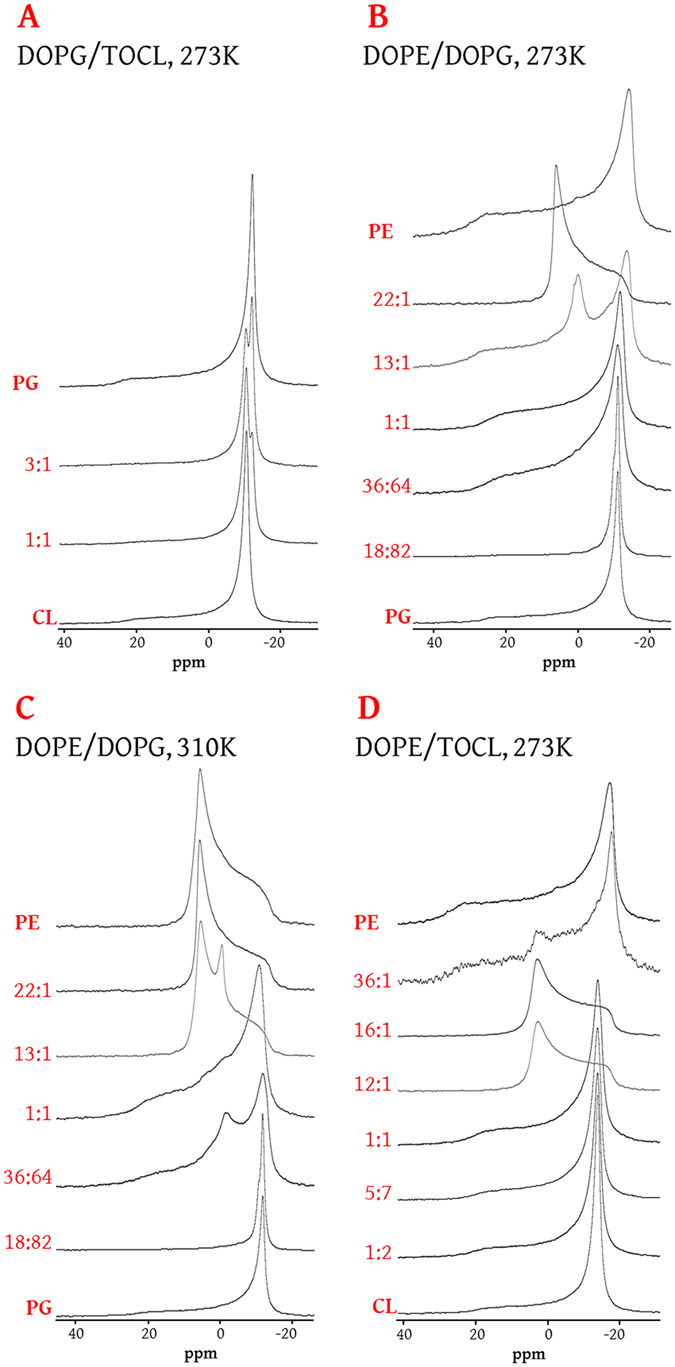
Stacked spectra showing the phase behaviour of two-lipid systems as a function of lipid head group composition. (A), DOPG/TOCL mixtures at 273 K; (B), DOPE/DOPG at 273 K; (C), DOPE/DOPG at 310 K; (D), DOPE/TOCL scan at 273 K. The 36:64 and 18:82 mixtures of DOPE:DOPG represent the ratio of these two lipids in the B period and at the C/D boundary, respectively. DOPG, dioleoylphosphatidylglycerol; TOCL, tetraoleoylcardiolipin; DOPE, phosphatidylethanolamine. Full temperature scans of each lipid mixture can be found in Figs S8–10.
However, it appears that doping DOPE with rather small quantities of DOPG (22-13:1, 4.3-7.1%) or TOCL (~36-12:1, 2.7-7.7%) allows the formation of curved phases at 273 K, particularly the HII phase (Figs 3B and S9, and Figs 3D and S10). Importantly, the phase transition from fluid lamellar to the HII phase occurs in DOPE around 10 K higher than this, at ∼ 281K60. This suggests that the presence of small quantities of these lipids can relieve the packing stress of the HII phase.
Data from a mixture of lysyl-DOPG with TOCL suggest that lysyl-DOPG has similar fluid properties to DOPG (Fig. S8C and D) but does not reduce the packing stress of DOPE systems as DOPG does (Fig. S9E and F). Notably, the behaviour of DOPE:lysyl-DOPG (22:1, Fig. S9F) is very similar to that of pure DOPE (Fig. S7C), and both are distinct from DOPE:DOPG (22:1, Fig. S9E). In the latter, the less abundant lipid appears to reduce the packing stress of the inverse hexagonal phase. This suggests that the role of lysyl-DOPG in the membrane differs from that of PG and may be related more to modulations in SCES and/or lipid packing than to fluidity.
Discussion
The evidence from this study indicates that the modulations in lipid composition in L. innocua that occur between the B period and C/D boundary are statistically significant. The behaviour of model systems suggests that the modulations in lipid composition on elongation of L. innocua represent a reduction in the SCES of the membrane (observed as a reduction in size of PE fraction, Fig. 3B) but without a reduction in its fluidity (with an increase in PG and despite a decrease in CL; Fig. 3A and D). This suggests that the physical properties of the membrane are linked to the cell cycle.
The behaviour of model lipid systems observed in this study supports the conclusion that changes in lipid composition favour changes in morphology of the membrane on a cellular scale. Comparison of the lipid profile and distribution between different rod-shaped bacteria (such as E. coli and B. subtilis) and between rod-shaped and spherical bacteria (such as Staphylococcus aureus) are therefore of interest in understanding the cellular structure of prokaryotes. However, comprehensive cell-cycle-based lipid profiling of prokaryotes is lacking at present. What is known, is that E. coli and B. subtilis have very different distributions of lipids in their membranes, which are linked to differences in proteins associated with cell division, and therefore cell shape is not expected to be the only factor that determines this change61.
We note that the modulations in lipid composition of the Gram-positive L. innocua are analogous to those reported for the Gram-negative bacterium, E. coli. In the latter organism, the abundance of the bilayer-forming PG increases during its elongation (from ~1:22 to ~1:16 against PE), with a reduction in the abundance of CL (from ~1:16 to ~1:36 against PE)15. Physical data from the present study suggest that there is also a reduction in the tendency to form the hexagonal phase (lower SCES) in E. coli, without a reduction in fluidity on progression from the B period to the C/D boundary. This indicates that E. coli has adopted the same pattern with respect to modulation of lyotropic phase behaviour between the B period and C/D boundary as seen in L. innocua.
This remarkable similarity between two bacteria that are quite separate in evolutionary terms raises the question of whether such modulations are important for the structure of rod-like cells in general. However, it is not clear from these data whether this trend is limited to cells with a cell wall; further work is required to establish what effects modulations in lipid composition have in other cell types.
It is appealing to speculate what further effects the modulations in lipid composition may have on membrane properties. The folding and function of proteins of both bacterial and eukaryotic origin have been shown to be sensitive to lipid membrane composition62–64, and to SCES and packing defects65, 66. This coupling between the membrane’s physical state and the proteins’ folding can also manifest itself in problematic rather than functional ways. For instance, there is evidence that charged head groups such as those of PG and phosphatidylserine enhance the membrane binding and mis-folding behaviour of α-Synuclein67, and that packing defects are involved in promoting mis-folding of a protein linked to Parkinson’s disease68, 69. Particular constellations of ganglioside and cholesterol have also been proposed to promote both membrane binding and mis-folding in α-Synuclein and β-amyloid peptide70.
It is clear that further work is required to fully characterise the relationship between lipid composition and distribution in bacteria71 and in eukaryotic72 cells. Data such as that acquired in this study indicate the extents to which the lipid composition and phase behaviour vary with the cell cycle. These alterations may in turn have an impact on the structural and functional properties of the membrane they comprise.
Experimental Methods
Reagents and chemicals
Growth medium, solvents and fine chemicals were purchased from SigmaAldrich (Gillingham, Dorset, UK) and used without purification. PhosSTOP tablets were purchased from Roche (Welwyn, Hertfordshire, UK) and stored at 4 °C. Purified lipids were purchased from Avanti Polar lipids Inc. (Alabaster, Alamaba, US) and used without further purification.
Cultivation of L. innocua
The growth kinetics of this bacterium in tryptic soya broth (TSB) was characterised over 24 h (Fig. 1). The preparation of one set (n = 1) of cultures is described. Each sample comprised three cultures in order to ensure that enough material was produced and to account for differences between populations. Thus for each biological replicate, 3 × 3 × 1 L cultures were grown in unmodified TSB that had been autoclaved. Mini-cultures (10 × 10 mL) were incubated (16 h, 37 °C, orbital incubator, 250 r.p.m.,) before inoculating 9 × 1 L to give cultures of OD620 ~0.03. After incubation (37 °C, 240 min, orbital, 250 r.p.m., average OD620 0.4), three cultures were mixed together to make the 4 h control time point and cells harvested (fixed-angle rotor, 6k × g, 10 min, 4 °C). Three cultures were treated with RIF (50 mg/L final concentration, added as methanolic solution of 50 mg/mL). After 360 min h, the three remaining control cultures were mixed and harvested together (making the 6 h control time point which is equivalent to the point at which the cultures treated with RIF are arrested), and after 480 min the remaining three cultures treated with RIF were harvested together.
Fluorescence and light microscopy
Cells were photographed after harvesting and before the administration of lipase inhibitors. Cells were stained with Nile Red as previously described73–75. Briefly, cells were collected by centrifugation (fixed-angle rotor, 6 k × g, 10 min, 4 °C), washed once with PBS and resuspended in PBS at OD620 ~2.0. Nile Red (2 µg/mL, sample volume 1 mL, dye stock solution in DMSO) was added and the sample (1 mL) allowed to stand at room temperature (20 min). Cells (60–80 per picture, 180–200 per sample) were measured from at least three images for each culture/time point each from four independent cultivations (i.e. (~70 × 3) × 3 × 4). The mean and standard deviation were calculated for each and an independent student t-test was used to determine statistical significance (p = < 0.01). The null hypothesis was that the lengths of the bacteria are the same for each sample. Images were taken using a Leica DMI6000 B microscope equipped with a Leica DFC350 FX camera and processed with (the 3D deconvolution function in) Leica Application Suite, Advanced fluorescence 1.7.0 software with AF6000 configuration and open source ImageJ software.
Isolation of lipid fraction
Fresh cell pellets were resuspended immediately (PBS, 5 mL) and treated with PhosSTOP (1 tab/10 mL final concentration, 10 × stock dissolved in PBS), 2-butoxyphenylboronic acid (BPBA, 2 mg/mL final concentration, ethanolic stock solution 100 mg/mL) and a stored mixture of mutanolysin (50 µg), lysozyme (10 mg), RNAase (200 µg) (volume of aliquots 1 mL, glycerol/PBS 1:1, stored at 193 K). The total volume of the resulting solutions was 10 mL per sample and they were agitated gently in air tight falcon tubes (50 mL, gel staining table, 16–20 h) before being frozen (193 K) and freeze-dried. The freeze-dried material was stored at 253 K until isolation of the lipid fraction.
The free-flowing powdered cellular matter was mixed with dichloromethane/methanol (20 mL, 3:1, 0.5 mg/mL triethylammonium chloride, TEAC) and sonicated (sonication bath, 5 min) before being centrifuged (5 min, 5.25 k × g). The organic solution was decanted and retained. The pellet was mixed with more of the same dichloromethane/methanol mixture (20 mL, 3:1, 0.5 mg/mL TEAC), agitated, centrifuged and the solvent decanted. The pellet was then resuspended in a mixture of dichloromethane and water (20 mL, 1:1, separating funnel) and diluted with sufficient methanol to make a stable uniphasic solution (~40 mL), and agitated briefly. The mixture was then made biphasic by addition of dichloromethane (20 mL). The dichloromethane solution was removed and the aqueous solution washed (dichloromethane, 20 mL). TEAC was added to the remaining aqueous solution (final concentration of 2 mM, 2 M stock) and the aqueous solution washed with dichloromethane (2 × 20 mL). The combined organic solutions (~130 mL) were filtered through filter paper and concentrated in vacuo before storage under nitrogen at 253 K.
Isolation of residual lipidic material
The aqueous phase and precipitate produced during the lipid isolation (above) were freeze-dried together and then treated with acid (H2SO4, 5%, 20 mL, gentle agitation, 24 h, r.t.p.). The reaction mixture was quenched (NaHCO3) and freeze-dried. The free-flowing powder was washed twice with chloroform. The organic phases were combined and concentrated in vacuo. The remaining material (~8 mg/120 mg LF isolated) was dissolved in deuterated chloroform (650 µL) before commencing 1H NMR. The integrals of each spectrum were calibrated according to the α-CH2 signal (2.3–2.4 ppm) present (Fig. S4).
Solution phase 31P NMR spectroscopy
Lipid films (10-15 mg) from cultures of L. Innocua were dissolved in the CUBO solvent system15, 35, 76 (500 µL/sample). Data acquisition was similar to published work15, 35 but using a Bruker 400 MHz Avance III HD spectrometer equipped with a 5 mm BBO S1 (smart) probe operating at 298 K for all data except that shown in Fig. S5. 31P spectra were acquired at 161.98 MHz using inverse gated proton decoupling, with 2048 scans per sample and a spectral width of 19.99 ppm. An overall recovery delay of 6.5 s was used which gave full relaxation. Data were processed using line broadening of 1.00 Hz prior to zero filling to 19428 points, Fourier transform and automatic baseline correction. The data for Fig. S5 were acquired on a Bruker Avance III 800 MHz spectrometer, equipped with a QCI cryoprobe probe. Acquisition used inverse gated proton decoupling. Spectra were averaged over 1312 transients with 3882 complex points with a spectral width of 14.98 ppm. An overall recovery delay of 8.4 s was used. Data were processed using an exponential line broadening window function of 1.5 Hz prior to zero filling to 32768 points, Fourier transform and automatic baseline correction. All spectra were processed and analysed using (the dcon function in) TopSpin 3.2.
Preparation of fatty acid isolates
Representative samples used for head group profiling were dried in vacuo (high-vac pump) before being diluted (H2SO4 97% in methanol, 1:20 v/v final concentration, 5 mL) and heated under intermittent agitation (4–6 h, 50 °C). The reaction mixture was quenched (NaHCO3, 1 g) before being diluted (H2O, 30 mL; diethyl ether, 20 mL). The mixture was shaken vigorously in a separating funnel before the organic fraction was collected. The aqueous solution was washed (diethyl ether, 20 mL). The organic solutions were combined and dried in vacuo. The resulting oil (5–10 mg) was dissolved (CDCl3, 500–600 µL) and subjected to 1H NMR spectroscopy. δH (500 MHz, 16 scans, CDCl3, calibrated to 7.24 ppm) 3.6 (singlet, RCH2CH2COOCH 3), 2.3 (triplet, J = 7.6 Hz, RCH2CH 2COOCH3), 1.6-1.5 (m, RCH 2CH2COOCH3), 1.4-1.0 (m, methylenes), 0.9-0.8 (m, methyl groups) ppm.
Mass spectrometry
Samples were prepared and measurements taken according to published methods15, 35. Accurate mass LC-MS and MS/MS was performed on an OrbiTrap Dionex 3000, (Waltham, MA, USA). Dry lipid mixtures were dissolved in dichloromethane/isopropanol 1:1 and an inject volume of 10 µL was used. Analytes were separated chromatographically on a UPLC C18 column (1.7 µm particle size, Waters) at 40 °C at a rate of 0.4 mL/min. Mobile phase A consisted of 0.1% formic acid in water at pH 6.0, and mobile phase B was 56% acetonitrile, 40% isopropanol and 5% water with 0.1% formic acid. Ions were monitored in positive mode (m/z range 300-2000, resolution 140,000). Selected peaks were fragmented (normalised collision energy set to 36) and MS2 collected with a resolution of 17,500.
Analyses consisted of searches for lipid isoforms in MS1 spectra, followed by fragmentation (MS2) for appropriate m/z values. Lipid isoform searches were restricted by head group (PG, PE, PA, PS and CL), FAR (chain length from 10 to 20 C, up to three double bonds) and adduct (H+, Na+, K+, NH4 +). The search was augmented to include lysyl and lyso- isoforms. Matches were compared (see Fig. S6 for example). The software-generated scores were checked manually. Analyses were performed using Thermo Xcalibur 3.0.63 and recently-developed software by Kochen et al.77. Original code for non-standard head groups was written by us in Matlab R2015b.
Broad line 31P NMR sample preparation
Lipid mixtures were made in solution (dichloromethane, 10–15 mg/mL) and dried to a lipid film in vacuo. Dried lipid films were dispersed in aqueous buffer (NaCl 100 mM, tris 50 mM, CaCl2 2.5 mM, MgCl2 2.5 mM, pH 7.4; 30:1 v/w) with sonication and agitation. Deuterium oxide (10% v/v against buffer) was added and the mixture agitated and then freeze-thawed 8–10 times. Samples were then stored at 193 K, transported at ~295 K and were stored at 253 K before running.
Broad line 31P NMR spectroscopy
A Bruker Avance III HD 400 MHz DRX spectrometer equipped with a Bruker BBO S1 (smart) probe was used for proton-decoupled broad line 31P NMR experiments. Experiments were performed at 161.98 MHz with an inverse-gated pulse sequence with proton decoupling during acquisition, spectral width of 200.44 ppm, acquisition time of 0.299 s, pre-scan delay (DE) time of 20 µs, receiver gain of 203 with 19428 data points and 2048 scans per spectrum. The duration of each scan, acquisition time and relaxation delay, was 2.332 s. Spectra were processed using TopSpin 3.2 and 3.5 with line broadening of 50 Hz, phase correction and automatic baseline correction (abs).
Broad line 31P NMR temperature scans
Samples were brought to and held at the desired temperature for 15 min before acquisition. Temperature scans consisted of acquisitions at the following temperatures: 298, 273, 293, 298, 310, 318, 273, 310 K. The latter two temperature points were used to assess the reliability of the equilibration time used (15 min). Acquisitions at 338 K were carried out separately with equilibration and acquisition at 310 K immediately beforehand. Stackplots of spectra were prepared manually from individual traces processed as above.
Electronic supplementary material
Acknowledgements
The Research Council of Norway is acknowledged for funding (grant application ES528542), and for financial support through the Norwegian NMR Platform, NNP (grant 226244 / F50). The authors wish to thank Drs Fedor Kryuchkov and Marc Niere for assistance with MS and microscopy, respectively, and SF would also like to thank Drs J. Loewe, S. Liddell and T. Allers for helpful discussions. The authors thank Drs Ø. Larsen and A.E. Lewis for critical review of the manuscript and acknowledge Dirk Petersen (NNP UiO node) and Olav-Audun Bjørkelund (NNP UiB node) for their help in facilitating the work.
Author Contributions
The laboratory work was carried out by S.F. and M.J., and supervised by S.F. and Ø.H. NMR data was collected by F.R. who also advised on acquisition parameters, and H.E.W. Data were analysed and interpreted by S.F., M.J., C.R. and Ø.H. C.R. supplied cultivation and handling protocols and Listeria innocua NCTC 11288 starting culture. S.F. and Ø.H. conceived the research questions and designed the experiments. The original grant proposal and the research programme were conceived, devised and written by Ø.H. The manuscript was written by S.F. and Ø.H., and figures prepared by S.F. and M.J. All authors commented on the manuscript and approved the final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06855-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bates D, et al. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferullo DJ, Lovett ST. The Stringent Response and Cell Cycle Arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haeusser DP, Levin PA. The Great Divide: Coordinating cell cycle events during bacterial growth and division. Current opinion in microbiology. 2008;11:94–99. doi: 10.1016/j.mib.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell LH. Nobel Lecture: Yeast and Cancer. Bioscience Reports. 2002;22:373–394. doi: 10.1023/A:1020918107706. [DOI] [PubMed] [Google Scholar]
- 6.Hunt T. Nobel Lecture: Protein Synthesis, Proteolysis, and Cell Cycle Transitions. Bioscience Reports. 2002;22:465–486. doi: 10.1023/A:1022077317801. [DOI] [PubMed] [Google Scholar]
- 7.Nurse P. Cyclin Dependent Kinases and Cell Cycle Control (Nobel Lecture) ChemBioChem. 2002;3:596–603. doi: 10.1002/1439-7633(20020703)3:7<596::AID-CBIC596>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Arends SJR, Weiss DS. Inhibiting Cell Division in Escherichia coli Has Little If Any Effect on Gene Expression. Journal of Bacteriology. 2004;186:880–884. doi: 10.1128/JB.186.3.880-884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rueda S, Vicente M, Mingorance J. Concentration and Assembly of the Division Ring Proteins FtsZ, FtsA, and ZipA during the Escherichia coli Cell Cycle. Journal of Bacteriology. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si F, Busiek K, Margolin W, Sun, Sean X. Organization of FtsZ Filaments in the Bacterial Division Ring Measured from Polarized Fluorescence Microscopy. Biophysical Journal. 2013;105:1976–1986. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taheri-Araghi S, et al. Cell-Size Control and Homeostasis in Bacteria. Current Biology. 2015;25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos M, et al. A Constant Size Extension Drives Bacterial Cell Size Homeostasis. Cell. 2014;159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghusinga KR, Vargas-Garcia CA, Singh A. A mechanistic stochastic framework for regulating bacterial cell division. Scientific Reports. 2016;6:30229. doi: 10.1038/srep30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robert L. Size sensors in bacteria, cell cycle control and size control. Frontiers in Microbiology. 2015;6:515. doi: 10.3389/fmicb.2015.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furse S, Wienk H, Boelens R, de Kroon AIPM, Killian JA. E. coli MG1655 modulates its phospholipid composition through the cell cycle. FEBS Letters. 2015;589:2726–2730. doi: 10.1016/j.febslet.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 16.Takada H, et al. An essential enzyme for phospholipid synthesis associates with the Bacillus subtilis divisome. Molecular Microbiology. 2014;91:242–255. doi: 10.1111/mmi.12457. [DOI] [PubMed] [Google Scholar]
- 17.Seddon, J. M. & Templer, R. H. in The Handbook of Biological Physics Vol. I (eds R. Lipowsky & E. Sackman) (Elsevier Science, 1995).
- 18.Cullis, P. R. & De Kruijff, B. Lipid polymorphism and the functional roles of lipids in biological membranes. ##Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes559, 399–420, doi:10.1016/0304-4157(79)90012-1 (1979). [DOI] [PubMed]
- 19.Cullis PR, De Kruijff B. The polymorphic phase behaviour of phosphatidylethanolamines of natural and synthetic origin. A 31P NMR study. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1978;513:31–42. doi: 10.1016/0005-2736(78)90109-8. [DOI] [PubMed] [Google Scholar]
- 20.Marsh D, Seddon JM. Gel-to-inverted hexagonal (L[beta]-HII) phase transitions in phosphatidylethanolamines and fatty acid-phosphatidylcholine mixtures, demonstrated by 31P-NMR spectroscopy and X-ray diffraction. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1982;690:117–123. doi: 10.1016/0005-2736(82)90245-0. [DOI] [PubMed] [Google Scholar]
- 21.Seddon JM. Structure of the Inverted Hexagonal (Hii) Phase, and Non-Lamellar Phase-Transitions of Lipids. Biochim Biophys Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-T. [DOI] [PubMed] [Google Scholar]
- 22.Seddon JM, Kaye RD, Marsh D. Induction of the lamellar-inverted hexagonal phase transition in cardiolipin by protons and monovalent cations. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1983;734:347–352. doi: 10.1016/0005-2736(83)90134-7. [DOI] [Google Scholar]
- 23.Ortiz A, Killian JA, Verkleij AJ, Wilschut J. Membrane Fusion and the Lamellar-to-Inverted-Hexagonal Phase Transition in Cardiolipin Vesicle Systems Induced by Divalent Cations. Biophysical Journal. 1999;77:2003–2014. doi: 10.1016/S0006-3495(99)77041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alley SH, Ces O, Barahona M, Templer RH. X-ray diffraction measurement of the monolayer spontaneous curvature of dioleoylphosphatidylglycerol. Chemistry and Physics of Lipids. 2008;154:64–67. doi: 10.1016/j.chemphyslip.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Findlay EJ, Barton PG. Phase behavior of synthetic phosphatidylglycerols and binary mixtures with phosphatidylcholines in the presence and absence of calcium ions. Biochemistry. 1978;17:2400–2405. doi: 10.1021/bi00605a023. [DOI] [PubMed] [Google Scholar]
- 26.Pazos M, et al. FtsZ Placement in Nucleoid-Free Bacteria. PLOS ONE. 2014;9:e91984. doi: 10.1371/journal.pone.0091984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Molecular Microbiology. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 28.Rowlett, V. W. & Margolin, W. The bacterial divisome: ready for its close-up. Philosophical Transactions of the Royal Society B: Biological Sciences370, doi:10.1098/rstb.2015.0028 (2015). [DOI] [PMC free article] [PubMed]
- 29.Perrin M, Bemer M, Delamare C. Fatal Case of Listeria innocua Bacteremia. Journal of Clinical Microbiology. 2003;41:5308–5309. doi: 10.1128/JCM.41.11.5308-5309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jemmi T, Stephan R. Listeria monocytogenes: food-borne pathogen and hygiene indicator. Rev Sci Tech. 2006;25:571–580. doi: 10.20506/rst.25.2.1681. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty T, Hain T, Domann E. Genome organization and the evolution of the virulence gene locus in Listeria species. International Journal of Medical Microbiology. 2000;290:167–174. doi: 10.1016/S1438-4221(00)80086-7. [DOI] [PubMed] [Google Scholar]
- 32.Boye E, Løbner-Olesen A. Bacterial growth control studied by flow cytometry. Research in Microbiology. 1991;142:131–135. doi: 10.1016/0923-2508(91)90020-B. [DOI] [PubMed] [Google Scholar]
- 33.Boye E, Løbner-Olesen A, Skarstad K. Timing of chromosomal replication in Escherichia coli. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1988;951:359–364. doi: 10.1016/0167-4781(88)90107-8. [DOI] [PubMed] [Google Scholar]
- 34.Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome-replication in individual Escherichia coli cells. Embo Journal. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furse S, et al. The lipidome and proteome of oil bodies from Helianthus annuus (common sunflower) Journal of chemical biology. 2013;6:63–76. doi: 10.1007/s12154-012-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldfine H, Johnston NC, Knob C. Non-specific phospholipase C of Listeria monocytogenes: activity on phospholipids in Triton X-100-mixed micelles and in biological membranes. Journal of Bacteriology. 1993;175:4298–4306. doi: 10.1128/jb.175.14.4298-4306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Netterling S, Vaitkevicius K, Nord S, Johansson J. A Listeria monocytogenes RNA Helicase Essential for Growth and Ribosomal Maturation at Low Temperatures Uses Its C Terminus for Appropriate Interaction with the Ribosome. Journal of Bacteriology. 2012;194:4377–4385. doi: 10.1128/JB.00348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filgueiras MH, den Kamp JAFO. Cardiolipin, a major phospholipid of Gram positive bacteria that is not readily extractable. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism. 1980;620:332–337. doi: 10.1016/0005-2760(80)90215-5. [DOI] [PubMed] [Google Scholar]
- 39.Tatituri RVV, Wolf BJ, Brenner MB, Turk J, Hsu FF. Characterization of polar lipids of Listeria monocytogenes by HCD and low-energy CAD linear ion-trap mass spectrometry with electrospray ionization. Anal Bioanal Chem. 2015;407:2519–2528. doi: 10.1007/s00216-015-8480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chihib N-E, Ribeiro da Silva M, Delattre G, Laroche M, Federighi M. Different cellular fatty acid pattern behaviours of two strains of Listeria monocytogenes Scott A and CNL 895807 under different temperature and salinity conditions. FEMS Microbiol. Lett. 2003;218:155–160. doi: 10.1111/j.1574-6968.2003.tb11512.x. [DOI] [PubMed] [Google Scholar]
- 41.Püttmann M, Ade N, Hof H. Dependence of fatty acid composition of Listeria spp. on growth temperature. Research in Microbiology. 1993;144:279–283. doi: 10.1016/0923-2508(93)90012-Q. [DOI] [PubMed] [Google Scholar]
- 42.Mastronicolis SK, et al. Alteration of the phospho- or neutral lipid content and fatty acid composition in Listeria monocytogenes due to acid adaptation mechanisms for hydrochloric, acetic and lactic acids at pH 5.5 or benzoic acid at neutral pH. Antonie van Leeuwenhoek. 2010;98:307–316. doi: 10.1007/s10482-010-9439-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metabolic Engineering. 2010;12:378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Ingram LO. Adaptation of membrane lipids to alcohols. Journal of Bacteriology. 1976;125:670–678. doi: 10.1128/jb.125.2.670-678.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grogan DW, Cronan JE. Cyclopropane ring formation in membrane lipids of bacteria. Microbiology and Molecular Biology Reviews. 1997;61:429–441. doi: 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y-Y, Cronan JE. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Molecular Microbiology. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 47.Furse S, Egmond MR, Killian JA. Isolation of lipids from biological samples. Molecular Membrane Biology. 2015;32:55–64. doi: 10.3109/09687688.2015.1050468. [DOI] [PubMed] [Google Scholar]
- 48.Bligh EG, Dyer WJ. A Rapid Method of Total Lipid Extraction and Purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 49.Murgia S, Mele S, Monduzzi M. Quantitative characterization of phospholipids in milk fat via P-31 NMR using a monophasic solvent mixture. Lipids. 2003;38:585–591. doi: 10.1007/s11745-003-1500-3. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh BK, Carroll KK. Isolation, Composition, and Structure of Membrane of Listeria monocytogenes. Journal of Bacteriology. 1968;95:688–699. doi: 10.1128/jb.95.2.688-699.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer W, Leopold K. Polar lipids of four listeria species containing L-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. International Journal of Systematic Bacteriology. 1999;49:653–662. doi: 10.1099/00207713-49-2-653. [DOI] [PubMed] [Google Scholar]
- 52.Mastronicolis SK, et al. Coordinated Regulation of Cold-Induced Changes in Fatty Acids with Cardiolipin and Phosphatidylglycerol Composition among Phospholipid Species for the Food Pathogen Listeria monocytogenes. Applied and Environmental Microbiology. 2008;74:4543–4549. doi: 10.1128/AEM.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seelig J, Gally HU. Investigation of phosphatidylethanolamine bilayers by deuterium and phosphorus-31 nuclear magnetic resonance. Biochemistry. 1976;15:5199–5204. doi: 10.1021/bi00669a001. [DOI] [PubMed] [Google Scholar]
- 54.Hauser H, Pascher I, Pearson RH, Sundell S. Preferred conformation and molecular packing of phosphatidylethanolamine and phosphatidylcholine. Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes. 1981;650:21–51. doi: 10.1016/0304-4157(81)90007-1. [DOI] [PubMed] [Google Scholar]
- 55.Kollmitzer B, Heftberger P, Rappolt M, Pabst G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/c3sm51829a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Killian JA, et al. Effect of divalent cations on lipid organization of cardiolipin isolated from Escherichia coli strain AH930. Biochimica et Biophysica Acta. 1994;1189:225–232. doi: 10.1016/0005-2736(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 57.Rietveld AG, Killian JA, Dowhan W, Dekruijff B. Polymorphic Regulation of Membrane Phospholipid composition in Escherichia coli. Journal of Biological Chemistry. 1993;268:12427–12433. [PubMed] [Google Scholar]
- 58.Krumova SB, et al. Phase behavior of phosphatidylglycerol in spinach thylakoid membranes as revealed by 31P-NMR. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2008;1778:997–1003. doi: 10.1016/j.bbamem.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 59.Murata N, Yamaya J. Temperature-Dependent Phase Behavior of Phosphatidylglycerols from Chilling-Sensitive and Chilling-Resistant Plants. Plant Physiology. 1984;74:1016–1024. doi: 10.1104/pp.74.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epand RM, et al. Formation of a new stable phase of phosphatidylglycerols. Biophysical Journal. 1992;63:327–332. doi: 10.1016/S0006-3495(92)81618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koynova R, Caffrey M. Phases and phase transitions of the hydrated phosphatidylethanolamines. Chemistry and Physics of Lipids. 1994;69:1–34. doi: 10.1016/0009-3084(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 62.Barák I, Muchová K. The Role of Lipid Domains in Bacterial Cell Processes. International Journal of Molecular Sciences. 2013;14:4050. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agasøster AV, et al. The Interaction of Peripheral Proteins and Membranes Studied with α-Lactalbumin and Phospholipid Bilayers of Various Compositions. Journal of Biological Chemistry. 2003;278:21790–21797. doi: 10.1074/jbc.M211466200. [DOI] [PubMed] [Google Scholar]
- 64.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proceedings of the National Academy of Sciences. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halskau Ø, et al. Large-scale modulation of thermodynamic protein folding barriers linked to electrostatics. Proceedings of the National Academy of Sciences. 2008;105:8625–8630. doi: 10.1073/pnas.0709881105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McDonald, C., Jovanovic, G., Ces, O. & Buck, M. Membrane Stored Curvature Elastic Stress Modulates Recruitment of Maintenance Proteins PspA and Vipp1. mBio6, doi:10.1128/mBio.01188-15 (2015). [DOI] [PMC free article] [PubMed]
- 67.Arduin A, Gaffney PRJ, Ces O. Regulation of PLCβ2 by the electrostatic and mechanical properties of lipid bilayers. Scientific Reports. 2015;5:12628. doi: 10.1038/srep12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stöckl M, Fischer P, Wanker E, Herrmann A. α-Synuclein Selectively Binds to Anionic Phospholipids Embedded in Liquid-Disordered Domains. Journal of Molecular Biology. 2008;375:1394–1404. doi: 10.1016/j.jmb.2007.11.051. [DOI] [PubMed] [Google Scholar]
- 69.Lokappa SB, et al. Sequence and Membrane Determinants of the Random Coil–Helix Transition of α-Synuclein. Journal of Molecular Biology. 2014;426:2130–2144. doi: 10.1016/j.jmb.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 70.Ghio S, Kamp F, Cauchi R, Giese A, Vassallo N. Interaction of α-synuclein with biomembranes in Parkinson's disease —role of cardiolipin. Progress in Lipid Research. 2016;61:73–82. doi: 10.1016/j.plipres.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Di Scala C, et al. Common molecular mechanism of amyloid pore formation by Alzheimer’s β-amyloid peptide and α-synuclein. Scientific Reports. 2016;6:28781. doi: 10.1038/srep28781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furse S, Scott DJ. Three-Dimensional Distribution of Phospholipids in Gram Negative Bacteria. Biochemistry. 2016;55:4742–4747. doi: 10.1021/acs.biochem.6b00541. [DOI] [PubMed] [Google Scholar]
- 73.van Meer G, de Kroon AIPM. Lipid map of the mammalian cell. Journal of Cell Science. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 74.Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbüchel A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Archives of Microbiology. 1999;171:73–80. doi: 10.1007/s002030050681. [DOI] [PubMed] [Google Scholar]
- 75.Donovan C, Schwaiger A, Krämer R, Bramkamp M. Subcellular Localization and Characterization of the ParAB System from Corynebacterium glutamicum. Journal of Bacteriology. 2010;192:3441–3451. doi: 10.1128/JB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Culeddu N, Bosco M, Toffanin R, Pollesello P. P-31 NMR analysis of phospholipids in crude extracts from different sources: improved efficiency of the solvent system. Magnetic Resonance in Chemistry. 1998;36:907–912. doi: 10.1002/(SICI)1097-458X(199812)36:12<907::AID-OMR394>3.0.CO;2-5. [DOI] [Google Scholar]
- 77.Strahl H, Bürmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nature Communications. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kochen MA, et al. Greazy: Open-Source Software for Automated Phospholipid Tandem Mass Spectrometry Identification. Analytical Chemistry. 2016;88:5733–5741. doi: 10.1021/acs.analchem.6b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


