Abstract
Cpefat/Cpefat mice have a naturally occurring point mutation within the carboxypeptidase E gene that inactivates this enzyme, leading to an accumulation of many neuroendocrine peptides containing C-terminal basic residues. These processing intermediates can be readily purified on an anhydrotrypsin affinity resin. Using MS to obtain molecular mass and partial sequence information, more than 100 peptides have been identified. These peptides represent fragments of 16 known secretory pathway proteins, including proenkephalin, proopiomelanocortin, protachykinins A and B, chromogranin A and B, and secretogranin II. Many of the identified peptides represent previously uncharacterized fragments of the precursors. For example, 12 of the 13 chromogranin B-derived peptides found in the present study have not been previously reported. Of these 13 chromogranin B-derived peptides, only five contain consensus cleavage sites for prohormone convertases at both the C and N termini. Two distinct chromogranin B-derived peptides result from cleavage at Trp-Trp bonds, a site not typically associated with neuropeptide processing. An RIA was used to confirm that one of these peptides, designated WE-15, exists in wild-type mouse brain, thus validating the approach to identify peptides in Cpefat/Cpefat mice. These “orphan” peptides are candidate ligands for orphan G protein-coupled receptors. In addition, the general technique of using affinity chromatography to isolate endogenous substrates from a mutant organism lacking an enzyme should be applicable to a wide range of enzyme-substrate systems.
Keywords: peptide processing, carboxypeptidase E, carboxypeptidase D, chromogranin, secretogranin
Most neuroendocrine peptides initially are produced from precursors by limited proteolysis. Proteolytic processing provides a mechanism for regulating the bioactivity of the resulting peptides and/or for controlling the stoichiometry of various peptide products (1–3). In general, the sites of proteolysis contain basic amino acids with the consensus site (basic)Xn(basic), where X is any amino acid(s) other than Cys and n is 0, 2, 4, or 6 (4, 5). The most frequent cleavage sites are Lys-Arg, Arg-Arg, and Arg-X-X-Arg, although cleavage also can occur at Lys-Lys and Arg-Lys sites (6). These sites are initially cleaved at the C-terminal side of the basic residues by a prohormone convertase (PC), such as PC1 (also known as PC3) or PC2 (4, 5). Following the action of the endopeptidase, a carboxypeptidase (CP) removes the C-terminal basic residues from the intermediate (7, 8).
The CP processing step is primarily catalyzed by CPE (EC3.4.17.10). CPE is present in peptide-containing secretory vesicles in neuroendocrine tissues and removes C-terminal Arg and Lys residues from a wide range of peptides (9, 10). A spontaneous mutation in the coding region of the CPE gene in Cpefat/Cpefat mice that causes a Ser202 to Pro substitution leads to inactivation of the enzyme and degradation of CPE protein (7, 11, 12). The absence of CPE activity causes the accumulation of C-terminally extended insulins, enkephalins, and other neuroendocrine peptides (11, 13–16). However, low levels of correctly processed peptides are detected in Cpefat/Cpefat tissues (11, 13–16), presumably due to processing by CPD in the trans-Golgi network (17–21).
The analysis of peptide processing has been limited by the tools that are available. Most studies on the in vivo processing of peptides rely on antisera to specific peptides and characterize the peptides primarily by their elution properties on various columns. The accumulation of neuroendocrine peptides with C-terminal basic residues in Cpefat/Cpefat mice provides a handle by which to purify these peptides using an immobilized anhydrotrypsin resin that specifically binds peptides with C-terminal basic groups. In the present study, we have used affinity chromatography in combination with MS to analyze peptides with C-terminal basic residues in brain and pituitary of Cpefat/Cpefat mice. This analysis has revealed a large number of novel fragments of known peptide precursors. One major finding is that processing is not limited to the conventional basic amino acid sites, but also occurs at a variety of other positions. In addition, the general approach described here can be easily modified to identify endogenous substrates of a variety of enzymes.
Materials and Methods
Purification of Peptides from Cpefat/Cpefat and Wild-Type Control Mice.
Cpefat/Cpefat mice and wild-type littermates (The Jackson Laboratory) were killed at 9–10 weeks of age. Tissues were removed, frozen on dry ice, and stored at −80°C. To confirm the genotyping of the mice, a portion of the cerebral cortex was removed before freezing and used to measure CPE enzymatic activity (22). Tissues from 10–20 mice were added to 10 vol of boiling 0.1 M acetic acid and homogenized (Polytron, Brinkmann). The homogenate was boiled for 10 min and centrifuged at 50,000 × g for 30 min at 4°C, and the supernatant was filtered through a Centriplus-10 membrane (Amicon). The flow through was adjusted to pH 5.0 with sodium acetate and combined with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (0.5% final) and CaCl2 (20 mM final). This mixture was applied to a column containing 0.5 ml of anhydrotrypsin-agarose (Panvera, Madison, WI) equilibrated in the same buffer. The flow through was recycled through the column several times. The column was washed first with 0.5% CHAPS in 100 mM Na acetate, pH 5.0 buffer and then with 10 mM Na acetate alone. Peptides were eluted with water (2 ml) and then 5 mM HCl (4 ml) and lyophilized.
Matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF) spectra of the samples were obtained on a Voyager DE-STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) in a positive linear mode using α-cyano-4-hydroxy-cinnamic acid saturated in 30% acetonitrile and 0.1% trifluoroacetic acid in water. Approximately 100 laser shots were summed per spectrum.
Liquid chromatography (LC) MS was performed by using a HP1100 HPLC (Hewlett–Packard) connected on-line to either a LCQ electrospray ionization ion trap mass spectrometer (Finnigan Thermoquest, San Jose, CA) or a Mariner electrospray ionization TOF mass spectrometer (PerSeptive Biosystems). In some experiments, disulfide bonds were reduced with 15 mM Tris(2-carboxyethyl)phosphine for 30 min at room temperature before MS. In a typical experiment, 10–20% of each sample was run on a reverse-phase C18 capillary column (Hypersil C18BDS, 3 μm, I.D. 300 μm, LC Packings) using a gradient from 7% to 58% acetonitrile in 0.1% formic acid over 70 min and a column flow rate of 4 μl/min. After this initial run and subsequent analysis of the MS data (see below), another 10–20% of the sample was run again to obtain collision-induced dissociated MS (MS/MS) spectra of selected ions that were highly enriched in the Cpefat/Cpefat mouse extract compared with the control extract.
Analysis of MS Data.
To determine the peptides enriched in Cpefat/Cpefat mouse tissues, the LCQ or Mariner MS data were arbitrarily divided into 2-min windows, and the ion profiles were compared between Cpefat/Cpefat and control extracts. Ions enriched in Cpefat/Cpefat extracts were further analyzed by examining the profile of elution versus time; those ions that appeared in a single scan were excluded as random noise in the detector. Ions that were present in a large number of scans also were excluded. The remaining ions were then selected for MS/MS analysis, as described above.
Several methods were used to analyze the MS/MS data. First, the program sequest was used to compare the observed MS/MS spectra to predicted fragmentation patterns of all known mouse proteins. Any positive match was further analyzed by comparing the MS/MS spectra to the fragmentation pattern of that peptide predicted by using the sherpa computer program. The MS/MS data were considered identified if the observed parent mass was within 0.04% of the predicted mass and at least 80% of the major MS/MS fragments were within 0.04% of predicted b- and y-series fragments. The MS/MS spectra that could not be identified with sequest were examined manually in one of two ways. One method used the ms-tag program to screen GenBank for peptides with predicted parent masses and fragments that matched the observed data. Alternatively, MS/MS data were examined for fragments that differed by the mass of an amino acid residue. In cases where several adjacent amino acid residues were predicted, these short sequences were compared with a database composed of all known mouse prohormones and secretory pathway proteins by using the genepro program. Any matches found from this analysis were further examined by comparing the observed parent mass to that of possible peptides that could arise from the protein (allowing for posttranslational modifications). Then, the observed MS/MS data were compared with the b- and y-series of fragments calculated by using the sherpa computer program.
Extraction, Purification, and Analysis of Peptides from Rat Brain.
Frozen rat brains (Pel-Freeze) were extracted in 10 vol of boiling water (containing 2.5 mM phenylmethylsulfonyl fluoride and 12.5 mM EDTA) followed by incubation at 100°C for 10 min. The homogenate was centrifuged (13,000 g for 30 min, 4°C). The supernatant was lyophilized, resuspended in 100 μl buffer A (150 mM sodium phosphate buffer, pH 7.5 containing 0.2% Triton X-100), and applied to a gel-exclusion Superdex Peptide HR 10/30 column (Amersham Pharmacia) in 0.1% trifluoroacetic acid containing 30% acetonitrile at 0.5 ml/min. One-minute fractions were collected, dried, resuspended in buffer A, and subjected to RIA as described below. The peak of immunoreactive peptides were pooled and applied to a HPLC C8 column (Vydac, Hesperia, CA). A gradient of acetonitrile in 0.1% trifluoroacetic acid was used. One-minute (0.3 ml) fractions were collected and analyzed by RIA. Retention times of PE-11 and mouse WE-15 were determined in a separate run. MALDI-TOF analysis of the HPLC fractions was performed as described above.
Immunoreactive PE-11 was determined by using anti-PE-11 antiserum (23), a gift from Reiner Fischer-Colbrie (Univ. of Innsbruck, Innsbruck, Austria). PE-11 was radioiodinated by using the chloramine-T method. The RIA was performed with antibody (1:4,000 dilution) in 150 mM sodium phosphate buffer, pH 7.5 containing 0.1% gelatin, 0.1% BSA (protease free, Sigma), 0.1% Triton X-100, and 0.02% sodium azide. The antigen-antibody complex was separated from the unbound radioligand by using goat-anti-rabbit globulin (Peninsula Laboratories).
Results
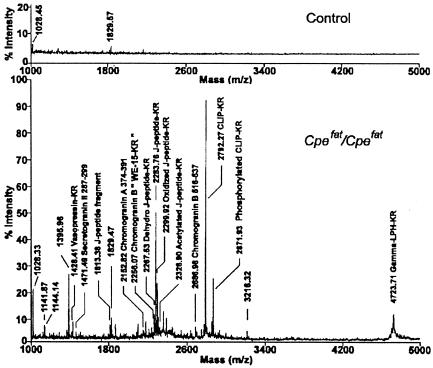
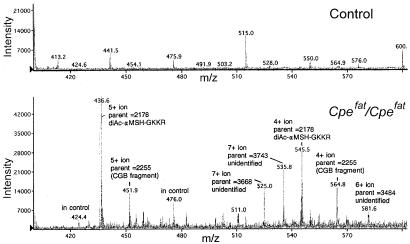
MALDI-TOF analysis of pituitary extracts after purification on the anhydrotrypsin column revealed a large number of peaks for the Cpefat/Cpefat extracts that are not present in the control extracts (Fig. 1). Many of the ions detected by MS correspond to the calculated mass of known peptides containing C-terminal basic amino acids. To detect additional peptides, the affinity-purified extracts were subjected to HPLC on a reverse-phase C18 column, and the eluent was analyzed with an on-line electrospray ionization mass spectrometer. Fig. 2 shows the ions in a portion of the eluate from 46 to 48 min and with a mass/charge of 400–600; the entire mass/charge range from 300 to 2,000 was analyzed from 0 to 70 min (not shown). Altogether, several thousand ions were found to be substantially enriched in extracts of Cpefat/Cpefat mouse brain and pituitary, compared with extracts from control mouse tissues.
Figure 1.
MALDI-TOF analysis of material extracted from 20 mouse pituitaries and purified on an anhydrotrypsin column. (Upper) Extract from control mouse pituitary. (Lower) Extract from Cpefat/Cpefat mouse pituitary. The observed [MH]+ mass of selected ions is indicated along with peptides that were subsequently identified by using MS/MS analysis. CLIP, corticotropin-like immunoreactive peptide; LPH, lipotropin.
Figure 2.
LC/MS analysis of affinity purified material from control (Upper) and Cpefat/Cpefat (Lower) mouse pituitary. A 2-min window is shown that represents the material that eluted from the reverse-phase column between 46 and 48 min. The predicted parent mass (uncharged form) of several of the ions is indicated. αMSH, α-melanocyte-stimulating hormone; CGB, chromogranin B.
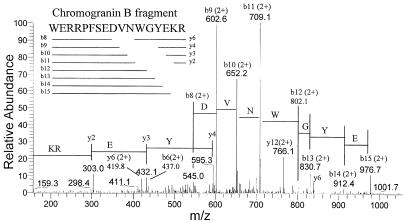
To obtain partial sequence information for selected ions, extracts from Cpefat/Cpefat mouse brain and pituitary were run on the HPLC C18 reverse-phase column under identical conditions as used in the previous step, and the eluent was analyzed by MS/MS. Representative data are shown in Fig. 3. For this peptide the mass/charge ratio of the input ion was 564.65 with a charge state of 4+ (parent mass 2254.6); this peptide corresponds to the 564.8 ion shown in Fig. 2. In this MS/MS spectrum, all of the major fragments match to predicted b- or y-series fragments of a region within chromogranin B (Fig. 3). This peptide contains C-terminal basic amino acids, as expected from its accumulation in Cpefat/Cpefat mice and affinity for the anhydrotrypsin column. However, the N-terminal cleavage site represents processing of a Trp-Trp bond.
Figure 3.
MS/MS analysis of the ion from Cpefat/Cpefat mouse pituitary with a m/z of 564.65 that eluted from the reverse-phase column at 46.8 min. In this example, more than 90% of the observed MS/MS ions are within 0.04% of the calculated b- or y-series fragments of a peptide derived from chromogranin B (Inset).
Similar MS/MS analysis was performed for a large number of spectra from multiple HPLC runs of Cpefat/Cpefat mouse brain and pituitary samples. Altogether, more than 100 peptides that were enriched in Cpefat/Cpefat mouse brain or pituitary were identified as fragments of one of 16 proteins (Table 1 and see Table 3, which is published as supplemental data on the PNAS web site, www.pnas.org). Most of the identified peptides arise from only six proteins: proopiomelanocortin (POMC), proenkephalin, chromogranin B, secretogranin II, provasopressin, and proSAAS. Many of the identified peptides correspond to expected fragments of these proteins, based on known or predicted cleavages at consensus sites. However, a number of novel fragments were detected. In the case of proenkephalin, the N-terminal side of two novel fragments (QLEDEAKELQKR and SDEEGENYSKEVPEIEKR) corresponds to cleavage following a Pro residue. In addition, a slightly shorter form of one of these peptides (LEDEAKELQKR) also was detected (Table 3). Many novel POMC fragments were identified along with the expected peptides (Table 3). Unlike the novel proenkephalin fragments, the N termini of many of the POMC-derived peptides required cleavages near acidic residues. In contrast, none of the observed fragments of chromogranin B resulted from cleavages at either Pro or acidic residues. Instead, four of these peptides had N termini that resulted from cleavages at hydrophobic residues and several of the N and/or C termini resulted from cleavages at basic residues that were not PC consensus sites (Table 2).
Table 1.
Protein precursors of peptides identified with basic C-terminal residues in Cpefat/Cpefat mouse brain and pituitary
| Precursor | Number of peptides found |
|---|---|
| Proopiomelanocortin | 39 |
| Proenkephalin | 13 |
| Chromogranin B | 13 |
| Secretogranin II | 10 |
| Provasopressin | 8 |
| ProSAAS | 7 |
| Chromogranin A | 3 |
| Prooxytocin | 3 |
| Growth hormone | 2 |
| Prodynorphin | 1 |
| Proneurokinin B | 1 |
| Procholecystokinin | 1 |
| Protachykinin A | 1 |
| Prothyrotropin-releasing hormone | 1 |
| Promelanin concentrating hormone | 1 |
| Propeptidylglycine-alpha-amidating monooxygenase | 1 |
Table 2.
Mouse chromogranin B fragments identified by MS/MS
| Region | Mass
|
Sequence of peptide (and adjacent sequence) | Found in | |
|---|---|---|---|---|
| Pred. | Obs. | |||
| 186–201 | 1859.0 | 1859.1 | (EEKK) HIEDSGEKPNTFSNKR (SE) | Pituitary |
| 186–201 | 1939.0 | 1939.2 | (EEKK) HIEDS*GEKPNTFSNKR (SE) *Phosphate | Pituitary |
| 357–374 | 2100.3 | 2101.0 | (RSYR) GLQYRGRGSEEDRAPRPR (SE) | Pituitary |
| 386–407 | 2566.6 | 2566.8 | (EYKR) NHPDSELESTANRHGEETEEER (SY) | Pituitary |
| 408–437 | 3378.6 | 3379.1 | (EEER) SYEGANGRQHRGRGREPGAHSALDTREEKR (LL) | Pituitary |
| 419–437 | 2122.3 | 2122.3 | (RQHR) GRGREPGAHSALDTREEKR (LL) | Pituitary |
| 438–456 | 2196.5 | 2196.2 | (EEKR) LLDEGHYPVRESPIDTAKR (YP) | Brain, pituitary |
| 482–515 | 4334.7 | 4334.4 | (QGRW) WQQEEQLGPEESREEVRFPDRQYEPYPITEKRKR (LG) | Pituitary |
| 516–537 | 2686.0 | 2686.3 | (KRKR) LGALFNPYFDPLQWKNSDFEKR (GN) | Brain, pituitary |
| 517–537 | 2572.9 | 2573.1 | (RKRL) GALFNPYFDPLQWKNSDFEKR (GN) | Pituitary |
| 520–536 | 2175.4 | 2175.8 | (LGAL) FNPYFDPLQWKNSDFEK (RG) | Pituitary |
| 571–587 | 2254.5 | 2254.6 | (NYDW) WERRPFSEDVNWGYEKR (SF) | Pituitary |
| 588–599 | 1401.6 | 1401.8 | (YEKR) SFARAPQLDLKR (QY) | Pituitary |
Regions in bold correspond to b- and/or y-series ions. Surrounding sequences are in parentheses.
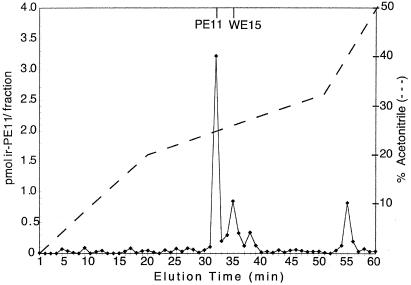
The novel fragment identified in Fig. 3 was chosen for further analysis by conventional methods. This 2,254.5-Da chromogranin B fragment corresponds to an N-terminally extended form of a peptide previously designated PE-11 (24). The peptide WERRPFSEDVNWGYE was synthesized and designated mouse WE-15; this peptide lacks the C-terminal Lys-Arg of the 2,254.5-Da peptide found in Cpefat/Cpefat mice because in wild-type animals these residues are expected to be removed by CPE. An antiserum directed against the C terminus of PE-11 (gift of Reiner Fischer-Colbrie) cross reacts (>90% efficiency) with mouse WE-15. When rat brain extracts were fractionated on a gel filtration column, a broad peak of immunoreactivity with an apparent molecular mass of 1–2 kDa was detected (data not shown). This peak of immunoreactivity was fractionated on a reverse-phase HPLC column. Approximately 10–20% of the immunoreactive peptide eluted in a position identical to that of synthetic mouse WE-15, and the majority eluted in a position identical to PE-11 (Fig. 4).
Figure 4.
RIA of rat brain extract after chromatography on gel filtration and reverse-phase HPLC columns. Extracts from three rat brains were pooled and subjected to gel-filtration chromatography on a Superdex Peptide 10/30 column. Fractions representing the 1- to 2-kDa peak of immunoreactive PE-11 were pooled, subjected to HPLC on a C8 column, and analyzed by RIA. The lines at the top denote the elution positions of synthetic mouse peptides.
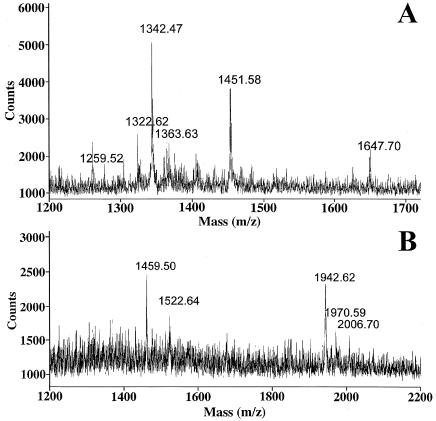
To obtain additional support that WE-15 is present in the fraction, MALDI-TOF MS was performed. Fraction 32 contains a peptide with a monoisotopic [MH]+ mass of 1,342.47 (Fig. 5A), which matches that of PE-11 (calculated monoisotopic [MH]+ mass 1,342.55). This peptide is not detectable in the adjacent HPLC fractions. Fraction 35 contains a peptide with an average [MH]+ mass of 1,942.62 (the monoisotopic peak is usually not detected in peptides close to or larger than 2 kDa due to the abundance of C13, and so the average mass value must be used). This observed mass matches the calculated average [MH]+ mass of 1,943.09 for rat WE-15; WEKRPFSEDVNWGYE (Fig. 5B). Taken together, these data support the data from the Cpefat/Cpefat mice and suggest that WE-15 is a significant product of chromogranin B processing.
Figure 5.
MALDI-TOF MS of rat brain extracts after chromatography on gel filtration and reverse-phase HPLC columns. (A) Spectrum from fraction 32, showing the presence of a peptide with a monoisotopic [MH]+ mass of 1342.47. (B) Spectrum from fraction 35, showing the presence of a peptide with an average [MH]+ mass of 1,942.62.
Discussion
There are several major findings of the present study. First, the large number of known peptides detected by using the affinity column approach with Cpefat/Cpefat mice validates this technique. Other methods to isolate neuropeptides require a large number of steps to purify each peptide and can detect only peptides for which antisera or receptor-based assay methods are available. In contrast, the present method used a single affinity column step to purify all peptides that are normally substrates of CPE, which includes most neuropeptides. Anhydrotrypsin has a high affinity for peptides with C-terminal basic residues and poorly binds peptides with nonbasic C-terminal residues (25), even if there are internal basic residues (unpublished results). Although proteins and peptides with C-terminal Lys or Arg normally exist in cells, these are present in both the Cpefat/Cpefat and control animals. The focus on peptides present in Cpefat/Cpefat mice and absent from controls eliminated all nonsecretory pathway proteins that are not normally substrates for CPE. Most of the identified proteins represent known neuropeptide precursors such as proenkephalin, POMC, and others (Table 1). Three of these proteins (chromogranin A, chromogranin B, and secretogranin II) have been proposed to function as precursors of neuroendocrine peptides, although other functions also have been proposed (26–30). ProSAAS, which was previously identified by using the technique described in this article, was found to potently inhibit PC1 (31). However, only partially processed peptides near the C-terminal region of proSAAS are effective PC1 inhibitors (32, 33), raising the possibility that the other peptide products have alternative functions, possibly as neuropeptides. Only one of the proteins identified in the present study (propeptidylglycine-α-amidating monooxygenase) has not been proposed to function as a neuropeptide precursor and was “found” because it is cleaved in the late secretory pathway by endopeptidases and CPE (34).
Another major finding of the present study is that a large number of peptides in brain and pituitary result from cleavage at nontraditional processing sites. Although postextraction degradation cannot be ruled out, there are several arguments against this. First, the tissues were frozen in dry ice immediately after dissection and were rapidly thawed by adding boiling 0.1 M acetic acid directly to the frozen sample. It is likely that the combination of heat and acid rapidly inactivated all of the endogenous peptidases. This concentration of acetic acid is not known to cause nonenzymatic hydrolysis of peptides, except Asp-Pro bonds that are acid labile (35). Most importantly, the nontraditional cleavage sites of the various prohormones are much different between proteins; proenkephalin is cleaved near Pro residues, POMC is cleaved near acidic residues, and chromogranin B is cleaved at hydrophobic residues (in addition to the PC consensus sites). This finding suggests cell-specific and/or protein-specific processing steps rather than general cleavages would be expected to arise from nonspecific mechanisms.
Other studies have detected neuroendocrine peptides that arise from nonbasic cleavages. Endothelin (ET) is produced by cleavage of big ET at a Trp-Val (big ET-1, big-ET-2) or Trp-Ile (big ET-3) site (36). The neuropeptide FF/SF precursor requires cleavage at an Ala-Phe site to produce neuropeptide FF and at a Trp-Ser site to produce neuropeptide SF (37). A previous study characterizing peptides in bovine pituitary using MS detected a number of nonbasic cleavage products of POMC, provasopressin, and other neuropeptide precursors (38). However, that study relied primarily on the molecular mass (and not MS/MS analysis, as done in the present study) so the identification of many of the fragments was only tentative. Previous studies have detected a number of chromogranin B-derived peptides in bovine adrenal medulla (27, 39), one of which corresponds to the 1,402-Da peptide found in the present study. Of 30 distinct fragments detected in two previous studies, 10 represent cleavage at nonbasic sites and six represent cleavage at basic sites that do not fit the PC consensus sequence (27, 39). Other studies detected an N-terminally extended form of PE-11 (23, 40, 41). However, there are no PC consensus sites within chromogranin B that would produce a peptide of the observed size, suggesting a novel cleavage site. It is possible that the N-terminally extended form of PE-11 detected in the previous studies is WE-15, which results from cleavage at Trp-Trp (Table 2).
Several enzymes that cleave at nonbasic sites are present in the secretory pathway of neuroendocrine tissues. In addition to cleaving big ET, purified ET-converting enzyme-1 efficiently cleaves a variety of neuroendocrine peptides including angiotensin I, bradykinin, neurotensin, and substance P at hydrophobic residues (42). An enzyme designated SKI-1/S1P is enriched in the Golgi apparatus and cleaves at nonbasic sites (43, 44). The consensus site for SKI1/S1P is similar to that of the PCs except that the residue in the P1 position can be either a basic residue, a Thr, or a Leu (44, 45). In yeast, insects, and frogs, processing of bioactive peptides involves aminopeptidases (46–48); it is possible that mammalian aminopeptidases or dipeptidylcarboxypeptidases are responsible for some of the peptides observed in the present study.
Virtually all of the peptides found in the Cpefat/Cpefat mice had both basic residues attached to the C terminus (see Table 3). Taken together with other studies that found low levels of the fully processed forms of several peptides (11, 13–16), this finding suggests that any CPD present after the endopeptidase step completely removes the C-terminal basic residues. Because the most abundant forms of the various peptides found in the present study contain both basic residues, CPD is presumably absent from the major intracellular site of peptide processing in brain and pituitary.
The detection of acetylated and phosphorylated peptides in the present study indicates that these steps can occur without the prior removal of C-terminal basic residues. In addition to confirming several known phosphorylation and acetylation sites, other modifications were detected. For example, Cpefat/Cpefat mouse pituitary contains a C-terminally extended form of α-melanocyte-stimulating hormone containing two acetyl groups (+84) on the N-terminal Ser in addition to another putative acetyl group (+42) near the C terminus of the peptide (Table 3). Unknown modifications that add 35 or 38 to the mass of POMC J-peptide also were detected. Although addition of 16 mass units was typically associated with oxidation of Met or Trp, in one case (CLIP peptide of POMC) this addition was found on a Phe. Further studies are needed to identify the exact nature of these unknown modifications.
The general method described in this study provides a rapid and efficient technique to identify substrates of CPE, many of which are likely to be bioactive neuropeptides. A large number of orphan receptors have been identified, implying that there is an equally large number of ligands for these receptors (49). Many ligands of known G protein-coupled receptors are peptides and so the ligands of orphan receptors also may be peptides, possibly some of the novel fragments of known proteins described in the present study.
The identification of endogenous neuropeptides is difficult using standard techniques. The method of detecting neuroendocrine peptides by the presence of C-terminal amide residues is not very sensitive and requires extensive purification of the peptides before sequencing (50). The approach described here for CPE substrates can easily be adapted to identify substrates for a range of enzymes. The key elements of this approach are a biological system lacking the enzyme (or with greatly reduced levels such that substrates accumulate), an affinity resin to isolate the substrates, and then a method of analysis of the affinity purified material. Initial studies on CPE substrates made use of a modified CPE in which the active site was mutated so that the enzyme would bind but not cleave substrates (51). Alternatively, inhibitors of CPE such as mercuric chloride completely eliminate enzyme activity but not substrate binding (unpublished work). Similar approaches presumably would work for other enzyme systems, with the basic need being a protein that tightly binds substrates. MS is well suited for analysis of the affinity-purified material. Alternatively, HPLC purification and conventional Edman sequencing also was used successfully to identify a proenkephalin-derived peptide in Cpefat/Cpefat mouse brain (data not shown). In addition, two-dimensional gel electrophoresis followed by MS of tryptic fragments also yielded prohormone fragments (data not shown). However, in this latter approach, the naturally occurring forms of the peptides could not be determined due to the trypsin digestion. Many variants on this basic procedure are possible and may be useful for other enzyme systems.
Supplementary Material
Acknowledgments
Thanks to Ruth Angeletti, Edward Nieves, Haiteng Deng, Scott Patterson, and Paul Courchesne for assistance with MS, Jim Douglass for helpful discussions, and Reiner Fischer-Colbrie for the PE-11 antiserum. This work was supported by a research grant from Amgen and National Institutes of Health Grants R01 DA-04494, K02 DA-00194 (to L.D.F.), R01 NS-26880, and K02 DA-00458 (to L.A.D.). The Laboratory for Macromolecular Analysis of the Albert Einstein College of Medicine is supported in part by Cancer Center Core Grant CA13330 and Diabetes Research Training Center Core Grant DK20541.
Abbreviations
- CP
carboxypeptidase
- ET
endothelin
- LC
liquid chromatography
- MALDI-TOF
matrix-assisted laser desorption ionization/time-of-flight
- MS/MS
collision-induced dissociated MS
- PC
prohormone convertase
- POMC
proopiomelanocortin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mains R E, Dickerson I M, May V, Stoffers D A, Perkins S N, Ouafik L H, Husten E J, Eipper B A. Front Endocrinol. 1990;11:52–89. [Google Scholar]
- 2.Steiner D F. In: Peptide Biosynthesis and Processing. Fricker L D, editor. Boca Raton, FL: CRC; 1991. pp. 1–16. [Google Scholar]
- 3.Seidah N G, Chretien M. Curr Opin Biotechnol. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- 4.Seidah N G, Chretien M. Methods Enzymol. 1994;244:175–188. doi: 10.1016/0076-6879(94)44015-8. [DOI] [PubMed] [Google Scholar]
- 5.Zhou A, Webb G, Zhu X, Steiner D F. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 6.Lindberg I, Hutton J C. In: Peptide Biosynthesis and Processing. Fricker L D, editor. Boca Raton, FL: CRC; 1991. pp. 141–174. [Google Scholar]
- 7.Fricker L D, Leiter E H. Trends Biochem Sci. 1999;24:390–393. doi: 10.1016/s0968-0004(99)01448-6. [DOI] [PubMed] [Google Scholar]
- 8.Fricker L D. In: The Enzymes, Volume 23: Co- and Posttranslational Proteolysis of Proteins. Dalbey R E, Sigman D S, editors. San Diego: Academic; 2001. , in press. [Google Scholar]
- 9.Fricker L D. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 10.Fricker L D. In: Handbook of Proteolytic Enzymes. Barrett A J, Rawlings N D, Woessner J F, editors. London: Academic; 1998. pp. 1341–1344. [Google Scholar]
- 11.Naggert J K, Fricker L D, Varlamov O, Nishina P M, Rouille Y, Steiner D F, Carroll R J, Paigen B J, Leiter E H. Nat Genet. 1995;10:135–142. doi: 10.1038/ng0695-135. [DOI] [PubMed] [Google Scholar]
- 12.Varlamov O, Leiter E H, Fricker L D. J Biol Chem. 1996;271:13981–13986. doi: 10.1074/jbc.271.24.13981. [DOI] [PubMed] [Google Scholar]
- 13.Rovere C, Viale A, Nahon J, Kitabgi P. Endocrinology. 1996;137:2954–2958. doi: 10.1210/endo.137.7.8770919. [DOI] [PubMed] [Google Scholar]
- 14.Fricker L D, Berman Y L, Leiter E H, Devi L A. J Biol Chem. 1996;271:30619–30624. doi: 10.1074/jbc.271.48.30619. [DOI] [PubMed] [Google Scholar]
- 15.Udupi V, Gomez P, Song L, Varlamov O, Reed J T, Leiter E H, Fricker L D, Greeley G H J. Endocrinology. 1997;138:1959–1963. doi: 10.1210/endo.138.5.5113. [DOI] [PubMed] [Google Scholar]
- 16.Cain B M, Wang W, Beinfeld M C. Endocrinology. 1997;138:4034–4037. doi: 10.1210/endo.138.9.5490. [DOI] [PubMed] [Google Scholar]
- 17.Song L, Fricker L D. J Biol Chem. 1995;270:25007–25013. doi: 10.1074/jbc.270.42.25007. [DOI] [PubMed] [Google Scholar]
- 18.Song L, Fricker L D. J Biol Chem. 1996;271:28884–28889. doi: 10.1074/jbc.271.46.28884. [DOI] [PubMed] [Google Scholar]
- 19.Fricker L D. In: Handbook of Proteolytic Enzymes. Barrett A J, Rawlings N D, Woessner J F, editors. London: Academic; 1998. pp. 1349–1351. [Google Scholar]
- 20.Varlamov O, Fricker L D. J Cell Sci. 1998;111:877–885. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- 21.Varlamov O, Eng F J, Novikova E G, Fricker L D. J Biol Chem. 1999;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- 22.Fricker L D. Methods Neurosci. 1995;23:237–250. [Google Scholar]
- 23.Kroesen S, Marksteiner J, Leitner B, Hogue-Angeletti R, Fischer-Colbrie R, Winkler H. Eur J Neurosci. 1996;8:2679–2689. doi: 10.1111/j.1460-9568.1996.tb01563.x. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen E, Welinder B S, Madsen O D. Endocrinology. 1991;129:3147–3156. doi: 10.1210/endo-129-6-3147. [DOI] [PubMed] [Google Scholar]
- 25.Kumazaki T, Terasawa K, Ishii S. J Biochem. 1987;102:1539–1546. doi: 10.1093/oxfordjournals.jbchem.a122202. [DOI] [PubMed] [Google Scholar]
- 26.Tatemoto K, Efendic S, Mutt V, Makk G, Feistner G J, Barchas J D. Nature (London) 1986;324:476–478. doi: 10.1038/324476a0. [DOI] [PubMed] [Google Scholar]
- 27.Strub J, Garcia-Sablone P, Lonning K, Taupenot L, Hubert P, Van Dorsselaer A, Aunis D, Metz-Boutigue M. Eur J Biochem. 1995;229:356–368. doi: 10.1111/j.1432-1033.1995.tb20476.x. [DOI] [PubMed] [Google Scholar]
- 28.Fischer-Colbrie R, Laslop A, Kirchmair R. Prog Neurobiol. 1995;46:49–70. doi: 10.1016/0301-0082(94)00060-u. [DOI] [PubMed] [Google Scholar]
- 29.Natori S, Huttner W B. Biochimie. 1994;76:277–282. doi: 10.1016/0300-9084(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 30.Huttner W B, Gerdes H H, Rosa P. Trends Biochem Sci. 1991;16:27–30. doi: 10.1016/0968-0004(91)90012-k. [DOI] [PubMed] [Google Scholar]
- 31.Fricker L D, McKinzie A A, Sun J, Curran E, Qian Y, Yan L, Patterson S D, Courchesne P L, Richards B, Levin N, et al. J Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian Y, Devi L A, Mzhavia N, Munzer S, Seidah N G, Fricker L D. J Biol Chem. 2000;275:23596–23601. doi: 10.1074/jbc.M001583200. [DOI] [PubMed] [Google Scholar]
- 33.Cameron A, Fortenberry Y, Lindberg I. FEBS Lett. 2000;473:135–138. doi: 10.1016/s0014-5793(00)01511-8. [DOI] [PubMed] [Google Scholar]
- 34.Kolhekar A S, Mains R E, Eipper B A. Methods Enzymol. 1997;279:35–43. doi: 10.1016/s0076-6879(97)79007-4. [DOI] [PubMed] [Google Scholar]
- 35.Allen G. Sequencing of Proteins and Peptides. 2nd Ed. Amsterdam: Elsevier; 1989. [Google Scholar]
- 36.Turner A J, Murphy L J. Biochem Pharmacol. 1995;51:91–102. doi: 10.1016/0006-2952(95)02036-5. [DOI] [PubMed] [Google Scholar]
- 37.Vilim F S, Aarnisalo A A, Nieminen M, Lintunen M, Karlstedt K, Kontinen V K, Kalso E, States B, Panula P, Ziff E. Mol Pharmacol. 1999;55:804–811. [PubMed] [Google Scholar]
- 38.Feistner G J, Hojrup P, Evans C J, Barofsky D F, Faull K F, Roepstorff P. Proc Natl Acad Sci USA. 1989;86:6013–6017. doi: 10.1073/pnas.86.16.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigafoos J, Chestnut W G, Merrill B M, Taylor L C E, Diliberto E J, Viveros O H. J Anat. 1993;183:253–264. [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Leitner B, Winkler H, Dahlstrom A. Neuroscience. 1998;84:281–294. doi: 10.1016/s0306-4522(97)00484-3. [DOI] [PubMed] [Google Scholar]
- 41.Marksteiner J, Bauer R, Kaufmann W A, Weiss E, Barnas U, Maier H. Neuroscience. 1999;91:1155–1170. doi: 10.1016/s0306-4522(98)00676-9. [DOI] [PubMed] [Google Scholar]
- 42.Johnson G D, Stevenson T, Ahn K. J Biol Chem. 1999;274:4053–4058. doi: 10.1074/jbc.274.7.4053. [DOI] [PubMed] [Google Scholar]
- 43.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 44.Seidah N G, Mowla S J, Hamelin J, Mamarbachi A M, Benjannet S, Toure B B, Basak A, Munzer J S, Marcinkiewicz J, Zhong M, et al. Proc Natl Acad Sci USA. 1999;96:1321–1326. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng D, Espenshade P J, Slaughter C A, Jaen J C, Brown M S, Goldstein J L. J Biol Chem. 1999;274:22805–22812. doi: 10.1074/jbc.274.32.22805. [DOI] [PubMed] [Google Scholar]
- 46.Fricker L D. In: Peptide Biosynthesis and Processing. Fricker L D, editor. Boca Raton, FL: CRC; 1991. pp. 199–230. [Google Scholar]
- 47.Anna-Arriola S S, Herskowitz I. Yeast. 1994;10:801–810. doi: 10.1002/yea.320100610. [DOI] [PubMed] [Google Scholar]
- 48.Kreil G. Trends Biochem Sci. 1990;15:23–26. doi: 10.1016/0968-0004(90)90126-v. [DOI] [PubMed] [Google Scholar]
- 49.Marchese A, George S R, Kolakowski L F, Lynch K R, O'Dowd B F. Trends Pharmacol Sci. 1999;20:370–375. doi: 10.1016/s0165-6147(99)01366-8. [DOI] [PubMed] [Google Scholar]
- 50.Tatemoto K, Mutt V. Proc Natl Acad Sci USA. 1978;75:4115–4119. doi: 10.1073/pnas.75.9.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian Y, Varlamov O, Fricker L D. J Biol Chem. 1999;274:11582–11586. doi: 10.1074/jbc.274.17.11582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.