Abstract
The ability of Thlaspi goesingense to hyperaccumulate Ni seems to be governed in part by enhanced accumulation of Ni within leaf vacuoles. We have characterized genes from T. goesingense encoding putative vacuolar metal ion transport proteins, termed metal tolerance proteins (TgMTPs). These proteins contain all of the features of cation-efflux family members, and evidence indicates they are derived from a single genomic sequence (TgMTP1) that gives rise to an unspliced (TgMTP1t1) and a spliced (TgMTP1t2) transcript. Heterologous expression of these transcripts in yeast lacking the TgMTP1 orthologues COT1 and ZRC1 complements the metal sensitivity of these yeast strains, suggesting that TgMTP1s are able to transport metal ions into the yeast vacuole in a manner similar to COT1 and ZRC1. The unspliced and spliced TgMTP1 variants differ within a histidine-rich putative metal-binding domain, and these sequence differences are reflected as alterations in the metal specificities of these metal ion transporters. When expressed in yeast, TgMTP1t1 confers the highest level of tolerance to Cd, Co, and Zn, whereas TgMTP1t2 confers the highest tolerance to Ni. TgMTP1 transcripts are highly expressed in T. goesingense compared with orthologues in the nonaccumulators Arabidopsis thaliana, Thlaspi arvense, and Brassica juncea. We propose that the high-level expression of TgMTP1 in T. goesingense accounts for the enhanced ability of this hyperaccumulator to accumulate metal ions within shoot vacuoles.
The genus Thlaspi contains numerous species that hyperaccumulate Ni. For example, field-collected specimens of Thlaspi goesingense Hálácsy from an ultramafic site in Redschlag, Austria, have been recorded with shoot Ni concentrations as high as 12,400 μg/g shoot dry biomass (1.2%) (1, 2). In the laboratory, we analyzed individuals from this T. goesingense population and confirmed their hyperaccumulator status (3). To determine the physiological basis of this Ni hyperaccumulation phenotype, we have investigated a number of physiological parameters in both the hyperaccumulator and the related nonaccumulator Thlaspi arvense. Production of Ni chelates in root exudates in the hyperaccumulator and nonaccumulator were found to be equivalent (4), as well as rates of Ni translocation to the shoots (3). However, the hyperaccumulator was found to be more Ni tolerant when compared with the nonaccumulator (3). Ni tolerance in the hyperaccumulator is related to its enhanced ability to compartmentalize Ni in shoot vacuoles (5) with 75% of the intracellular leaf Ni in the hyperaccumulator being localized to the vacuole (5). Furthermore, vacuoles from the hyperaccumulator contain approximately double the Ni of the nonaccumulator, even though protoplasts from each species contained equal amounts of Ni (5). We conclude that vacuolar compartmentalization of Ni in the hyperaccumulator plays a major role in Ni tolerance and hyperaccumulation, although little is known about its molecular mechanism (6). Here we present data characterizing the functional properties and role of T. goesingense metal tolerance protein (TgMTP)-1 in the mechanism of Ni hyperaccumulation, and propose a role for TgMTP1 in enhanced vacuolar compartmentalization of Ni in the hyperaccumulator T. goesingense.
Materials and Methods
Plant Material and Culture.
Seeds of the Ni hyperaccumulator T. goesingense were collected from an ultramafic site in Redschlag, Austria (3). Seeds of Arabidopsis thaliana (Col-3) were purchased from Lehle Seeds (Round Rock, TX), and seeds of Thlaspi arvense and Brassica juncea (426308) were obtained from the Crucifer Genetics Cooperative (Univ. of Wisconsin, Madison, WI). Plants were grown and harvested as described (7).
Plasmids, Bacterial, and Yeast Strains.
The Escherichia coli strain TOP10 F′ [F′ [lacIq Tn10(TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZ-Δ-M15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG] was used for PCR and reverse transcription (RT)-PCR product cloning and plasmid DNA production. The TgMTP1 genes were cloned into the pYES plasmid (Invitrogen) and subcloned into p425 GAL1 (ATCC no. 87331). For yeast expression, the following strains were transformed by means of a chemical method (8): cot1 (CYP514), (MATα ura3–52 leu2–3,112 trp1-Δ1 cot1-Δ1∷URA3); zrc1 (CYP502), (MATα ura3–52 leu2–3,112 trp1-Δ1 zrc1-Δ100∷LEU2; ref. 9); and Δpep5 (BJ7964), (MATα ura3–52 leu2Δ1 his3Δ200 trp1Δ101 Δpep5∷TRP1; ref. 10).
RNA and DNA Isolation.
Total RNA and genomic DNA were isolated as described (7).
PCR and RT-PCR Cloning of TgMTP1 Genes.
For isolation of the TgMTP1 genomic clone, 5 μg of genomic DNA from T. goesingense was PCR-amplified, using 100 pmol of 5′ MTP primer (5′-GCAAGCTTATGGAGTCTTCAAGTCCCCATCATGAGGTTAATG-3′) and 100 pmol of 3′ MTP primer (5′-ATGAATTCTTAGCGC TCGATTTG TAT-3′) with 35 cycles of 95°C denaturation for 1 min, 50°C annealing for 2 min, 72°C extension for 2 min, and a final 5 min 72°C extension step. PCR products were cloned into pYES by using standard techniques (11). Putative clones were sequenced (Univ. of Arizona, Tucson, AZ) and identified by comparison with ZAT (12). For isolation of TgMTP1 transcript cDNA, 100 ng of shoot mRNA isolated from T. goesingense plants treated with 50 μM Ni for 48 h were reverse transcribed by using 4 units of avian myeloblastosis virus (AMV) reverse transcriptase (Promega), 100 pmol of 5′ MTP primer, and 100 pmol of 3′ MTP for 30 min at 42°C. First-strand cDNA products were PCR amplified with 5 units of Pfu polymerase (Promega) and 25 cycles (see above). PCR products were cloned into pYES (11) and TgMTP1 transcripts identified by sequencing. TgMTP1t1 was identified and used to screen the remaining RT-PCR clones by means of colony hybridization after the Northern blot hybridization protocol. Resulting positive clones were sequenced and identified as a homolog of TgMTP1t1 and named TgMTP1t2.
Alignments were performed by using clustal w (13). Analysis of conserved domains was performed by using reversed position-specific blast (14), and analysis of transmembrane domains was performed by using tmpred (http://www.ch.embnet.org/software/TMPRED_form.html). Sequences were organized and analyzed at the Biology Workbench (http://workbench.sdsc.edu).
Yeast Metal Tolerance Assay.
Yeast strains were grown to 1.5 OD600 in 10 ml of yeast nitrogen base (YNB) minimal medium SC amino acid supplement + adenine (SCA)/2% glucose − uracil − leucine for the cot1 and zrc1 mutants or YNB SCA/2% glucose − uracil for the Δpep5 mutant (YNB SCA = 6.7 g/liter yeast nitrogen base/2 g/liter SC amino acid supplement/100 mg/liter adenine). One milliliter of yeast culture was used to inoculate 50 ml of YNB SCA/4% galactose-uracil or -leucine (cot1 and zrc1) or -uracil only (Δpep5), containing 1.5% top agar (plant tissue-culture grade) and mixed well. This solution then was poured into 150 × 15-mm Petri dishes and allowed to cool. Sterile paper filter disks (6 mm) were placed at regular intervals on the surface of the top agar, and solutions containing various metal ions were spotted on the disks. Plates were incubated at 30°C for 4–7 days, and the area of the zone of inhibition was measured.
RNA and DNA Blots.
For Northern analysis, 30 μg of total shoot RNA per sample was analyzed by using described protocols (7). For genomic Southern analysis, 20 μg of genomic DNA per sample was digested with 60 units of each of the restriction enzymes (SacI, PvuII, and EcoRI) for 6–8 h at 37°C and was analyzed by using protocols as described (7). For probe preparation, cDNAs were digested with the appropriate restriction enzyme for 1 h at 37°C. Digested DNA was separated on a 1.5% agarose gel, and the appropriate size fragments were excised, recovered by electroelution, and labeled with 32P following standard protocols (7).
Results and Discussion
Members of the cation-efflux (CE) family (15), also known as the cation diffusion facilitator (CDF) family, are known to be involved in Zn efflux across the plasma membrane and into intracellular vacuoles in mammals (16). Recent evidence suggests ZAT, an A. thaliana CE family member, also possibly effluxes Zn into the vacuole, although direct evidence is lacking (12). We initiated an investigation of the role of CE family members in the mechanism of vacuolar metal ion accumulation in plants by cloning cDNAs encoding putative vacuolar metal ion transporter from the Ni hyperaccumulator T. goesingense. This species is known to have an enhanced capacity to accumulate Ni within vacuoles in the shoot as part of its hyperaccumulation mechanism (5).
By using PCR, we isolated an ortholog of the A. thaliana ZAT gene from the hyperaccumulator T. goesingense and named it TgMTP1 (T. goesingense metal tolerance protein; Fig. 1). Although ZAT holds prior authority, to allow for expansion of the CE plant family, we propose that MTP would be a better base name for ZAT-related proteins (see ref. 15 for discussion).
Figure 1.
Alignment of TgMTP1 DNA sequences. The open box graphically illustrates the full-length alignment. The filled box (bp 481–720) is expanded to detail the alternatively spliced sequence. Nucleotides shaded in dark gray are conserved in all three sequences, and nucleotides in light gray are conserved only in two of the sequences. Intron 1 is illustrated with a dark line below the detailed sequence alignment. SNP positions are shown as asterisks.
To establish the presence, and estimate the copy number of TgMTP1 in the T. goesingense genome, we performed genomic Southern blot analyses by using three restriction enzymes, SacI, PvuII, and EcoRI (Fig. 2). As predicted for a single-gene copy, SacI and PvuII (internal cutters) produced two major bands, with EcoRI (external cutter) producing a single major band (Fig. 2A). As a control, we also performed an A. thaliana genomic Southern blot of ZAT, which is known to be a single-copy gene [The Arabidopsis Genome Initiative (AGI) http://www.arabidopsis.org/]. As predicted for the single-copy ZAT, SacI (internal cutter) produced two major bands, and PvuII and EcoRI (external cutters) produced a single major band each (Fig. 2B). The lighter bands observed on the A. thaliana Southern blot represent cross-hybridization of the ZAT probe with the known ZAT homologs AtMTPa1 (AGI no. AT3 g61940), AtMTPa2 (AGI no. AT3 g58810), and AtMTPb1 (AGI no. AT2 g29410; ref. 15). The lighter bands observed in the T. goesingense Southern blot (Fig. 2A) also represent cross-hybridization of the TgMTP1 probe with orthologs of the A. thaliana AtMTPa1, AtMTPa2, and AtMTPb1 sequences. We confirmed the presence of such orthologs in T. goesingense by cloning and sequencing TgMTPa1 and TgMTPb1 (data not shown).
Figure 2.
Genomic Southern blot analysis of TgMTP1 and AtZAT. (A) T. goesingense genomic DNA was cut with SacI and PvuII (internal cutters) and EcoRI (external cutter) and probed with TgMTP1. (B) A. thaliana genomic DNA was cut with SacI (internal cutter) and PvuII and EcoRI (external cutters) and probed with AtZAT. Major DNA fragments are marked with asterisks.
To determine the intronic gene structure of TgMTP1, we cloned transcript cDNA from total shoot RNA by using RT-PCR. Two transcript sequences, TgMTP1t1 and TgMTP1t2, were cloned. Sequence alignments (Fig. 1) revealed 14 single-nucleotide polymorphisms (SNPs) between the sequences, and a 96-bp deletion between nucleotides 541 and 638 in TgMTP1t2 (Fig. 1).
Because the hyperaccumulator seed used in our studies was collected from a wild T. goesingense population, the SNPs observed in the TgMTP1 sequences most likely reflect different TgMTP1 alleles in this population and not multiple TgMTP1 genes within the T. goesingense genome. Such a conclusion is supported by the genomic Southern blot analysis that shows TgMTP1 to be a single-copy gene (Fig. 2).
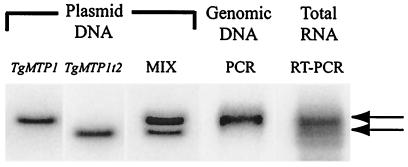
There are two alternative explanations for the origin of the 96-bp deletion in TgMTP1t2. Either it represents an intron that is spliced from TgMTP1 or TgMTP1t2 represents the transcript of an unidentified allele of TgMTP1. To distinguish between these two possibilities, we performed PCR and RT-PCR on genomic and total RNA by using primers that amplify full-length TgMTP1. The PCR products were run on an agarose gel, blotted, and probed with TgMTP1 (Fig. 3). Amplification of TgMTP1 from genomic DNA gave rise to a single product equal in size to full-length TgMTP1. Such a conclusion is consistent with the genome containing only a full-length TgMTP1 sequence. However, amplification of TgMTP1 from RNA produced two products that hybridized with TgMTP1. The largest and most abundant fragment was equal in size to TgMTP1 (Fig. 3), whereas the second smaller product was equal in size to TgMTP1t2 (Fig. 3). Amplification of this smaller second product exclusively from RNA is consistent with the 96-bp deletion in TgMTP1t2 being an intron and confirms that TgMTP1t2 represents an alternatively spliced transcript of TgMTP1.
Figure 3.
Southern blot analysis of TgMTP1 PCR and RT-PCR products. TgMTP1 and TgMTP1t2 cDNAs and a mixture of both (MIX) were prepared by digestion of pYES containing these clones. Full-length TgMTP1 cDNA products were also amplified from both genomic DNA (PCR) and RNA (RT-PCR). All cDNAs were separated on an agarose gel, blotted, and probed with TgMTP1. Two major cDNA products were amplified from RNA (marked with arrows).
The boundary sequences of plant nuclear mRNA introns are defined by a GU 5′ donor site and an AG 3′ acceptor site; although in rare instances, 5′ GC/AG 3′ or 5′ AU/AC 3′ also occur (17). The 5′ donor site in TgMTP1 is GC (Fig. 1), a known but rare donor site. However, the 3′ acceptor site in TgMTP1 seems to be UG, an unknown acceptor site. Our evidence (Figs. 1–3) strongly supports TgMTP1t2 being produced by splicing of TgMTP1. However, because the TgMTP1 sequence shown in Fig. 1 does not contain complete splice sites, we speculate that another as yet unidentified TgMTP1 allele is present in the Redschlag T. goesingense population that contains the appropriate splice sites, and it is this allele that gives rise to the spliced TgMTP1t2 transcript. In support of this, we have recently isolated two other genomic TgMTP1 alleles that contain a GU 5′ donor site, created by a C to U polymorphism (data not shown). We note that the SNPs in TgMTP1 cluster around both the 5′ and 3′ intron border sites (Fig. 1), making the generation of allelic splice variant likely. Instances of such SNPs creating new splice sites are well established in plants (18).
A search for conserved domains (http://workbench.sdsc.edu) classifies TgMTP1 as CE proteins, with both TgMTP1 splice variants possessing all features of CE family members (Fig. 4). These include the N-terminal signature sequence (19), CE domain (http://pfam.wustl.edu/), six predicted membrane-spanning domains (19), and a variable histidine-rich region near the middle of the protein sequence (20) that is predicted to project into the cytoplasm. It has been suggested that this type of histidine-rich domain may be involved in metal binding or transport (19). The CE family includes yeast COT1 and ZRC1 proteins (9, 21), mammalian ZNT1–4 proteins (20, 22), and bacterial CzD proteins (23). All these proteins are known to be involved in transport of metal ions including Ni, Zn, Co, and Cd, and there is strong evidence that COT1 and ZRC1 are responsible for transport of Co/Ni and Zn/Cd into the yeast vacuole (24).
Figure 4.
Alignment of TgMTP1 and ZAT predicted amino acid sequences. The cation diffusion facilitator (CDF) signature sequence, CE domains, histidine-variable region, and transmembrane (TM) domains are shown. Amino acid residues shaded in dark gray are conserved; residues in light gray are present in two of the three protein sequences.
Alignment of TgMTP1 with A. thaliana ZAT shows these proteins to be orthologs, with unspliced TgMTP1t1 showing 87% identity to ZAT (Fig. 4). Alignment of the predicted amino acid sequence of the unspliced and spliced variants of TgMTP1 shows that splicing differences are located exclusively in the central histidine-rich variable region of these proteins (Fig. 4). To investigate whether these sequence differences confer different functional properties to the proteins, we heterologously expressed them in yeast mutants deficient in the TgMTP1 evolutionary orthologs COT1 and ZRC1 (9, 21). Both COT1 and ZRC1 are members of the CE family, and yeast mutants lacking these genes are sensitive to Co, Ni, Cd, and Zn (9, 21). COT1 and ZRC1 are localized to the yeast vacuolar membrane (24) and are thought to be involved in transporting metal ions into the vacuole.
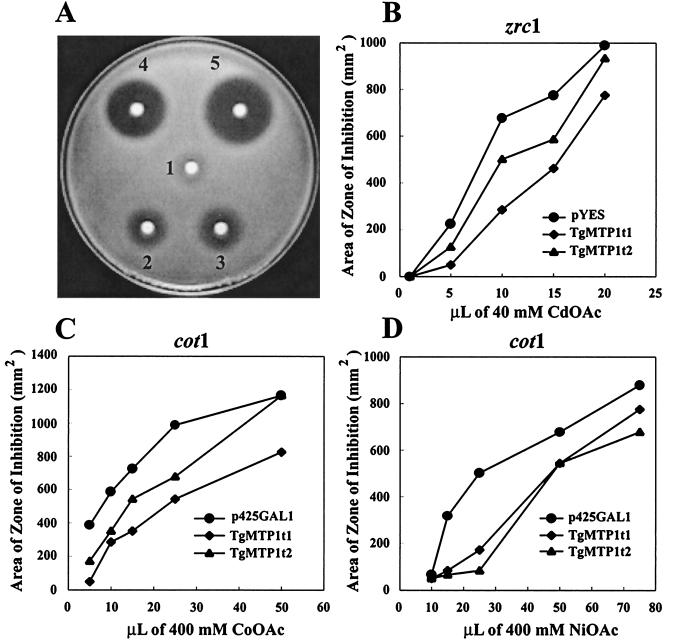
Yeast mutants cot1 and zrc1 heterologously expressing TgMTP1t1 and TgMTP1t2 were assayed for metal tolerance. Fig. 5A shows a representative metal-tolerance assay plate, with zones of yeast growth inhibition being clearly visible around the metal ion-soaked discs. Expression of TgMTP1t1 and TgMTP1t2 in cot1 and zrc1 yeast complemented the Cd, Co, and Ni sensitivity of these strains (Fig. 5 B–D), and we conclude that the T. goesingense TgMTP1 proteins act as functional orthologs of the yeast vacuolar metal ion transport proteins COT1 and ZRC1.
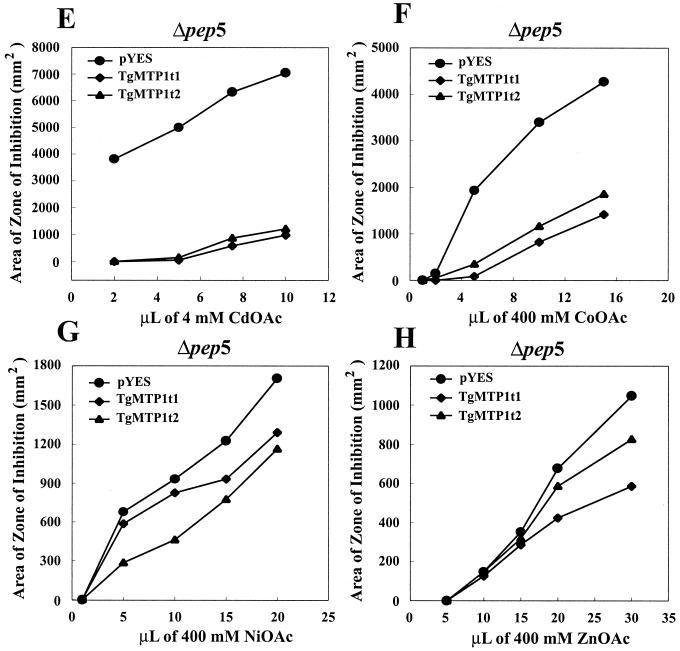
Figure 5.
Quantification of yeast metal tolerance. (A) An example of a yeast metal tolerance-assay plate showing the zones of cot1 growth inhibition around Co-soaked filter discs, with discs 1–5 containing 2, 4, 6, 10, and 20 μmol of Co, respectively. (B–H) Quantification of Cd, Co, Ni, and Zn tolerance of cot1, zrc1, and Δpep5 yeast strains expressing TgMTP1t1 and TgMTP1t2. Metal tolerance was quantified by using the assay system described in A.
To further confirm the ability of TgMTP1 to confer metal tolerance, we heterologously expressed both TgMTP1t1 and TgMTP1t2 in the metal-sensitive yeast mutant Δpep5. This mutant is metal-sensitive because it forms numerous small immature vacuoles instead of properly enlarged mature vacuoles (10). Both variants of TgMTP1 were able to suppress the Cd, Co, Ni, and Zn sensitivity of the Δpep5 mutant (Fig. 5 E–H). The suppression of metal sensitivity in Δpep5 may be explained in two ways. Either TgMTP1 proteins are targeted to the small immature vacuoles in this yeast mutant where they are involved in sequestering metal ions, or TgMTP1 proteins are targeted to the plasma membrane where they efflux metal ions from the yeast. Further studies are required to distinguish these mechanisms.
In all of the yeast strains tested, the variant proteins TgMTP1t1 and TgMTP1t2 showed consistent differences in their ability to confer metal tolerance (Fig. 5). TgMTP1t1 derived from the unspliced transcript had the highest capacity to confer tolerance to Cd, Co, and Zn (Fig. 5 B, C, E, F, and H), whereas TgMTP1t2 derived from the spliced transcript had the highest capacity to confer tolerance to Ni (Fig. 5 D and G). The amino acid sequences of the two variant TgMTP1s differ exclusively in the central histidine-rich domain of the proteins, with TgMTP1t2 lacking this region almost entirely (Fig. 4). Loss of this histidine-rich domain in TgMTP1t2 seems to confer increased Ni specificity to this protein.
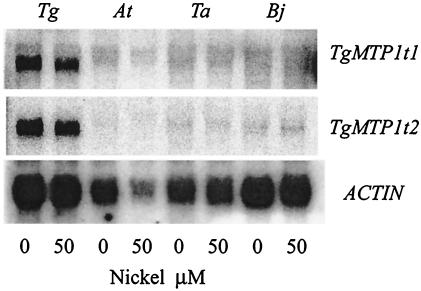
To determine whether the steady-state levels of TgMTP1 mRNAs are induced by Ni or are constitutively higher in the hyperaccumulator, we performed Northern analysis on both the hyperaccumulator T. goesingense and the nonaccumulators T. arvense, A. thaliana, and B. juncea. This analysis showed that TgMTP1 transcripts are more abundantly expressed in shoots of the hyperaccumulator compared with the nonaccumulators (Fig. 6). Probing of the Northern blot with a full-length T. arvense ortholog of TgMTP1 confirmed that these results were not caused by differential hybridization of the TgMTP1 probe (data not shown) but genuinely reflect increased levels of the TgMTP1 transcript in T. goesingense. Increased expression of TgMTP1 transcripts in the hyperaccumulator is constitutive and not effected by exposure to either high concentration of Ni for short periods (100, 250, or 500 μM for 24 or 48 h) or lower concentrations for extended periods (100 μM for 1–4 weeks; data not shown). Ni hyperaccumulation in T. goesingense is a known constitutive trait (25), which supports the observation that TgMTP1 is constitutively expressed. Increased expression of TgMTP1 in T. goesingense is consistent with this protein playing an important role in detoxification of Ni by vacuolar compartmentalization, a known mechanism of Ni tolerance in this hyperaccumulator (5).
Figure 6.
Northern blot of TgMTP1 expression in hyperaccumulator and nonaccumulator species. Total RNA was isolated from shoots of the hyperaccumulator T. goesingense and the nonaccumulators A. thaliana, T. arvense, and B. juncea after exposure to Ni for 48 h. Northern blots equally loaded with 30 μg of total RNA were probed with TgMTP1t1, stripped, and reprobed with TgMTP1t2, and finally stripped and reprobed with an A. thaliana actin probe as an RNA loading control.
In summary, we present an analysis of the functional activity and role of the TgMTP1 proteins in Ni hyperaccumulation in T. goesingense. Compared with nonaccumulator species, these proteins are constitutively highly expressed in the hyperaccumulator as two variant forms, TgMTP1t1 and TgMTP1t2, that are derived from the alternative splicing of the single-copy genomic clone TgMTP1. Heterologous yeast expression of these proteins complemented the metal sensitivity of the yeast mutants cot1 and zrc1 that lack functional vacuolar metal ion transport proteins. The complementation of metal sensitivity in these yeast mutants confirms that the TgMTP1 proteins can act as function orthologs of the yeast COT1 and ZRC1 vacuolar metal ion transporters. Sequence differences between the two splice variants of TgMTP1 cause altered metal ion selectivity in the proteins, with TgMTP1t1 conferring the highest tolerance to Cd, Co, and Zn, whereas TgMTP1t2 confers the highest tolerance to Ni. We propose that the overexpression and alternate splicing of these TgMTP1 metal ion transport proteins in T. goesingense are involved in the enhanced ability of this hyperaccumulator to compartmentalize Ni in its shoot vacuoles; a process critical to the ability of this plant to hyperaccumulate Ni (3, 5).
Acknowledgments
We thank Drs. Douglas Conklin and Carol Woolford for the kind gift of the yeast strains used in this study.
Abbreviations
- CE
cation efflux
- RT
reverse transcription
- SNP
single-nucleotide polymorphism
Footnotes
References
- 1.Wenzel W W, Jockwer F. Environ Pollut. 1999;104:145–155. [Google Scholar]
- 2.Baker A J M, Brooks R R. Biorecovery. 1989;1:81–126. [Google Scholar]
- 3.Krämer U, Smith R D, Wenzel W W, Raskin I, Salt D E. Plant Physiol. 1997;115:1641–1650. doi: 10.1104/pp.115.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salt D E, Kato N, Kramer U, Smith R D, Raskin I. In: Phytoremediation of Contaminated Soil and Water. Terry N, Banuelos G S, editors. Boca Raton: CRC; 1999. pp. 191–202. [Google Scholar]
- 5.Krämer U, Pickering I J, Prince R C, Raskin I, Salt D E. Plant Physiol. 2000;122:1343–1353. doi: 10.1104/pp.122.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persans M W, Salt D E. Biotechnol Genet Eng Rev. 2001;17:385–409. doi: 10.1080/02648725.2000.10647999. [DOI] [PubMed] [Google Scholar]
- 7.Persans M W, Yan X, Patnoe J-M, Krämer U, Salt D E. Plant Physiol. 1999;121:1117–1126. doi: 10.1104/pp.121.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz R D, Woods R A. In: Molecular Genetics of Yeast: A Practical Approach. Johnston J A, editor. New York: Oxford Univ. Press; 1994. pp. 121–134. [Google Scholar]
- 9.Conklin D S, Culbertson M R, Kung C. Mol Gen Genet. 1994;244:303–311. doi: 10.1007/BF00285458. [DOI] [PubMed] [Google Scholar]
- 10.Woolford C A, Dixon C K, Manolson M F, Wright R, Jones E W. Genetics. 1990;125:739–752. doi: 10.1093/genetics/125.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 12.Van der Zaal B J, Neuteboom L W, Pinas J E, Chardonnes A N, Schat H, Verkleji J A C, Hooykaas P J J. Plant Physiol. 1999;119:1047–1055. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mäser, P., Thomine, S., Schroeder, J. I., Hirschi, K., Ward, J., Sze, H., Amtmann, A., Maathuis, F. J. M., Talke, I. N., Sanders, D., et al. (2001) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 16.McMahon R J, Cousins R J. J Nutr. 1998;128:667–670. doi: 10.1093/jn/128.4.667. [DOI] [PubMed] [Google Scholar]
- 17.Simpson G G, Filipowicz W. Plant Mol Biol. 1996;32:1–41. doi: 10.1007/BF00039375. [DOI] [PubMed] [Google Scholar]
- 18.Frances H, Bligh J, Larkin P D, Roach P S, Jones C A, Fu H, Park W. Plant Mol Biol. 1998;38:407–415. doi: 10.1023/a:1006021807799. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen J T, Saier M H. J Membr Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 20.Palmiter R D, Cole T B, Findley S D. EMBO J. 1996;15:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 21.Conklin D S, McMaster J A, Culbertson M R, Kung C. Mol Cell Biol. 1992;12:3678–3688. doi: 10.1128/mcb.12.9.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmiter R D, Findley S D. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nies D H. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Kaplan J. J Biol Chem. 1998;273:22181–22187. doi: 10.1074/jbc.273.35.22181. [DOI] [PubMed] [Google Scholar]
- 25.Reeves R D, Baker A J M. New Phytol. 1984;98:191–204. doi: 10.1111/j.1469-8137.1984.tb06108.x. [DOI] [PubMed] [Google Scholar]