Abstract
Quality evaluation of mesenchymal stem cells (MSCs) based on efficacy would be helpful for their clinical application. In this study, we aimed to find the factors of human bone marrow MSCs relating to cartilage repair. The expression profiles of humoral factors, messenger RNAs (mRNAs), and microRNAs (miRNAs) were analyzed in human bone marrow MSCs from five different donors. We investigated the correlations of these expression profiles with the capacity of the MSCs for proliferation, chondrogenic differentiation, and cartilage repair in vivo. The mRNA expression of MYBL1 was positively correlated with proliferation and cartilage differentiation. By contrast, the mRNA expression of RCAN2 and the protein expression of TIMP-1 and VEGF were negatively correlated with proliferation and cartilage differentiation. However, MSCs from all five donors had the capacity to promote cartilage repair in vivo regardless of their capacity for proliferation and cartilage differentiation. The mRNA expression of HLA-DRB1 was positively correlated with cartilage repair in vivo. Meanwhile, the mRNA expression of TMEM155 and expression of miR-486-3p, miR-148b, miR-93, and miR-320B were negatively correlated with cartilage repair. The expression analysis of these factors might help to predict the ability of bone marrow MSCs to promote cartilage repair.
1. Introduction
Mesenchymal stem cells (MSCs) have the capacity for self-renewal [1] and differentiation into several mesoderm-type lineages, including osteoblasts, chondrocytes, adipocytes, myocytes, and vascular cells [2] and are considered to be nonimmunogenic [3, 4]. Therefore, MSCs are one of the most promising cellular sources of stem cells that can be studied without using any immunosuppressive drugs, for both research and clinical purposes. Clinical studies using MSCs are widely available. For example, MSCs have been used in the therapy of diseases such as extended cartilage [5, 6] and osseous defects [7], intervertebral disc [8], acute myocardial infarction [9], leukemia [10], and diabetes [11]. We have started two clinical trials of intra-articular injection of autologous bone marrow MSCs for articular cartilage repair based on our previous animal experiments [5, 12, 13]. However, the functional quality of MSCs for cartilage regeneration might be diversified depending on the donor due to the heterogeneity of MSCs. There have been reports that differentiation and proliferation capacity decrease with age [14, 15] and, consequently, the use of autologous MSCs for tissue repair, which in some indications concerns elderly patients, has certain limits [16]. Quality evaluation confirming the properties of MSCs has been established and is based on cell surface markers (negative for CD14 or CD11b, CD19, CD34, CD45, CD79α, and HLA-DR and positive for CD73, CD90, CD105, CD166, and CD44 [17–19]) and differentiation capacity [20–22]. However, quality evaluation based on the efficacy of MSCs has been not established. The evaluation criteria for quality of MSCs based on the efficacy would be required for the practical application of MSC transplantation. In this study, we aimed to devise a method for functional quality assessment of MSCs for cartilage regeneration by examining the relationships among the following data in human bone marrow MSCs (hMSCs): profiles of cartilage anabolic and catabolic factors, messenger RNAs (mRNAs), and microRNAs (miRNAs), and the capacity for cell proliferation, chondrogenic differentiation in vivo, and cartilage regeneration in vivo.
2. Materials and Methods
All procedures were approved and performed by the Guide for Animal Experimentation, Hiroshima University, and the Committee of Research Facilities for Laboratory Animal Sciences, Graduate School of Biomedical Sciences, Hiroshima University.
In this study, hMSCs were purchased from Lonza Walkersville Inc. (PT-2501, Walkersville, MD, USA). All these hMSCs passed the quality inspection conducted by Lonza company using cell viability (more than 75%), adipogenic and osteogenic differentiation (Oil Red O staining and calcium deposition staining), and FACS analysis of cell surface markers (more than 90% were positive for CD29, CD44, CD105, and CD166 and negative for CD14, CD34, and CD45). Assays of cell growth rate (GR) and colony formation (CF), pellet cultures in vitro, and transplantation of hMSCs into cartilage defect models in vivo were performed using hMSCs from five different donors. The age, race, and sex of the five different donors were as follows: 22-year-old black man, 20-year-old white man, 39-year-old black man, 29-year-old white woman, and 41-year-old white woman.
2.1. Cultures of hMSCs and Human Skin Fibroblasts
The hMSCs at passage two were centrifuged at 500g for 5 min. The cells were resuspended in culture medium composed of Dulbecco's modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA, USA), 15% fetal bovine serum (FBS; SAFC Biosciences Inc., Irvine, CA, USA), 20 mmol/ml of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Life Technologies), 50 μg/ml gentamycin (Gentacin®, MSD, Tokyo, Japan), and 0.25 μg/ml amphotericin (Fungizon®, Bristol-Myers Squibb, New York, NY, USA). The suspension was then plated into culture dishes. The dishes were incubated in an atmosphere of 95% relative humidity and 5% CO2 at 37°C until 70–80% confluency, and the cells were then harvested with Trypsin (TrypLE™ Express; Life Technologies, Carlsbad, CA, USA). After adding high-glucose DMEM® with 10% FBS and 1% antibiotics, the cells were neutralized and harvested by centrifugation at 200g for 5 min and the resulting supernatant frozen at −80°C with Cellbanker® 1 (LSI Medience, Tokyo, Japan) until further testing. The cells were defined as passage three (P3). The hMSCs at P3 were reseeded under high-glucose DMEM with 10% FBS and 1% antibiotics to grow the hMSCs. These adherent cells have been referred to as hMSCs at passage four (P4). The hMSCs at P4 were used in the current study. Adult normal human dermal fibroblasts (hFBs; Lonza Japan, Tokyo, Japan) at P2 were cultured with Fibroblast Cell Growth Medium-2 BulletKit (FGM™-2 BulletKit™; Lonza Japan, Tokyo, Japan) until P4 by a similar method and the cells at P4 were used.
2.2. Assay of Growth Rate for hMSCs
The hMSCs at P4 were seeded onto culture dishes at 5.0 × 103 cells/cm2 in Mesenchymal Stem Cell Basal Medium (MSCBM™; Lonza Japan, Tokyo, Japan) and incubated in an atmosphere of 95% relative humidity and 5% CO2 at 37°C. Cell division rate was assessed after 5 days.
2.3. Colony-Forming Assay
In a pilot study, we confirmed a positive correlation (P < 0.001, R = 0.992) between hMSC seeding density and capacity for CF. Because the CF measurement did not reflect the seeding density, the capacity for CF did not have an effect of the secretor factor and could measure quality of cells by CF measurement. The hMSCs at P4 were plated at 1.5 × 103 cells/T75 flask in MSCBM (Lonza Japan) for 14 days. The medium was changed at three- to four-day intervals. After embedding the plate in paraffin, the cells were stained by 1.0% crystal violet solution (Wako, Osaka, Japan) for 10 min. Aggregates of cells differentiated than 50 cells were counted as one colony, and we calculated the ratio of these colonies among all seeded cells.
2.4. Analysis of Protein Derived from Culture Supernatant
MSCs at 70–80% confluency were refed with culture media. After 48 h incubation, the media were collected. To remove debris, the media were centrifuged at 600g for 5 min and the supernatants were collected as MSC-conditioned media (MSC-CMs). They were stored at −80°C until they were used for the following assays.
The MSC-CMs were analyzed by enzyme-linked immunosorbent assay (ELISA). Sandwich ELISA kits purchased from R&D Systems (Minneapolis, MN, USA) were used for bone morphogenetic protein- (BMP-) 2, BMP-7, fibroblast growth factor- (FGF-) 2, insulin-like growth factor- (IGF-) 1, transforming growth factor- (TGF-) α, TGF-β1, TGF-β2, tissue inhibitor of metalloproteinase- (TIMP-) 1, TIMP-2, platelet-derived growth factor- (PDGF-) AA, interleukin- (IL-) 2, IL-17, monocyte chemotactic protein- (MCP-) 1, matrix metalloproteinase- (MMP-) 1, MMP-3, MMP-9, MMP-13, RANTES, stromal cell-derived factor- (SDF-) 1α, macrophage inflammatory protein- (MIP-) 1α, MIP-1β, and MIP-3α. Kits from Life Technologies (Carlsbad, CA, USA) were used for hepatocyte growth factor (HGF), IL-1β, IL-4, IL-8, IL-10, TNF-α, interferon- (IFN-) γ, and vascular endothelial growth factor (VEGF). Assays were performed according to the manufacturer's instructions in duplicate. As a control, culture media were also analyzed. The hFBs at P4 were cultured at the same time and were compared with hMSCs as a control.
2.5. The Assessment of Chondrogenic Differentiation Using Pellet Culture for hMSCs
The capacity for chondrogenic differentiation of the hMSCs from each donor was evaluated using pellet culture.
A pellet culture system for chondrogenesis was used [23–26]. About 2.5 × 105 hMSCs at P4 were placed in a 15 ml polyethyleneterephthalate tube (Sumitomo Bakelite) and centrifuged at 450g for 10 min. The pellet was cultured at 37°C with 5% CO2 in 500 μl of chondrogenic medium containing 500 ng/ml BMP-6 (27) (R&D Systems, Minneapolis, MN, USA) in addition to high-glucose DMEM supplemented with 10 ng/ml TGF-β3 (R&D Systems, Minneapolis, MN, USA), 10−7 M dexamethasone, 50 μg/ml ascorbate-2-phosphate, 40 μg/ml proline, 100 μg/ml pyruvate (Sigma-Aldrich, St. Louis, MO, USA), and 50 mg/ml ITS+ Premix (Becton Dickinson; 6.25 μg/ml insulin, 6.25 μg/ml transferrin, 6.25 ng/ml selenous acid, 1.25 mg/ml BSA, and 5.35 mg/ml linoleic acid). The medium was replaced every 3 to 4 days for 21 days. For microscopy, the pellets were embedded in paraffin, cut into 5 μm sections, and stained with 0.05% toluidine blue solution (Muto Pure Chemicals Co., Ltd., Tokyo, Japan). The production of extracellular matrix was evaluated by measuring the percentage of metachromasy in pellets (PMP) derived from hMSCs stained with toluidine blue by image processing software (WinROOF®, Mitani, Tokyo, Japan). We calculated the PMP for all areas in pellets.
2.6. Real-Time Quantitative Polymerase Chain Reaction (qPCR) of Pellets
Total RNA was isolated from pellets using a Qiagen RNeasy Micro Kit (Qiagen, Valencia, CA, USA). cDNA was synthesized from RNA using Super Script VILO Master Mix (Life Technologies). As a control, total RNA was obtained from normal knee cartilage dissected from skeletally matured cadaveric donors (Articular Engineering, Northbrook, IL, USA). qPCR was performed using Power SYBR Green Master Mix (Life Technologies). cDNA samples were analyzed for both collagen type II (COL II) and the reference gene (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)). Assays were performed according to the manufacturer's instructions. The primer sequences used for the experiment are shown in Table 1. The amount of COL II mRNA expressed was normalized to GAPDH expression. In addition, the amount of mRNA expressed in hMSCs from donors (hMSC-1 to hMSC-5) was normalized to expression in hMSC-1 for the purpose of comparing the mRNA expressions among five donors.
Table 1.
Primer sequences used for qPCR.
| Gene | Primer sequence (5′ → 3′) |
|---|---|
| Type II collagen | GGCAATAGCAGGTTCACGTACA |
| CGATAACAGTCTTGCCCCACTT | |
| Type X collagen | CAAGGCACCATCTCCAGGAA |
| AAAGGGTATTTGTGGCAGCATATT | |
| Aggrecan | TACGAAGACGGCTTCCACCA |
| CTCATCCTTGTCTCCATAGC | |
| SOX9 | GTACCCGCACTTGCACAAC |
| GTAATCCGGGTGGTCCTTCT | |
| CD44 | AAGACACATTCCACCCCAGT |
| GGTTGTGTTTGCTCCACCTT | |
| GAPDH | ATGGGGAAGGTGAAGGTCG |
| TAAAAGCAGCCCTGGTGACC |
2.7. The Assessment of Capacity for Cartilage Regeneration Using Cartilage Repair
In this study, male nude rats aged 9 to 10 weeks were used and were anesthetized with an intraperitoneal administration of 1.0 ml/kg sodium pentobarbital before surgery. The medial parapatellar approach was used to expose the knee joint. We created full thickness articular cartilage defects of 2 mm in diameter and 1 mm in depth at the patellar groove of the distal femur using a power drill, and the joint capsule and skin incision were closed with 6-0 nylon sutures. Rats were divided into two groups. In the control group, phosphate-buffered saline (PBS, 25 μl) was injected into the five operated joints (n = 5). This group indicated the natural course of healing of the osteochondral defect. In the hMSC group, 3 × 105 hMSCs from the five donors were injected into each operated joint (n = 5/donor). After transplantation, all nude rats were allowed to move freely in their cages.
2.8. Histological Evaluation
All nude rats were sacrificed by intraperitoneal injection of a lethal dose of pentobarbital sodium at 12 weeks after the injection. The patellar groove was resected and fixed in 4% paraformaldehyde for 24 h. The specimens were then decalcified with 0.5 M ethylenediaminetetraacetic acid. After that, the specimens embedded in paraffin block were cut into 5 μm sections serially along the sagittal plane that included the center of the defect. For histological assessment, these sections were stained with Safranin-O/Fast green. The specimens were graded semiquantitatively. The grading scale was based on a histological grading scale for cartilage regeneration as previously described [27].
2.9. 3D-Gene® Human Oligo Chip 25k for mRNA and TaqMan® Low-Density Array for miRNA Expression Profiling
The hMSCs and hFBs at P4 were homogenized on plate using TRIzol Reagent (Life Technologies), and total RNAs were isolated according to the manufacturer's instructions. For mRNA microarray analysis, 3D-Gene Human Oligo chip 25k (3D-Gene; Toray Industries, Tokyo, Japan) was used (24,460 distinct genes). The gene expression of hFBs at P4 was used as a control for normalization. Experimental procedures for TaqMan low-density array analysis (TLDA®; Life Technologies) were performed using TaqMan Array Human microRNA Cards® (card A v2.0 and card B v3.0) according to the manufacturer's instructions to identify differentially expressed miRNAs in hMSCs of each donor.
2.10. Statistical Analysis
All assays were performed in triplicate. The results are shown as mean values and standard deviations. Pearson's correlation coefficient calculated using software of the statistic program for Windows by Statcel4® of Excel Statistics (Statcel4: Nebula Company, Bunkyo-ku, Tokyo, Japan) was used to evaluate associations among capacity for cell proliferation, chondrogenic differentiation, and cartilage regeneration, and expression pattern of proteins, mRNAs, and miRNAs in hMSCs. Multiple comparison was performed for the evaluation of histological scores of specimens from the knee of nude rats between each group using Bartlett's test and one-way analysis of variance. When a significant p value was found, the Tukey-Kramer method was used to identify significant differences among the groups. The significance level was defined at P < 0.05 for all tests.
3. Results
3.1. Growth Rate and Colony Forming
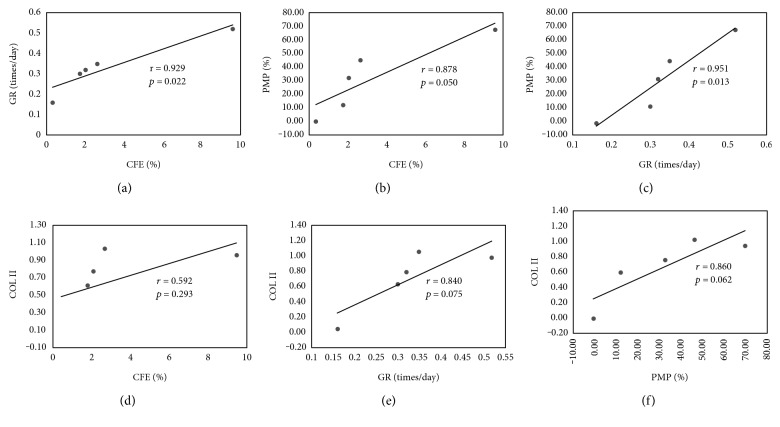
The hMSCs from five different donors were ranked from hMSC-1 to hMSC-5 in order of GR, and the hMSC-1 to hMSC-5 were from a 22-year-old black man, 20-year-old white man, 39-year-old black man, 29-year-old white woman, and 41-year-old white women, respectively. GR and colony-forming efficiency (CFE) for hMSC-1 to hMSC-5 and hFBs were 0.52 times/day and 9.6%, 0.35 times/day and 2.6%, 0.32 times/day and 2.0%, 0.30 times/day and 1.7%, 0.16 times/day and 0.3%, and 0.73 times/day and 0.0%, respectively. CFE showed a positive correlation with GR (r = 0.929, P = 0.022) (Figure 1(a)).
Figure 1.
Correlations between (a) CFE and GR, (b) CFE and PMP, (c) GR and PMP, (d) CFE and COLII, (e) GR and COLII, and (f) PMP and COLII were r = 0.929 and P = 0.022, r = 0.878 and P = 0.050, r = 0.951 and P = 0.013, r = 0.592 and P = 0.293, r = 0.840 and P = 0.075, and r = 0.860 and P = 0.062, respectively.
3.2. Proteins Derived from Culture Supernatant
To evaluate the protein secretion from MSCs relating to cartilage repair, antianabolic and catabolic factors for cartilage in culture supernatant were chosen for assessment. In the assessment of protein expression using ELISA, the anabolic factors TIMP-1, TIMP-2, TGF-β1, TGF-β2, PDGF-AA, HGF, and IGF-1 and the catabolic factors IL6, IL8, SDF-1a, MMP13, VEGF, MCP-1, MMP1, and MMP-3 were detected in the culture supernatant for each donor (Table 2). On the other hand, we were not able to validate protein expression for BMP-2, BMP-7, IL-4, IL-10, FGF-2, IFNγ, IL1β, IL2, IL17, MMP9, RANTES, TNFα, MIP1α, MIP1β, MIP3α, and TGFα. The 15 factors out of 31 could be detected using ELISA.
Table 2.
Secretional capacity of proteins per 10,000 hMSCs (pg/10,000 hMSCs).
| TIMP-1 | TIMP-2 | TGF-β1 | TGF-β2 | PDGF-AA | HGF | IGF-1 | ||
|---|---|---|---|---|---|---|---|---|
| hMSC-1 | 5841.6 | 2790.4 | 192.3 | 19.8 | 1.1 | 2.5 | 17.7 | |
| hMSC-2 | 12129.6 | 3714.5 | 283.6 | 31.8 | 2.0 | 4.3 | 44.3 | |
| hMSC-3 | 14791.6 | 4227.6 | 333.2 | 40.5 | 2.1 | 1.4 | 64.8 | |
| hMSC-4 | 23319.9 | 4394.7 | 367.0 | 38.5 | 1.7 | 85.8 | 44.4 | |
| hMSC-5 | 41621.9 | 7640.3 | 479.5 | 54.6 | 4.2 | 60.3 | 80.8 | |
| hFB | 5879.2 | 3176.6 | 128.9 | 11.6 | 0.5 | 0.0 | 27.4 | |
|
| ||||||||
| IL6 | IL-8 | SDF-1a | MMP13 | VEGF | MCP-1 | MMP-1 | MMP-3 | |
|
| ||||||||
| hMSC-1 | 123.9 | 0.4 | 150.1 | 1.7 | 449.8 | 30.0 | 55.6 | 10.4 |
| hMSC-2 | 355.8 | 1.4 | 301.8 | 2.4 | 513.7 | 54.8 | 7.5 | 8.7 |
| hMSC-3 | 606.8 | 1.1 | 278.8 | 3.1 | 742.0 | 59.0 | 32.1 | 22.8 |
| hMSC-4 | 314.7 | 5.0 | 378.6 | 16.1 | 789.3 | 133.0 | 42.8 | 12.6 |
| hMSC-5 | 966.5 | 8.4 | 284.5 | 4.1 | 1253.7 | 322.0 | 99.3 | 75.5 |
| hFB | 58.4 | 1.6 | 380.5 | 0.5 | 49.5 | 81.5 | 1096.2 | 1279.9 |
3.3. Percentage of Metachromasy in Pellets
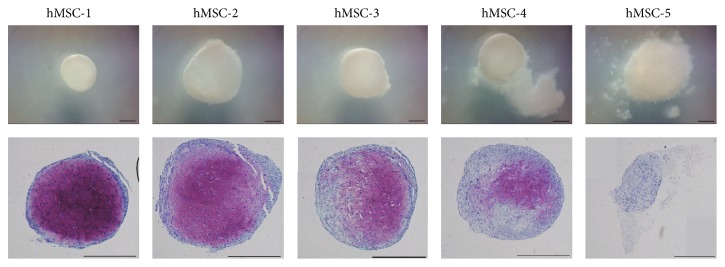
PMP of hMSC-1 to hMSC-5 were 69.3%, 46.16%, 32.8%, 12.4%, and 0.0%, respectively (Figure 2). The capacity for the production of extracellular matrix, demonstrating chondrogenic differentiation, was positively correlated with GR and CFE (GR: r = 0.951, P = 0.013; CFE: r = 0.878, P = 0.050) (Figures 1(b) and 1(c)).
Figure 2.
Percentages of metachromasy in pellets of hMSC-1 to hMSC-5 were 69.3%, 46.16%, 32.8%, 12.4%, and 0.0%, respectively. Scale bars, 500 μm.
3.4. Real-Time PCR Assays of Pellets
COLII gene expressions in hMSC-1 to hMSC-5 were 1.00, 1.08, 0.80, 0.63, and 0.00, respectively. Gene expression of COL II showed a correlative trend with CFE (r = 0.592, P = 0.293) and a correlation with GR (r = 0.840, P = 0.075) and PMP (r = 0.860, P = 0.062) (Figures 1(d), 1(e), and 1(f)). These findings indicate that chondrogenic capacity of hMSCs is positively correlated with proliferation capacity.
3.5. Histological Evaluation of Cartilage Repair
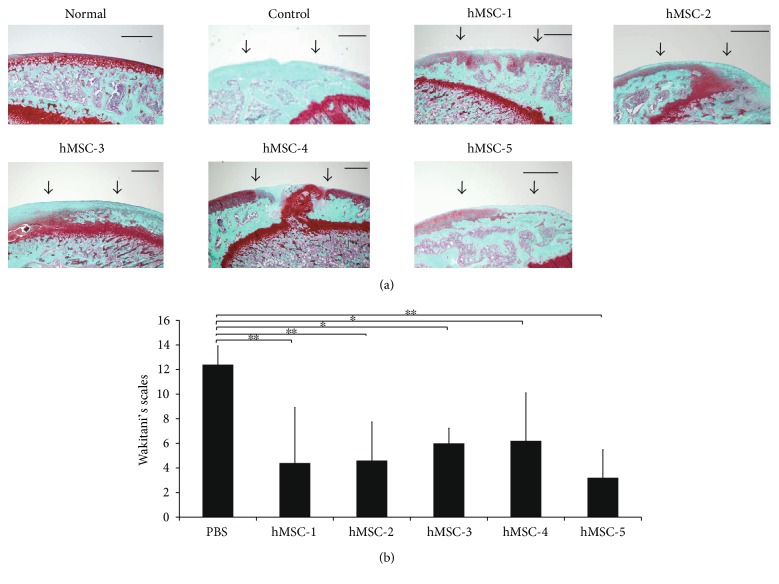
In the control group, the chondral defect area was not stained with Safranin-O. The mean histological score of the control samples was 12.40 ± 1.52 (SD) points. In hMSC-1, hMSC-2, and hMSC-5, the chondral defect area was partially stained with Safranin-O. However, in hMSC-3 and hMSC-4, the margins of the defect were irregular and the repair cartilage was composed of fibrous tissue. In hMSC-1 to hMSC-5 groups, the mean histological score was 4.4 ± 4.51 (SD), 4.6 ± 3.13 (SD), 6.0 ± 1.22 (SD), 6.2 ± 3.90 (SD), and 3.2 ± 2.28 (SD), respectively (Figure 3).
Figure 3.
(a) Histological findings with Safranin-O/Fast green staining at 12 weeks after injection of hMSCs into the cartilage defect models. Scale bars, 500 μm. (b) Assessment of the five different donors using Wakitani's scales (∗∗P < 0.01, ∗P < 0.05).
3.6. Gene Expression of hMSCs Assessed by 3D-Gene Human Oligo Chip 25k and TaqMan Low-Density Array
Of 24,460 mRNAs analyzed by 3D-Gene, we detected mRNA expressions of MYBL1 (MYB proto-oncogene like 1) and RCAN2 (regulator of calcineurin 2) related to proliferation capacity in vitro and HLA-DRB1 (major histocompatibility complex, class II, DR beta 1) and TMEM155 (transmembrane protein 155) related to cartilage repair in vivo (Table 3). Of 768 miRNAs analyzed by TLDA, we detected miR-486-3p, miR-148b, miR-93, and miR-320B in cartilage repair in vivo (Table 4). The gene expression in hMSCs from each donor was evaluated for fold changes compared with the gene expression in hFBs as a control for normalization. In the assessment of genes that were not detected in hFBs, the lowest gene expression in hMSCs was used as a control for normalization.
Table 3.
Changes in mRNA expression in hMSCs.
| Gene | hMSC-1 | hMSC-2 | hMSC-3 | hMSC-4 | hMSC-5 | |
|---|---|---|---|---|---|---|
| In vitro | MYBL1 | 5.44 | 4.38 | 4.76 | 3.33 | 2.97 |
| RCAN2 | 0.07 | 0.07 | 0.08 | 0.14 | 0.15 | |
|
| ||||||
| In vivo | HLA-DRB1 | 0.07 | 0.15 | 0.28 | 0.45 | 0.03 |
| TMEM155 | 4.25 | 4.16 | 3.37 | 2.03 | 4.86 | |
Fold changes.
Table 4.
Changes in miRNA expression in hMSCs.
| Gene | hMSC-1 | hMSC-2 | hMSC-3 | hMSC-4 | hMSC-5 | |
|---|---|---|---|---|---|---|
| In vivo | miR-486-3p | 2.12 | 1.55 | 0.88 | 1.00 | 3.11 |
| miR-148b | 2.37 | 3.85 | 1.00 | 1.98 | 6.57 | |
| miR-93 | 4.16 | 4.27 | 3.60 | 2.76 | 4.55 | |
| miR-320B | 2.34 | 3.46 | 1.99 | 0.89 | 4.53 |
Fold changes.
3.7. Relation In Vitro
The hMSCs from the five donors were divided into two groups of hMSC-1 to hMSC-3 and hMSC-4 to hMSC-5 according to their results in chondrogenic differentiation. In the toluidine blue staining and RT-PCR assays of pellets from hMSCs, hMSC-1 to hMSC-3 showed rich, whereas hMSC-4 and hMSC-5 showed poor, production of extracellular matrix. In addition, hMSC-1 to hMSC-3 showed good capacity, whereas those of hMSC-4 and hMSC-5 showed poor capacity, for cell proliferation. The mRNAs and miRNAs that could be divided into two groups of hMSC-1 to hMSC-3 and hMSC-4 to hMSC-5 according to their results in chondrogenic differentiation were selected and assessed for their correlation with chondrogenic differentiation.
In the assessment of mRNAs and miRNAs, the expression of MYBL1 was higher and that of RCAN2 lower in hMSCs from the three donors with good cell proliferation and production of extracellular matrix than in hMSCs from the other two donors. In addition, protein expression levels of both TIMP-1 and VEGF were negatively correlated with cell proliferation and production of extracellular matrix (Table 5).
Table 5.
Correlation analysis of specific factors in vitro.
| mRNA | CFE | GR | PMP | |
|---|---|---|---|---|
| MYBL1 | P | 0.098 | 0.042∗ | 0.014∗ |
| r | 0.808 | 0.891 | 0.947 | |
| RCAN2 | P | 0.271 | 0.122 | 0.032∗ |
| r | −0.614 | −0.777 | −0.909 | |
| Protein | ||||
| TIMP1 | P | 0.164 | 0.026∗ | 0.025∗ |
| r | −0.727 | −0.923 | −0.923 | |
| VEGF | P | 0.179 | 0.030∗ | 0.034∗ |
| r | −0.711 | −0.914 | −0.906 |
P = probability, r = correlation coefficient, ∗P < 0.05.
3.8. Relation In Vivo
The hMSCs from the five donors were divided into two groups of hMSC-1, hMSC-2, and hMSC-5 and hMSC-3 to hMSC-4 according to their results in evaluation of cartilage regeneration using Wakitani's scales. The mRNAs and miRNAs that could be divided into two groups of hMSC-1, hMSC-2, and hMSC-5 and hMSC-3 to hMSC-4 according to their results in cartilage regeneration were selected and assessed for their correlation with cartilage regeneration.
The expressions of MYBL1, RCAN2, TIMP-1, and VEGF showing relevance with cell proliferation and chondrogenic differentiation had no correlation with cartilage repair in vivo. On the other hand, the expression of HLA-DRB1 was higher and that of TMEM155, miR-486-3p, miR-148b, miR-93, and miR-320b lower in hMSCs from the three donors that showed good cartilage repair in vivo than in hMSCs from the other two donors (Table 6). However, protein expression levels of anabolic and catabolic factors for cartilage were not correlated with cartilage repair in vivo.
Table 6.
Correlation analysis of specific factors in vivo.
| mRNA and miRNA | CFE | GR | PMP | Wakitani | |
|---|---|---|---|---|---|
| HLA-DRB1 | P | 0.605 | 0.918 | 0.666 | 0.024∗ |
| r | −0.315 | −0.065 | −0.265 | 0.726 | |
| TMEM155 | P | 0.779 | 0.937 | 0.763 | 0.031∗ |
| r | 0.175 | −0.050 | 0.187 | −0.911 | |
|
| |||||
| miR-486-3p | P | 0.932 | 0.645 | 0.766 | 0.008∗ |
| r | 0.054 | −0.282 | −0.185 | −0.966 | |
| miR-148b | P | 0.567 | 0.301 | 0.444 | 0.045∗ |
| r | −0.347 | −0.584 | 0.453 | −0.887 | |
| miR-93 | P | 0.799 | 0.950 | 0.735 | 0.037∗ |
| r | 0.158 | −0.039 | 0.209 | −0.900 | |
| miR-320B | P | 0.725 | 0.490 | 0.784 | 0.032∗ |
| r | −0.218 | −0.413 | −0.171 | −0.910 | |
P = probability, r = correlation coefficient, ∗P < 0.05.
Finally, there was no correlation between the cell proliferation and production of extracellular matrix in vitro and the cartilage repair in vivo.
4. Discussion
This study demonstrated that the mRNA expression of MYBL1 was positively correlated with proliferation and cartilage differentiation of hMSCs and that the mRNA expression of RCAN2 and the protein expressions of TIMP-1 and VEGF were negatively correlated with proliferation and cartilage differentiation of MSCs. However, we also showed that MSCs from all five donors had the capacity to promote cartilage repair in vivo regardless of their capacity for proliferation and cartilage differentiation. The mRNA expression of HLA-DRB1 was positively correlated with cartilage repair in vivo, and the mRNA expression of TMEM155 and expressions of miR-486-3p, miR-148b, miR-93, and miR-320b were negatively correlated with cartilage repair.
In vitro, the capacity for chondrogenic differentiation, which is an index of extracellular matrix production, was high in cells with high proliferation, as indicated by CFE and the GR. However, we found that the cell proliferation and the chondrogenic differentiation cannot be used for the quality assessment of the MSCs based on the efficacy of cartilage repair in vivo. Thus, CFE, the gene expressions of MYBL1 and RCAN2, and the protein expressions of TIMP-1 and VEGF can be used for the quality assessment of the MSCs based on the capacity of proliferation and chondrogenic regeneration, but not for the quality assessment of the MSCs based on the efficacy of cartilage repair. Previous studies reported that the tissue regeneration promoted by MSC transplantation might be mediated predominantly through the indirect paracrine mechanisms rather than the direct regeneration from transplanted MSCs [28–31]. Our previous study of intra-articular injection of green fluorescent protein (GFP) expressing rat MSCs into a rat cartilage defect model also showed that GFP-positive cell could be observed at the injured site at four weeks after the treatment but could not be detected in the posttreatment specimens at eight and 12 weeks [5]. This might be the reason for discrepancy between the in vitro chondrogenic capacity of MSCs and the cartilage repair in vivo. On the other hand, the expression levels of HLA-DRB1, TMEM155, miR-486-3p, miR-148b, miR-93, and miR-320b might be used for the quality assessment of MSCs based on the efficacy of cartilage repair. HLA-DRB1 is part of a family of genes called the human leukocyte antigen (HLA) complex that has a critical role in the immune system. HLA-DRB1 was reported to have participated in the pathopoiesis of rheumatoid arthritis [32, 33]. However, the function of HLA-DRB1 in MSCs has not been reported. The function of TMEM in MSCs is also unknown. MiR-93 has been implicated in multiple cell processes, including proliferation, apoptosis, invasion, and extracellular matrix degradation [34–37]. Jing and Jiang reported that miR-93 is lower in human degenerative nucleus pulposus tissues and that its level is associated with disc degeneration grade. In addition, overexpression of miR-93 increases expression of type II collagen by targeting MMP3 and might thereby promote cartilage repair [38]. On the other hand, the functions of miR-486-3p, miR-148b, and miR-320b relating to MSCs or cartilage have not been previously studied. We found expression of specific mRNAs and miRNAs in hMSCs to be related to the capacity for cartilage regeneration. These genes might be used for the quality assessment of hMSCs before their use in treatment for cartilage repair.
In this study, the cartilage repair in vivo after MSC transplantation was incomplete. The xenograft of human MSCs to nude rats might be the reason of insufficient repair of articular cartilage. Another possible reason for insufficient repair of articular cartilage is the use of purchased hMSCs. The commercialized hMSCs were extremely expanded and frozen. This might have an undesirable influence on the quality of hMSCs for cartilage repair. In the next step, the qualities of hMSCs from the patients who take part in clinical trials should be assessed in the same way.
5. Conclusions
The cell proliferation and chondrogenic differentiation of hMSCs in vitro have no correlation with cartilage regeneration in vivo. On the other hand, we found expression of HLA-DRB1, TMEM155, miR-486-3p, miR-148b, miR-93, and miR-320B in hMSCs to be related to the capacity for cartilage regeneration. These factors might be useful for the quality assessment of hMSCs before their use in treatment for cartilage repair.
Acknowledgments
This work was supported by a grant-in-aid for scientific research to Mitsuo Ochi from the Highway Program for Realization of Regenerative Medicine.
Disclosure
An earlier version of this work was presented as a poster at ORS 2017 Annual Meeting, March 19–22, 2017.
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Horwitz E. M., Blanc K. L., Dominici M., et al. Clarification of the nomenclature for MSC: the International Society for Cellular Therapy position statement. Cytotherapy. 2005;7(5):393–395. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 2.Vater C., Kasten P., Stiehler M. Culture media for the differentiation of mesenchymal stromal cells. Acta Biomaterialia. 2011;7(2):463–477. doi: 10.1016/j.actbio.2010.07.037. [DOI] [PubMed] [Google Scholar]
- 3.Chamberlain G., Fox J., Ashton B., Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25(11):2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 4.Niemeyer P., Seckinger A., Simank H. G., Kasten P., Sudkamp N., Krause U. Allogenic transplantation of human mesenchymal stem cells for tissue engineering purposes: an in vitro study. Orthopade. 2004;33(12):1346–1353. doi: 10.1007/s00132-004-0731-3. [DOI] [PubMed] [Google Scholar]
- 5.Nishimori M., Deie M., Kanaya A., Exham H., Adachi N., Ochi M. Repair of chronic osteochondral defects in the rat. A bone marrow-stimulating procedure enhanced by cultured allogenic bone marrow mesenchymal stromal cells. Journal of Bone and Joint Surgery. British Volume (London) 2006;88(9):1236–1244. doi: 10.1302/0301-620X.88B9.17810. [DOI] [PubMed] [Google Scholar]
- 6.Meng Q., Man Z., Dai L., et al. A composite scaffold of MSC affinity peptide-modified demineralized bone matrix particles and chitosan hydrogel for cartilage regeneration. Scientific Reports. 2015;5, article 17802 doi: 10.1038/srep17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein P., Bornhauser M., Gunther K. P., Stiehler M. Bone tissue engineering in clinical application: assessment of the current situation. Orthopade. 2009;38(11):1029–1037. doi: 10.1007/s00132-009-1493-8. [DOI] [PubMed] [Google Scholar]
- 8.Arkesteijn I. T., Smolders L. A., Spillekom S., et al. Effect of coculturing canine notochordal, nucleus pulposus and mesenchymal stromal cells for intervertebral disc regeneration. Arthritis Research & Therapy. 2015;17:p. 60. doi: 10.1186/s13075-015-0569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlic D., Kajstura J., Chimenti S., et al. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10344–10349. doi: 10.1073/pnas.181177898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S. T., Jang J. H., Cheong J. W., et al. Treatment of high-risk acute myelogenous leukaemia by myeloablative chemoradiotherapy followed by co-infusion of T cell-depleted haematopoietic stem cells and culture-expanded marrow mesenchymal stem cells from a related donor with one fully mismatched human leucocyte antigen haplotype. British Journal of Haematology. 2002;118(4):1128–1131. doi: 10.1046/j.1365-2141.2002.03767.x. [DOI] [PubMed] [Google Scholar]
- 11.Urban V. S., Kiss J., Kovacs J., et al. Mesenchymal stem cells cooperate with bone marrow cells in therapy of diabetes. Stem Cells. 2008;26(1):244–253. doi: 10.1634/stemcells.2007-0267. [DOI] [PubMed] [Google Scholar]
- 12.Kamei G., Kobayashi T., Ohkawa S., et al. Articular cartilage repair with magnetic mesenchymal stem cells. The American Journal of Sports Medicine. 2013;41(6):1255–1264. doi: 10.1177/0363546513483270. [DOI] [PubMed] [Google Scholar]
- 13.Harada Y., Nakasa T., Mahmoud E. E., et al. Combination therapy with intra-articular injection of mesenchymal stem cells and articulated joint distraction for repair of a chronic osteochondral defect in the rabbit. Journal of Orthopaedic Research. 2015;33(10):1466–1473. doi: 10.1002/jor.22922. [DOI] [PubMed] [Google Scholar]
- 14.Zaim M., Karaman S., Cetin G., Isik S. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells. Annals of Hematology. 2012;91(8):1175–1186. doi: 10.1007/s00277-012-1438-x. [DOI] [PubMed] [Google Scholar]
- 15.Fossett E., Khan W. S., Pastides P., Adesida A. B. The effects of ageing on proliferation potential, differentiation potential and cell surface characterisation of human mesenchymal stem cells. Current Stem Cell Research & Therapy. 2012;7(4):282–286. doi: 10.2174/157488812800793027. [DOI] [PubMed] [Google Scholar]
- 16.Reppel L., Schiavi J., Charif N., et al. Chondrogenic induction of mesenchymal stromal/stem cells from Wharton’s jelly embedded in alginate hydrogel and without added growth factor: an alternative stem cell source for cartilage tissue engineering. Stem Cell Research & Therapy. 2015;6:p. 260. doi: 10.1186/s13287-015-0263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominici M., Blanc K. L., Mueller I., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 18.Pittenger M. F., Mackay A. M., Beck S. C., et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 19.Segawa Y., Muneta T., Makino H., et al. Mesenchymal stem cells derived from synovium, meniscus, anterior cruciate ligament, and articular chondrocytes share similar gene expression profiles. Journal of Orthopaedic Research. 2009;27(4):435–441. doi: 10.1002/jor.20786. [DOI] [PubMed] [Google Scholar]
- 20.Antebi B., Zhang Z., Wang Y., Lu Z., Chen X. D., Ling J. Stromal-cell-derived extracellular matrix promotes the proliferation and retains the osteogenic differentiation capacity of mesenchymal stem cells on three-dimensional scaffolds. Tissue Engineering. Part C, Methods. 2015;21(2):171–181. doi: 10.1089/ten.TEC.2014.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leferink A. M., Santos D., Karperien M., Truckenmuller R. K., van Blitterswijk C. A., Moroni L. Differentiation capacity and maintenance of differentiated phenotypes of human mesenchymal stromal cells cultured on two distinct types of 3D polymeric scaffolds. Integrative Biology (Camb) 2015;7(12):1574–1586. doi: 10.1039/c5ib00177c. [DOI] [PubMed] [Google Scholar]
- 22.Lindner U., Kramer J., Behrends J., et al. Improved proliferation and differentiation capacity of human mesenchymal stromal cells cultured with basement-membrane extracellular matrix proteins. Cytotherapy. 2010;12(8):992–1005. doi: 10.3109/14653249.2010.510503. [DOI] [PubMed] [Google Scholar]
- 23.Johnstone B., Hering T. M., Caplan A. I., Goldberg V. M., Yoo J. U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Experimental Cell Research. 1998;238(1):265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 24.Sekiya I., Vuoristo J. T., Larson B. L., Prockop D. J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukura Y., Muneta T., Tsuji K., Koga H., Sekiya I. Mesenchymal stem cells in synovial fluid increase after meniscus injury. Clinical Orthopaedics and Related Research. 2014;472(5):1357–1364. doi: 10.1007/s11999-013-3418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koga H., Muneta T., Ju Y. J., et al. Synovial stem cells are regionally specified according to local microenvironments after implantation for cartilage regeneration. Stem Cells. 2007;25(3):689–696. doi: 10.1634/stemcells.2006-0281. [DOI] [PubMed] [Google Scholar]
- 27.Wakitani S., Goto T., Pineda S. J., et al. Mesenchymal cell-based repair of large, full-thickness defects of articular cartilage. The Journal of Bone and Joint Surgery-American Volume. 1994;76(4):579–592. doi: 10.2106/00004623-199404000-00013. [DOI] [PubMed] [Google Scholar]
- 28.Psaltis P. J., Zannettino A. C., Worthley S. G., Gronthos S. Concise review: mesenchymal stromal cells: potential for cardiovascular repair. Stem Cells. 2008;26(9):2201–2210. doi: 10.1634/stemcells.2008-0428. [DOI] [PubMed] [Google Scholar]
- 29.Buxton A. N., Bahney C. S., Yoo J. U., Johnstone B. Temporal exposure to chondrogenic factors modulates human mesenchymal stem cell chondrogenesis in hydrogels. Tissue Engineering Part A. 2011;17(3-4):371–380. doi: 10.1089/ten.TEA.2009.0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monsarrat P., Vergnes J. N., Nabet C., et al. Concise review: mesenchymal stromal cells used for periodontal regeneration: a systematic review. Stem Cells Translational Medicine. 2014;3(6):768–774. doi: 10.5966/sctm.2013-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang X., Ding Y., Zhang Y., Tse H. F., Lian Q. Paracrine mechanisms of mesenchymal stem cell-based therapy: current status and perspectives. Cell Transplantation. 2014;23(9):1045–1059. doi: 10.3727/096368913X667709. [DOI] [PubMed] [Google Scholar]
- 32.Nepom G. T., Byers P., Seyfried C., et al. HLA genes associated with rheumatoid arthritis. Identification of susceptibility alleles using specific oligonucleotide probes. Arthritis and Rheumatism. 1989;32(1):15–21. doi: 10.1002/anr.1780320104. [DOI] [PubMed] [Google Scholar]
- 33.Firestein G. S. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 34.Fang L., Deng Z., Shatseva T., et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene. 2011;30(7):806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 35.Yu X. F., Zou J., Bao Z. J., Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World Journal of Gastroenterology. 2011;17(42):4711–4717. doi: 10.3748/wjg.v17.i42.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du L., Zhao Z., Ma X., et al. miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;33(34):4307–4315. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., He X. X., Chang Y., Sun S. Z., Xu C. R., Lin J. S. Downregulation of MiR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepato-Gastroenterology. 2012;59(120):2367–2373. doi: 10.5754/hge12458. [DOI] [PubMed] [Google Scholar]
- 38.Jing W., Jiang W. MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells. Cell Proliferation. 2015;48(3):284–292. doi: 10.1111/cpr.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]