Abstract
Finding a good predictive model for a high-dimensional data set can be challenging. For genetic data, it is not only important to find a model with high predictive accuracy, but it is also important that this model uses only few features and that the selection of these features is stable. This is because, in bioinformatics, the models are used not only for prediction but also for drawing biological conclusions which makes the interpretability and reliability of the model crucial. We suggest using three target criteria when fitting a predictive model to a high-dimensional data set: the classification accuracy, the stability of the feature selection, and the number of chosen features. As it is unclear which measure is best for evaluating the stability, we first compare a variety of stability measures. We conclude that the Pearson correlation has the best theoretical and empirical properties. Also, we find that for the stability assessment behaviour it is most important that a measure contains a correction for chance or large numbers of chosen features. Then, we analyse Pareto fronts and conclude that it is possible to find models with a stable selection of few features without losing much predictive accuracy.
1. Introduction
In many applications of bioinformatics, the goal is to find a good predictive model for high-dimensional data. To avoid overfitting and to discover the relevant features, feature selection should be integrated into the model fitting process [1]. The feature selection should be stable; that is, the sets of chosen features should be similar for similar data sets, as an unstable feature selection would question the reliability of the results [2].
Over the past decade, a variety of frameworks for stability evaluation have been proposed. Overviews of existing stability measures are given in [3, 4]. The theoretical properties of different measures of stability are studied in [5]. Pitfalls with respect to interpreting the values of stability measures are discussed in [6] and experimental setups for stability evaluation are presented in [7]. Ensemble methods for making feature selection more stable than a single feature selection method are proposed in [8–10]. The research that has been done in all of the aforementioned aspects of stability assessment is reviewed in [11] and various feature selection methods including ensemble methods are analysed in [12–18]. It is shown that conducting a stable feature selection before fitting a classification model can increase the predictive performance of the model [19]. Most of these works consider both high stability and high predictive accuracy of the resulting classification model as target criteria but do not consider the number of selected features as a third target criterion.
In this paper, we pursue two goals. Firstly, we compare a variety of stability measures empirically. We aim at finding out which of the measures assess the stability similarly in practical applications. Also, we aim at choosing stability measures that are suitable for finding desirable models for a given data set. Secondly, we suggest a strategy for finding a desirable model for a given data set with respect to the following criteria:
The predictive accuracy must be high.
The feature selection must be stable.
Only a small number of features must be chosen.
The predictive power of a predictive model is obviously important and is usually the only criterion considered in model selection. However, when trying to discover relevant features, for example, to understand the underlying biological process, it is also necessary to keep the set of selected features both small and stable. To reach all three targets simultaneously, we combine feature selection and classification methods. For these “augmented” methods, we measure the three target criteria jointly during hyperparameter tuning and we choose configurations which perform well considering all three target criteria.
The rest of the paper is organised as follows. In Section 2, we describe the measures of stability, filter methods, and classification methods which are considered in this paper. In Section 3, the data sets used in our experiments are presented. Section 4 contains the empirical comparison of stability measures. Section 5 covers our second experiment, where we search for desirable configurations with respect to the three target criteria explained above. Section 6 summarizes the conclusions of our work.
2. Methods
In this section, we explain different measures of stability, filter methods for feature selection and classification methods. We also describe the concept of Pareto optimality.
2.1. Measures of Stability
We use the following notation: assume that there is a data set containing n observations of the p features X1,…, Xp. Resampling is used to split the data set into m subsets. The feature selection method is then applied to each of the m subsets. Let Vi ⊂ {X1,…, Xp}, i = 1,…, m, denote the set of chosen features for the i-th subset of the data set and let |Vi| be the cardinality of this set.
2.1.1. Intersection Based Stability Measures
The following intersection based stability measures consider a feature selection to be stable if the cardinalities of all pairwise intersections |Vi∩Vj| are high. The measures standardise the cardinalities of the intersections in different ways. Three simple stability measures based on stability indices are defined as
Extending SJ in a way that different but highly correlated variables count towards stability gives the stability measure:
- Zucknick et al. [23]:
(4) - with
(5)
where Cor(x, y) is the Pearson correlation between x and y. 𝕀A denotes the indicator function for a set A, and
| (6) |
The idea of a stability measure that is corrected for chance was first proposed in [27]. The reason for a correction for chance is that |Vi∩Vj| necessarily becomes large if |Vi| and |Vj| are large. The idea is made applicable in situations in which the numbers of chosen features vary:
- Lustgarten et al. [24]:
(7)
2.1.2. Frequency Based Stability Measures
Let hj, j = 1,…, p, denote the number of sets Vi that contain feature Xj so that hj is the absolute frequency with which feature Xj is chosen. Frequency based stability measures evaluate such situations as stable in which the features are chosen for either most of the subsets or not at all. The entropy-based measure of stability relies on hj and is given by
Novovicová et al. [25]:
| (8) |
with q = ∑j=1phj and V = ⋃i=1mVi.
- Davis et al. [13]:
(9)
with α ≥ 0 and V like before is a stability measure, where the minuend rewards frequent choices of variables, while the subtrahend penalises large sets of chosen features.
The relative weighted consistency is defined as
- Somol and Novovicová [26]:
(10) - with
(11)
and q is like before. Calculating (hj − 1)/(m − 1) scales the positive absolute frequencies to [0,1]. All scaled frequencies with hj > 0 are assigned the weight hj/q. The correction terms cmin and cmax cause the measure to lie within the range [0,1]. As the correction terms depend on q, this measure contains a correction for chance.
2.1.3. Correlation
The Pearson correlation can be used as a stability measure. To do so, Nogueira and Brown [5] define a vector zi ∈ {0,1}p for each set of selected features Vi to indicate which features are chosen. The j-th component of zi is equal to 1 if Vi contains Xj; that is, zij = 𝕀Vi(Xj), j = 1,…, p. The resulting stability measure is
- Correlation [5]:
(12)
with Cor(zi, zj) denoting the Pearson correlation between zi and zj. The Pearson correlation measures the linear association between continuous variables. When applied to binary data like the vectors z1,…, zm, the Pearson correlation is equivalent to the phi coefficient for the contingency table of each two of these vectors.
2.1.4. Theoretical Properties
Nogueira and Brown [5] define four properties which are desirable for stability measures:
Fully defined (fulfilled if the measure does not require the cardinalities |V1|,…, |Vm| to be identical)
Upper/lower bounds (fulfilled if the upper and lower bounds of a measure are both finite)
Maximum (fulfilled if a deterministic selection of the same k features achieves the maximum value and if the maximum value is only achieved by a deterministic selection)
Correction for chance (fulfilled if the expected value of the stability measure for a random feature selection is constant, that is, does not depend on the number of chosen features). When features are chosen entirely at random, uncorrected measures usually attain the higher values the more features are selected.
For SJ, SD, SL, SS, and SC, these properties are analysed in [5]. We report these results in Table 1 and add the results for SO, SZ, SD-α, and SN. Additionally, the theoretical ranges of the stability measures are given in Table 1. High values indicate high stability and low values indicate low stability for all measures. Note that the upper bound for SD-α depends on α.
Table 1.
Theoretical properties proposed in [5] and theoretical ranges of the stability measures.
| SJ | SD | SO | SZ | SL | SD-α | SN | SS | SC | |
|---|---|---|---|---|---|---|---|---|---|
| Jaccard [20] | Dice [21] | Ochiai [22] | Zucknick et al. [23] | Lustgarten et al. [24] | Davis et al. [13] | Novovicová et al. [25] | Somol and Novovicová [26] | Correlation [5] | |
| Fully defined | × | × | × | × | × | × | × | × | × |
| Bounds | × | × | × | × | × | × | × | × | × |
| Maximum | × | × | × | × | — | — | × | — | × |
| Correction for chance | — | — | — | — | × | — | — | × | × |
| Range | [0,1] | [0,1] | [0,1] | [0,1] | [−1,1] | [0,1] | [0,1] | [−1,1] |
SO, SZ, SD-α, and SN are fully defined and have finite bounds. SO, SZ, and SN fulfil the maximum property. Note that for each two sets Vi and Vj the value of cij + dij is always smaller than |Vi∖Vj| + |Vj∖Vi|. SD-α fulfils the maximum property only for α = 0. A deterministic feature selection of k > 1 features only reaches a value of 1 − k · (α/p). None of the four measures is corrected for chance. SD-α with α > 0 does contain a correction for large numbers of chosen features but this is not a correction for chance as the expected value for a random feature selection still depends on the number of chosen features. If V1,…, Vm are randomly chosen features sets of size k, then the expected value of SD-α is E(SD-α) = k · E(1/|V|) − k · (α/p) because ∑j=1phj = mk and median(|V1|,…, |Vm|) = k in this scenario. So, E(SD-α) could only be some function f(m, p) which does not depend on k if E(1/|V|) = (1/k) · f(m, p) + α/p. The latter is not possible as E(1/|V|) cannot include a term depending on α.
2.2. Filter Methods
Filter methods select a subset of the features {X1,…, Xp}. For each feature, a score is calculated and then the best features, that is, those with the highest scores, are chosen. One can specify either the number of features to be selected or a threshold and choose all features whose scores exceed the threshold. For the filter methods explained below, all features need to be scaled metrically.
2.2.1. Variance Filter
For each feature, its variance is calculated and used as a score. High variance therefore means a high score.
2.2.2. AUC Filter
The score of the AUC filter represents the classification accuracy when each feature is used directly and separately for target value prediction. For each feature Xi, we use the following prediction rule for the target variable Y: , i = 1,…, p, with 𝕀 denoting the indicator function. The Receiver Operating Curve displays the sensitivity and specificity of a classification rule for all choices of a threshold c; see [28]. We use the area under the Receiver Operating Curve (AUC) of the classification rule to measure how well each feature separates the target variable. An AUC value of 1 means that there is a threshold c for which the prediction rule is perfectly accurate. An AUC value of 0 means that there is a threshold c for which the rule predicts all labels wrongly which implies that Xi can achieve perfect classification with the rule . A value of 0.5 is the worst possible in this application. We therefore use |0.5 − AUC| as the AUC filter score.
2.2.3. MRMR Filter
The idea of the maximum relevance minimum redundancy (MRMR) filter is to include the most relevant features for class prediction while making sure that no redundant features are chosen [29]. The MRMR filter is an iterative procedure which chooses the features one after another in a greedy forward fashion. In each step, the feature that maximizes the quotient (relevance/redundancy) among all features which have not been chosen at that point is selected. We use the score of our AUC filter as a measure of relevance for each feature. To quantify the redundancy of a feature for a given set of already chosen features, we sum the absolute Pearson correlations of that feature with all features in the set. If selecting a given number of features, there is no need to calculate the filter score (relevance/redundancy) for all features because the score decreases monotonically with the number of iterations. Therefore, the first score values are the highest.
2.3. Classification Methods
Classification methods are well understood. In this work, we use the following classification methods: GLM Boosting [30, 31], Lasso Logistic Regression [32], Random Forest [33], and Support Vector Machine (SVM) [33]. Note that Lasso Logistic Regression, GLM Boosting, and Random Forest conduct an embedded feature selection, while SVMs use all features. The features selected by Lasso Logistic Regression are the ones whose corresponding regression parameters are not equal to 0. GLM Boosting models are weighted sums of base learners and each base learner uses only one feature. The number of boosting iterations limits the upper number of base learners and thereby the upper number of features that can be part of the classification rule. So, the feature selection is performed by the selection of the corresponding base learners in the boosting update iterations. The features that are included in a Random Forest model can be assessed by checking which features are assigned variable importance values greater than 0.
2.4. Terminology
In this paper, we use the term “model” when talking about a classification rule which is already fitted to the data and we use the term “method” when referring to a classification or filter method. Combinations of filter and classification methods where a filter method is applied first and a classification rule is learned on the remaining features in a second step are called “augmented methods.” To talk about augmented methods with fixed hyperparameter values, we use the term “configuration.”
2.5. Pareto Optimality
Let M be some finite set and let f : M → ℝt be an objective function to minimize. Note that each maximization problem can be transformed into a minimization problem by multiplication with −1. If t = 1 holds, all points in the image f(M) = {y ∈ ℝt : ∃x ∈ M with f(x) = y} are comparable and therefore f(M) has a distinct minimum. However, when t ≥ 2 holds, some of the elements of f(M) may not be comparable: they may be smaller in one component and larger in another one. The set f(M) thus does not necessarily have a distinct minimum. Instead, there will likely be a set of incomparable minimal points which is called the Pareto front. A point y ∈ ℝt Pareto dominates another point z ∈ ℝt if ∀i = 1,…, t : yi ≤ zi and ∃j ∈ {1,…, t} : yj < zj. The Pareto front is the subset of f(M) which contains no dominated points: For a more detailed introduction in Pareto optimality, see [34].
3. Data Sets
In our analyses, we use three data sets. Two data sets contain microarray data and the other one contains RNASeq data. The two microarray data sets, AP_Breast_Ovary and AP_Colon_Kidney, contain the same features but compare different types of cancer. The data sets have been used for stability analysis [35]. They are available on the online platform OpenML [36], data IDs 1165 and 1137. Before conducting our experiments, we have removed the ID column in both data sets.
The RNASeq data set, Stomach, is created from supplementary material of [37]. We use the level 4 data matrix about RNA Expression from IlluminaGA RNASeq and IlluminaHiseq RNASeq. We consider only patients with fundus ventriculi (C 16.1) and antrum pyloricum (C 16.3) because these two cancer types form the largest two classes. To normalise the features, we transform all values of x into log(x + 1). The original data set consists of 29,699 features which makes it too large for an analysis using our framework on our high performance computing cluster. For feasibility reasons, we have to reduce its number of features. As most of the features have little variation, we prefilter the original data set by keeping only the 10,000 features with the largest variances.
Information about the dimensions of the three preprocessed data sets is shown in Table 2. AP_Breast_Ovary and AP_Colon_Kidney contain more observations than Stomach. The class sizes in AP_Colon_Kidney and Stomach are roughly balanced; the ones in AP_Breast_Ovary are not. Figures 1, 2, and 3 show PCA plots of the data sets. It seems that the classes in AP_Colon_Kidney and AP_Breast_Ovary are easier to separate than the ones in Stomach. However, one should note that only up to 25.62% of the data variation is explained by the respective first two principal components.
Table 2.
Information about the data sets: number of features and observations in each data set and sizes of the two classes for each data set.
| Data set | # features | # observations | # class 0 | # class 1 |
|---|---|---|---|---|
| AP_Breast_Ovary | 10,935 | 542 | 344 | 198 |
| AP_Colon_Kidney | 10,935 | 546 | 286 | 260 |
| Stomach | 10,000 | 215 | 110 | 105 |
Figure 1.
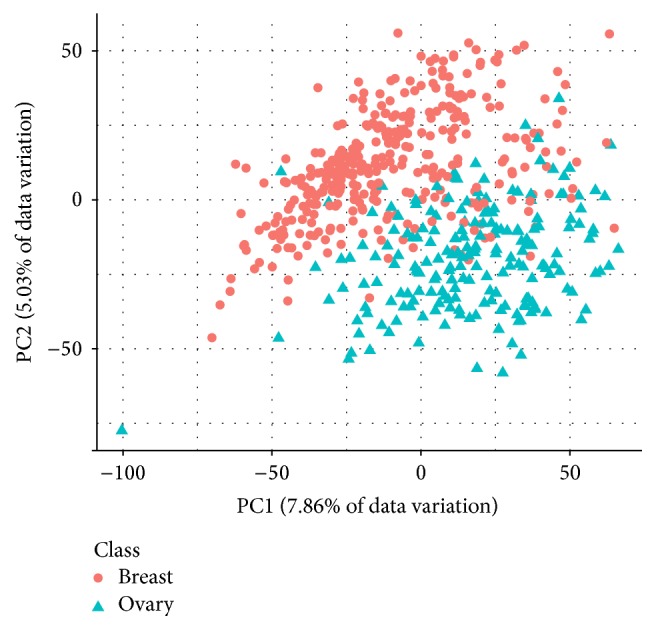
PCA plot of the data set AP_Breast_Ovary. The first two principal components explain 12.89% of the data variation.
Figure 2.

PCA plot of the data set AP_Colon_Kidney. The first two principal components explain 23.89% of the data variation.
Figure 3.

PCA plot of the data set Stomach. The first two principal components explain 25.62% of the data variation.
4. Empirical Comparison of Stability Measures
In this section, we compare the stability measures given in Section 2.1 empirically. We analyse the stability assessment behaviour of the stability measures, finding groups of similar measures. We investigate the impact of the number of chosen features on the stability measures. Furthermore, we compare the stability measures regarding their Pareto optimal configurations with respect to maximal accuracy, stability, and sparsity. Based on our observations, we conclude which stability measures are most suitable for stability analysis. The results of this study are used to select a subset of the stability measures for our study in Section 5.
4.1. Experimental Setup
We fit “augmented” methods to the three data sets and evaluate the performances of the resulting models. Augmented methods combine filter and classification methods; that is, the filter is applied first and the classification rule is learned on the remaining features. Note that, for classification methods which perform an embedded feature selection, this results in a cascading feature selection process. We combine each filter method from Section 2.2 with each classification method from Section 2.3. For each of the resulting 12 augmented methods, we choose 1,000 hyperparameter configurations which leaves us with 12,000 configurations to be analysed per data set. We draw the values for the hyperparameters randomly and independently from the sets given in Table 3. The values for the hyperparameters λ, σ, and C are drawn by randomly selecting x ∈ [−15,15] and then calculating 2x. Note that setting n.feats to all (i.e., 10,935 for data sets AP_Breast_Ovary and AP_Colon_Kidney or 10,000 for data set Stomach) is equivalent to applying the classification method without the filter method.
Table 3.
Hyperparameter ranges for all methods.
| Method | Parameter | Range |
|---|---|---|
| Filter | n.feats | {1,2,…, all} |
| GLM Boosting | m stop | {1,2,…, 215} |
| Lasso Log. Reg. | λ | {2x : x ∈ [−15,15]} |
| Random Forest | num.trees | {1,2,…, 215} |
| Random Forest | min.node.size | {1,2,…, 25} |
| SVM | σ | {2x : x ∈ [−15,15]} |
| SVM | C | {2x : x ∈ [−15,15]} |
The effect of the hyperparameters on the classification performance or the feature selection stability depends on the data. Therefore, we only discuss the effect of the hyper parameters on the sparsity of the resulting models. All filter methods have only one hyperparameter named n.feats specifying how many features should be selected. A small value of n.feats will result in a sparse model. For GLM Boosting, we have one hyperparameter named mstop which denotes the number of boosting iterations. A small value of mstop will result in a sparse model. A large value of mstop can lead to a large model. Note that the same base learner can be used in multiple iterations. As each base learner is a linear model using only one feature, this means that a large value of mstop can also result in a sparse model. The Lasso Logistic Regression has the hyperparameter λ which determines the trade-off between likelihood minimization and regression parameter minimization. A large value of λ will force the L1 norm of the regression parameter to be small which will be achieved by many components of the regression parameter being equal to 0. A large value of λ therefore will result in a sparse model. For Random Forest, we vary two hyperparameters: num.trees is the number of classification trees in the forest and min.node.size is the minimum number of observations in terminal nodes. A large value of min.node.size will cause the classification trees to be small. A very small value of num.trees and a large value of min.node.size therefore will result in a sparse model. Furthermore, we fit SVM with RBF kernel with kernel width parameter σ and regularisation parameter C. As SVM does not perform embedded feature selection, none of the hyperparameter configurations will lead to a sparse model.
For configuration evaluation, we perform 10-fold cross validation; that is, we fit 10 models, each based on 90% of the observations. For each model, we predict the class on the 10% of the observations that were not used for fitting and we calculate the misclassification rate. Additionally, for each model, we determine the set of chosen features by the combined feature selection of the filter method and the embedded feature selection of the classification method. To evaluate the predictive performance of the configuration, we calculate the mean value of the 10 misclassification rates (mean misclassification rate). To assess the mean size of the models, we determine the mean value of the cardinalities of the sets of chosen features (mean number of chosen features). We evaluate the stability of the configuration based on the 10 features sets obtained from the 10 models. We use all stability measures defined in Section 2.1. For SD-α, we employ 0, 1, 2, and 10 as values for α.
Performing 10-fold cross validation means setting m in Section 2.1 to 10. The advantage of cross validation is that we know that the training data sets in each two iterations are very similar: they share 80/90 of their observations. See [6] for details about stability and data similarity. The size of the subsamples influences the stability values [35]. But since the stability values allow only a comparison of configuration on the same data set and do not permit a data-independent conclusion [6], we can choose an arbitrary value of m here.
4.2. Software
For our studies, we use R version 3.3.1 [38]. The package mlr [39] provides the machine learning framework, and batchtools [40] is used to roll out the experiments on a high performance computing cluster. For the classification and filter methods, we additionally rely on the R packages fmrmr [41], kernlab [42], LiblineaR [43], mboost [44], ranger [45], and ROCR [46].
4.3. Results and Discussion
We compare the measures of stability empirically on the data set AP_Colon_Kidney. For the two other data sets, we have obtained very similar results which lead to the same conclusions.
4.3.1. Overview
We compare the measures of stability given in Section 2.1 empirically. Figure 4 displays the values of all considered stability measures for all 12,000 configurations. This general overview shows that most stability measures take on values between 0 and 1, only SC and SL take on some barely negative values. Keep in mind that SC and SL have the theoretical range [−1,1], while the other theoretical ranges are [0,1] or [0,1 − α/10935]; see Table 1. The values of the stability measures differ a lot in location and dispersion. For the given configurations, SN has the largest and SD-10 hast the smallest median value among all considered measures of stability. SD-0 has the largest interquartile range, followed by SJ. SL has the smallest interquartile range.
Figure 4.

Boxplots of the values of all considered stability measures for all 12,000 configurations. A small value indicates low stability and a large value indicates high stability.
4.3.2. Similarity of Stability Measures
We are interested in whether the different measures of stability consider the same configurations to be stable or if some measures evaluate a configuration as stable and others as unstable. Among the 12,000 configurations that we have analysed, there are both stable and unstable ones and we therefore assume that each measure assigns its maximum value to a very stable configuration and its minimum value to a very unstable configuration.
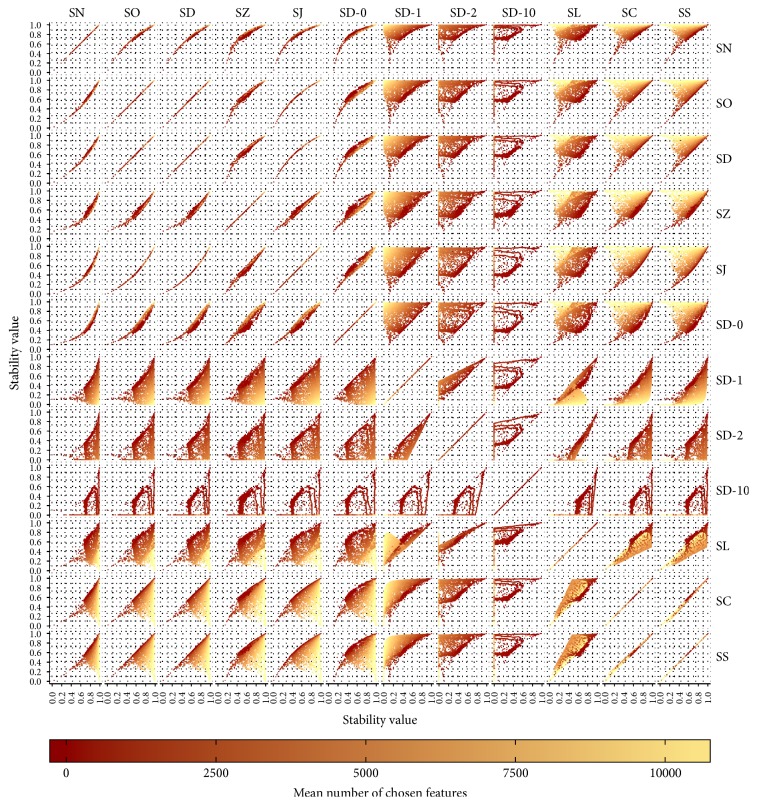
Figure 5 shows scatter plots for all pairs of stability measures. In all scatter plots, each of the 12,000 configurations is represented as one dot. The colour of the dot represents the mean number of chosen features. For SN, SO, SD, SZ, SJ, and SD-0, all dots lie close to a line or curve. This means that these stability measures evaluate the stability of all configurations very similarly, independent of the mean size of the fitted models. This group consists of all the stability measures which are not corrected for chance. SD-1 and SD-2 are similar to each other considering their stability evaluation behaviour. However, their similarity is not as strong as the similarity among the aforementioned group of uncorrected measures. SD-10 is not similar to any other of the considered stability measures. SC and SS evaluate the stability very similarly. SL is similar to both of them but not as similar as SC and SS are to each other. Except for SC and SS, the groups of corrected measures are more heterogeneous than the group of uncorrected measures.
Figure 5.
Scatter plot for all pairs of stability measures based on all 12,000 configurations. The order of the stability measures allows identifying groups of similar measures easily.
Figure 5 not only allows finding groups of measures which assess the stability of the configurations similarly but also demonstrates that the stability of configurations which lead to small models is evaluated very similarly by all stability measures. For SD-10, this only applies to very small models. Keep in mind that this measure penalises large numbers of chosen features very much. The differences in stability assessment behaviour only exist for configurations which lead to larger models. For larger models, the uncorrected measures assign higher stability values than the corrected measures. While it was expected that the stability measures behave differently for large models, it was not clear that they would all behave so similarly for small models.
4.3.3. Connection to Number of Chosen Features
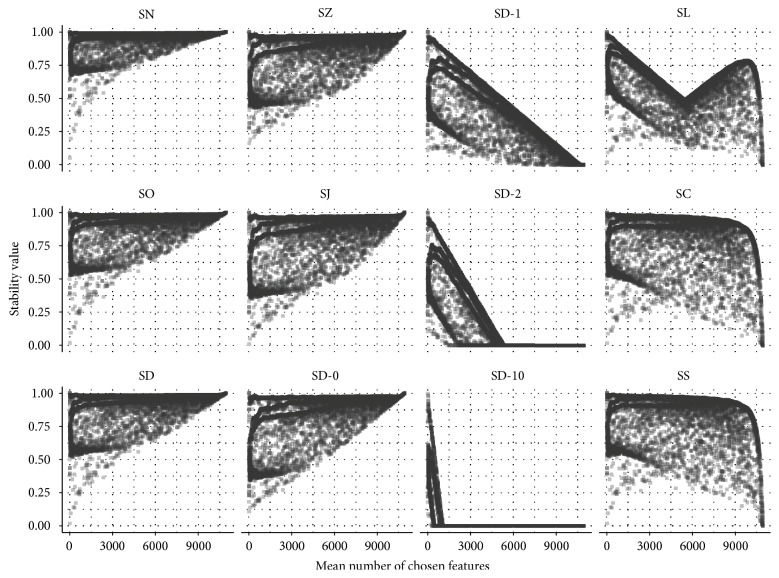
In the previous paragraph, we have observed that the similarity of the stability measures depends on the sizes of the fitted models. Now we analyse the dependence of each stability measure on the mean number of chosen features. Figure 6 displays the stability values and the mean number of chosen features for all configurations. Each plot shows the stability values assessed by one stability measure.
Figure 6.
Scatter plot of the stability values and mean number of chosen features for all 12,000 configurations.
For all stability measures, it is possible to take on small or large values if the mean sizes of the resulting models of the configurations are small. The uncorrected measures SN, SO, SD, SZ, SJ, and SD-0 assign the higher stability values the larger the mean model sizes of the configurations are. For the measures SD-1, SD-2, and SD-10, it is the other way round: they assign the lower stability values the larger the mean model sizes of the configurations are. SD-2 and SD-10 constantly give a stability value of 0 if the mean number of chosen features is large. For SC and SS, the assessed stability values also decrease for increasing mean number of chosen features. This decrease is not linear like for SD-1, SD-2, and SD-10. Instead, it allows high stability values to be assigned for most mean model sizes. Only for very large models, none of the corresponding configurations is given high stability values. For SL, the maximally achieved stability values first decrease and then increase again with the mean number of chosen features. This property is due to the max term in the denominator in the definition of SL and has been discussed in [5].
4.3.4. Comparison of Pareto Optimal Configurations
As we have stated in Section 1, we think that for many domains desirable models should have high classification accuracy, high stability, and a small number of chosen features. Therefore, it is of interest whether the use of different stability measures leads to different Pareto optimal configurations with respect to the three criteria. For each stability measure, we assess the Pareto optimal configurations. The results of this analysis are displayed in Figure 7. There are some configurations that are Pareto optimal for all stability measures. However, most of the configurations in Figure 7 are just Pareto optimal for some stability measures. This means that the set of Pareto optimal configurations depends on the selected stability measure. The groups of similar stability measures which we have identified by analysing Figure 5 also lead to roughly similar sets of Pareto optimal configurations here.
Figure 7.
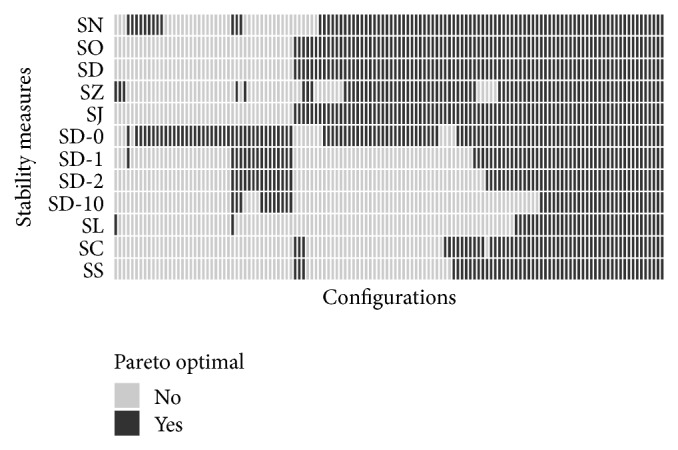
Pareto optimal configurations per stability measure. The colors indicate whether a configuration is Pareto optimal considering classification performance, mean size of chosen features, and stability assessed by one of the 12 different stability measures. Only the 132 configurations which are Pareto optimal for at least one stability measure are displayed here.
4.3.5. Empirical Properties
To summarize the empirical properties of the stability measures which we have found out in this section, we display the properties which we consider to be advantageous or disadvantageous in Table 4. We think that a large empirical spread is important because this allows distinguishing stable from unstable configurations easily. With “overall spread,” we refer to the spreads which we have observed in Figure 4. The statements about the spreads for different model sizes are taken from the analysis of Figure 6. The empirical ranges of the stability measures are comparable because most theoretical ranges are [0,1] and the measures with theoretical ranges [−1,1] barely attain any values below 0 in our empirical analysis. For a comparison of the theoretical properties of stability, see [5].
Table 4.
Advantages and disadvantages of all stability measures.
| Stability measure | Advantages | Disadvantages |
|---|---|---|
| SN | — | Small spread for large models |
| SO | — | Small spread for large models |
| SD | — | Small spread for large models |
| SZ | — | Small spread for large models |
| SJ | Large overall spread | Small spread for large models |
| SD-0 | Large overall spread | Small spread for large models |
| SD-α, α > 0 | — | The value of α is arbitrary but has a strong impact |
| SL | — | Medium sized models cannot achieve as high stability values as small or large models can |
| SC | Large spread for most model sizes | — |
| SS | Large spread for most model sizes | — |
Based on their empirical properties, SC and SS are the most desirable stability measures. SC is the only measure which fulfils all the theoretical properties proposed in [5] and displayed in Table 1. Based on both the theoretical and the practical aspects, we think that SC is the most suitable stability measure. If an uncorrected measure is desired, we think that SJ is a good choice because it has the largest overall spread among the uncorrected measures which fulfil all theoretical properties except for the correction for chance.
5. Finding Desirable Configurations
In this section, we propose a strategy for finding desirable configurations. We analyse Pareto fronts looking for configurations with a stable selection of few features without losing much predictive accuracy compared to model fitting only based on predictive performance. In contrast to Section 4, the focus of this section is not to compare the stability measures but to analyse the proposed strategy for finding desirable configurations.
5.1. Experimental Setup
In this study, we conduct a random search for configurations which lead to sparse and stable models with high prediction accuracy. We use the three data sets presented in Section 3 and the same augmented methods and software as in Section 4. Like in Section 4, for each of the 12 augmented methods, we analyse 1,000 hyperparameter configurations which we determine at random from the sets given in Table 3. This gives us 12,000 configurations per data set.
We split each data set in two halves. We use the first half (training data) to find desirable configurations. To do so, we evaluate the 12,000 configurations on the training data. We perform 10-fold cross validation to determine the mean misclassification rate, the mean number of chosen features, and the values of the stability measures SJ, SD-1, SD-10, and SC (see Section 4.1). For each stability measure, we choose the best configurations with respect to predictive performance, sparsity, and stability. Then we evaluate these configurations on the other half (testing data). For the evaluation on the testing data, we also perform 10-fold cross validation and determine the mean misclassification rate, the mean number of chosen features, and the value of the respective stability measure.
Evaluating the chosen configurations on data which has not been used for choosing the configurations allows us to assess unbiased estimates for the three target criteria. It is necessary to perform resampling on both halves of the data in order to be able to evaluate the stability on both halves. By using this procedure, the number of observations is roughly equal for all model fits on both training and testing data.
We choose the four stability measures SJ, SD-1, SD-10, and SC as representatives of the four groups of measures identified in Section 4.3. SJ is not corrected; the other three measures are corrected either for chance or for large numbers of chosen features. The results for all stability measures are available as Supplementary Material available online at https://doi.org/10.1155/2017/7907163.
5.2. Results and Discussion
We propose considering the predictive accuracy, the stability, and the number of chosen features jointly when searching for desirable configurations. Based on three data sets, we will show that it is possible to find configurations that perform a stable selection of few features without losing much predictive accuracy compared to model fitting only considering the predictive performance. For visualisation reasons in this publication, we do not analyse all configurations which are Pareto optimal considering all three criteria. Instead, we focus on the stability and sparsity only taking into account the best configurations accuracy-wise. As we are looking for a predictive model, the predictive performance can be considered the most important criterion. Therefore, we consider only those configurations whose mean misclassification rate on the training data does not exceed c∗ + 0.05. c∗ denotes the best mean misclassification rate on the training data achieved by any of the configurations on the same data set. Keep in mind that our results will not necessarily be optimal with respect to all three target criteria because we possibly sacrifice some accuracy in favour of increased stability and sparsity. Throughout this section, we transform the stability values into “1 − stability,” so that all target criteria are to be minimized.
Figure 8 gives an overview of the configurations fitted on data set AP_Breast_Ovary whose mean misclassification rate does not exceed the c∗ + 0.05 threshold. The mean number of chosen features and the stability values (both on the training data) are displayed. The colours indicate the augmented methods used for model fitting. We can see how the different augmented methods generally perform in comparison to each other on this data set. GLM Boosting combined with any of the three filters always leads to sparse models. The mean sizes of the models for the other three classification methods vary a lot. For all classification methods, the most stable models are fitted using the variance filter.
Figure 8.
Mean number of chosen features and stability of the 6,997 configurations for data set AP_Breast_Ovary whose mean misclassification rate is within 0.05 of the best classification accuracy.
To find desirable configurations for the three data sets AP_Breast_Ovary, AP_Colon_Kindey, and Stomach, we analyse the Pareto optimal configurations with respect to their stability and size which do not exceed the c∗ + 0.05 threshold. We compare their performances to the performances of the configurations with the best classification accuracy. The latter are the configurations chosen if one does not take into account the stability or the size.
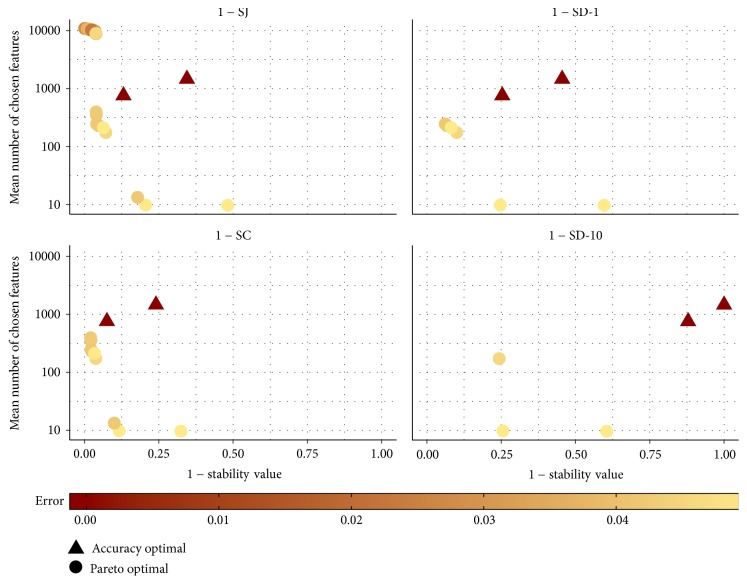
Figure 9 displays the Pareto front for the Pareto optimal configurations with respect to their stability and size (mean number of chosen features) for data set AP_Breast_Ovary. Additionally, the best configurations considering only the predictive accuracy are shown. They are marked by triangles. The colour represents the error (mean misclassification rate) of the respective configuration. The plots are based on the training data. Note that the ordinate is scaled logarithmically.
Figure 9.
Pareto front for the Pareto optimal configurations for data set AP_Breast_Ovary considering the mean number of chosen features and the stability value. Only configurations whose mean misclassification rate is within 0.05 of the best classification accuracy are taken into account. Additionally, the accuracy optimal configurations are shown.
Only if SJ is used as stability measure are configurations which lead to large models considered Pareto optimal. This is because SJ is the only uncorrected measure and hence assigns high stability values if almost all features are chosen. The accuracy optimal configurations possess a perfect predictive behaviour on the training data but they use many features and their stability is smaller than the stability of many Pareto optimal configurations.
The Pareto optimal configurations which result in models using less than 500 features on the training data as well as the accuracy optimal configurations are shown in Tables 5 and 6. The 20 Pareto optimal configurations whose models in fact averagely use more than 8,800 features are omitted. The tables state the configurations and their performances on the training and testing data. The performances on the training data permit identifying the configurations in Figure 9. The performances on the testing data are used to evaluate the configurations unbiasedly. SJ leads to the highest number of Pareto optimal configurations. All the configurations which are Pareto optimal for SJ and use less than 500 features are Pareto optimal for SC, too. Only some of the configurations that are Pareto optimal for SC are also Pareto optimal for SD-1 or SD-10. For all Pareto optimal configurations, the predictive performance, mean model size, and stability on the testing data are very similar to the values achieved on the training data. For the accuracy optimal configurations, the predictive performance on the testing data is noticeably worse than that on the training data. The accuracy optimal configurations overfit the training data much more than the Pareto optimal configurations. On the testing data, the predictive performance of the accuracy optimal configurations is only as good as the one of the Pareto optimal configurations. Most Pareto optimal configurations possess higher stability on the testing data than the accuracy optimal configurations considering all four stability measures. The configurations which are Pareto optimal for the three corrected measures lead to averagely smaller models on the testing data than the accuracy optimal configurations.
Table 5.
Pareto optimal configurations in Figure 9 whose resulting models averagely use less than 500 features (above the horizontal line) and accuracy optimal configurations (below the horizontal line). The ID column references Table 6.
| ID | Classifier | Parameters | Filter | n.feats | Error | Error | Size | Size |
|---|---|---|---|---|---|---|---|---|
| (train) | (test) | (train) | (test) | |||||
| 1 | Random Forest | num.trees = 20577, | Variance | 397 | 0.041 | 0.048 | 397.0 | 397.0 |
| min.node.size = 2 | ||||||||
| 2 | SVM | C = 81.668, σ = 0.0001 | Variance | 355 | 0.040 | 0.044 | 355.0 | 355.0 |
| 3 | Lasso Log. Reg. | λ = 2.060 | Variance | 246 | 0.041 | 0.059 | 246.0 | 245.9 |
| 4 | Lasso Log. Reg. | λ = 0.133 | Variance | 232 | 0.037 | 0.044 | 232.0 | 232.0 |
| 5 | Random Forest | num.trees = 4742, | Variance | 212 | 0.048 | 0.044 | 212.0 | 212.0 |
| min.node.size = 2 | ||||||||
| 6 | Lasso Log. Reg. | λ = 0.085 | Variance | 174 | 0.044 | 0.048 | 174.0 | 174.0 |
| 7 | GLM Boosting | m stop = 27 | Variance | 728 | 0.041 | 0.041 | 13.3 | 15.1 |
| 8 | GLM Boosting | m stop = 25 | Variance | 363 | 0.048 | 0.074 | 9.8 | 7.2 |
| 9 | GLM Boosting | m stop = 13 | AUC | 232 | 0.048 | 0.077 | 9.7 | 11.0 |
|
| ||||||||
| 10 | Random Forest | num.trees = 1062, | MRMR | 1756 | 0 | 0.055 | 1466.0 | 1611.9 |
| min.node.size = 3 | ||||||||
| 11 | Random Forest | num.trees = 4671, | AUC | 760 | 0 | 0.051 | 759.7 | 760.0 |
| min.node.size = 11 | ||||||||
Table 6.
Stability values for the configurations in Table 5. The ID column references Table 5. “—” means that the configuration is not Pareto optimal for the corresponding stability measure.
| ID | 1 − SJ | 1 − SJ | 1 − SC | 1 − SC | 1 − SD-1 | 1 − SD-1 | 1 − SD-10 | 1 − SD-10 |
|---|---|---|---|---|---|---|---|---|
| (train) | (test) | (train) | (test) | (train) | (test) | (train) | (test) | |
| 1 | 0.039 | 0.046 | 0.021 | 0.025 | — | — | — | — |
| 2 | 0.040 | 0.054 | 0.021 | 0.029 | — | — | — | — |
| 3 | 0.041 | 0.060 | 0.021 | 0.032 | 0.062 | 0.091 | — | — |
| 4 | 0.047 | 0.060 | 0.025 | 0.032 | 0.066 | 0.097 | — | — |
| 5 | 0.063 | 0.054 | 0.034 | 0.028 | 0.081 | 0.073 | — | — |
| 6 | 0.072 | 0.057 | 0.038 | 0.030 | 0.100 | 0.090 | 0.243 | 0.234 |
| 7 | 0.178 | 0.340 | 0.100 | 0.208 | — | — | — | — |
| 8 | 0.206 | 0.283 | 0.117 | 0.169 | 0.247 | 0.447 | 0.255 | 0.453 |
| 9 | 0.482 | 0.503 | 0.325 | 0.341 | 0.597 | 0.561 | 0.605 | 0.570 |
|
| ||||||||
| 10 | 0.345 | 0.284 | 0.240 | 0.195 | 0.455 | 0.441 | 1.000 | 1.000 |
| 11 | 0.131 | 0.158 | 0.075 | 0.092 | 0.253 | 0.277 | 0.879 | 0.903 |
For most configurations displayed in Table 5, n.feats, the number of features selected by the filter equals the mean number of features used in the models. This means that among these configurations only GLM Boosting noticeably performs an embedded feature selection, even though Lasso Logistic Regression and Random Forest are able to do that, too.
Figure 10(a) shows the results for data set AP_Colon_Kidney. For all four stability measures, there is only one dot, indicating the performance of 67 Pareto optimal configurations. The Pareto optimal configurations are more stable and result in smaller models than the accuracy optimal configurations on the training data. Figure 11 displays the target criteria values achieved by the 67 Pareto optimal configurations. On the testing data, they still result in small models but the feature selection is not as stable as on the training data. The accuracy optimal configurations are shown in Tables 7 and 8. They perform a bit better than most of the Pareto optimal configurations considering the accuracy on the testing data. The Pareto optimal configurations averagely achieve a mean misclassification rate of 0.060. The accuracy optimal configurations are less stable and result in larger models on the testing data than most Pareto optimal configurations.
Figure 10.
Pareto front for the Pareto optimal configurations for data set AP_Colon_Kidney (a) and Stomach (b) considering the mean number of chosen features and the stability value. Only configurations whose mean misclassification rate is within 0.05 of the best classification accuracy are taken into account. Additionally, the accuracy optimal configurations are shown.
Figure 11.
Mean misclassification rate, mean number of chosen features, and stability values for the 67 Pareto optimal configurations in Figure 10(a).
Table 7.
Accuracy optimal configurations in Figure 10(a). The ID column references Table 8. The 67 Pareto optimal configurations are not displayed here.
| ID | Classifier | Parameters | Filter | n.feats | Error (train) |
Error (test) |
Size (train) |
Size (test) |
|---|---|---|---|---|---|---|---|---|
| 12 | Lasso Log. Reg. | λ = 0.0001 | Variance | 10812 | 0 | 0.026 | 10811.9 | 10812.0 |
| 13 | Lasso Log. Reg. | λ = 20.559 | MRMR | 9927 | 0 | 0.025 | 4546.2 | 4728.2 |
| 14 | GLM Boosting | mstop = 28692 | Variance | 972 | 0 | 0.026 | 51.0 | 55.4 |
| 15 | GLM Boosting | mstop = 213 | Variance | 1586 | 0 | 0.040 | 25.7 | 42.5 |
Table 8.
| ID | 1 − SJ | 1 − SJ | 1 − SC | 1 − SC | 1 − SD-1 | 1 − SD-1 | 1 − SD-10 | 1 − SD-10 |
|---|---|---|---|---|---|---|---|---|
| (train) | (test) | (train) | (test) | (train) | (test) | (train) | (test) | |
| 12 | 0.004 | 0.004 | 0.188 | 0.168 | 0.993 | 0.991 | 1.000 | 1.000 |
| 13 | 0.563 | 0.557 | 0.671 | 0.680 | 0.934 | 0.944 | 1.000 | 1.000 |
| 14 | 0.426 | 0.553 | 0.274 | 0.387 | 0.468 | 0.601 | 0.510 | 0.646 |
| 15 | 0.440 | 0.519 | 0.286 | 0.355 | 0.543 | 0.566 | 0.565 | 0.602 |
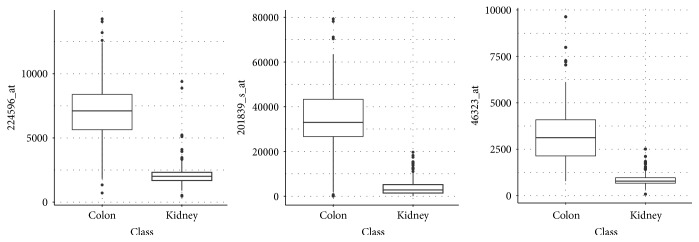
For this data set, the Pareto optimal configurations result in easily interpretable models: the ten fitted models of all Pareto optimal configurations on the training data use the same feature. This feature, however, is not the same for all configurations. For 57 configurations, the gene 224596_at (SLC44A1) is used, for 9 configurations, the gene 201839_s_at (EPCAM) is used, and for one configuration, the gene 46323_at (CANT1) is used. Figure 12 shows boxplots for the gene expression values of the three genes. The plots are based on all observations. For all three genes, low values indicate class “Kidney” and high values indicate class “Colon.” Considering the score of the AUC filter, gene 46323_at is the best of all genes in this data set for predicting the target variable when using only one variable and one cut point. Gene 224596_at is third best and gene 201839_s_at is on position 72. This ranking is based on all observations, too.
Figure 12.
Gene expression values for the genes 224596_at, 201839_s_at, and 46323_at for data set AP_Colon_Kidney.
Figure 10(b) shows the results for data set Stomach. The range of the mean misclassification rate of the Pareto optimal configurations shows that this data set poses a much more difficult classification problem. Like in Figure 9, most Pareto optimal configurations are found for SJ, including also configurations which result in larger models, and SD-10 only leads to few Pareto optimal configurations.
All Pareto optimal configurations as well as the accuracy optimal configuration are displayed in Tables 9 and 10. The mean misclassification rate on the testing data is much worse than that on the training data for all configurations in Table 9. The configuration with ID 18, which has the smallest classification error rate on the training data out of all Pareto optimal configurations for this data set, has by far the smallest classification error rate on the testing data. The average sizes of the fitted models do not differ much between training and testing data; the stability values differ a bit. The best configuration considering classification accuracy only is less stable than the Pareto optimal configurations. On the testing data, its predictive performance is similar to the Pareto optimal configurations.
Table 9.
Pareto optimal configurations in Figure 10(b) (above the horizontal line) and accuracy optimal configurations (below the horizontal line). The ID column references Table 10.
| ID | Classifier | Parameters | Filter | n.feats | Error | Error | Size | Size |
|---|---|---|---|---|---|---|---|---|
| (train) | (test) | (train) | (test) | |||||
| 16 | Lasso Log. Reg. | λ = 0.0001 | AUC | 2730 | 0.215 | 0.466 | 2726.9 | 2726.1 |
| 17 | Lasso Log. Reg. | λ = 0.00004 | MRMR | 1972 | 0.219 | 0.491 | 1971.1 | 1970.2 |
| 18 | GLM Boosting | mstop = 153 | MRMR | 461 | 0.193 | 0.360 | 35.5 | 46.8 |
| 19 | GLM Boosting | mstop = 17 | AUC | 8330 | 0.221 | 0.494 | 11.8 | 14.0 |
|
| ||||||||
| 20 | GLM Boosting | mstop = 228 | Variance | 8526 | 0.175 | 0.461 | 48.7 | 60.9 |
Table 10.
Stability values for the Pareto optimal configurations in Table 9. The ID column references Table 9. “—” means that the configuration is not Pareto optimal for the corresponding stability measure.
| ID | 1 − SJ | 1 − SJ | 1 − SC | 1 − SC | 1 − SD-1 | 1 − SD-1 | 1 − SD-10 | 1 − SD-10 |
|---|---|---|---|---|---|---|---|---|
| (train) | (test) | (train) | (test) | (train) | (test) | (train) | (test) | |
| 16 | 0.352 | 0.441 | — | — | — | — | — | — |
| 17 | 0.368 | 0.457 | 0.281 | 0.369 | 0.609 | 0.694 | — | — |
| 18 | 0.594 | 0.747 | 0.426 | 0.601 | 0.656 | 0.749 | — | — |
| 19 | 0.596 | 0.864 | 0.428 | 0.767 | 0.673 | 0.815 | 0.684 | 0.827 |
|
| ||||||||
| 20 | 0.664 | 0.739 | 0.502 | 0.591 | 0.708 | 0.739 | 0.752 | 0.794 |
Summarizing the results for all three data sets, we have seen that it is possible to choose configurations with a stable selection of few features without losing much predictive accuracy compared to model fitting only based on predictive performance.
6. Conclusion and Outlook
We compared a variety of different stability measures empirically using microarray and RNASeq data. We employed “augmented” methods consisting of a filter and a classification method. The feature selection process of these methods is cascading: the filter method selects a certain number of features and the embedded feature selection of the classification method chooses a subset of the remaining features.
We found out that the stability of small models (few features) is assessed similarly by all stability measures. For the behaviour of the stability measures concerning large models, it is most important if a measure contains a correction term for chance or large numbers of chosen features. The measures without a correction term tend to assign large stability values to models that contain many features. In these situations, the measures with a correction term assign small stability values. The different stability assessment behaviours for large models can result in different decisions concerning optimal configurations. The group of uncorrected measures evaluate the stability very similarly to each other. This means that even though the definitions of the uncorrected stability measures look quite different, it does not matter which of the uncorrected measures is chosen for stability assessment as they will all lead to very similar results. The group of corrected measures is more heterogeneous than the group of uncorrected measures.
We also conducted a random search for desirable configurations considering their predictive performance, stability, and size (number of chosen features) using augmented methods. We analysed the Pareto optimal configurations considering their stability and size taking only into account those configurations whose classification performance did not exceed c∗ + 0.05 with c∗ denoting the best mean misclassification rate achieved by any of the configurations on the data set. The resulting Pareto fronts gave several options to choose sparse and stable configurations which at the same time have high classification accuracy. In comparison to model fitting considering only the predictive performance, the Pareto optimal configurations are more stable and most of them use less features. On independent testing data, the Pareto optimal configurations are just as accurate for two of the data sets and only a bit less accurate for one data set. That means that although on the training data it looks like configurations are picked at the expense of a decreased accuracy, configurations are chosen which excel with respect to all three criteria simultaneously on the testing data. Nevertheless, in our future work, we will tackle this optimization problem three-dimensionally, accepting the drawback of visualisation difficulties.
It is important to keep in mind what kind of model is desired. If you are absolutely sure that you only want to choose a small percentage of the features in your data set, you should pick a corrected stability measure. We suggest using SC because of its theoretical and empirical properties. In some applications, it might be necessary to select many or even all features, for example, because sufficient predictive accuracy is only possible if using most of the features. In such situations, we suggest picking an uncorrected measure because the corrected measures are not defined if all features are selected. Additionally, an uncorrected measure allows pushing the Pareto front towards larger models. We recommend SJ based on its theoretical and empirical properties.
Although we have considered only genetic data sets in our analysis, we think that our conclusions are valid for many other applications as well. In our future work, we will employ model based optimization instead of random search for finding desirable configurations efficiently.
Supplementary Material
Results of the study for finding desirable configurations for all stability measures.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG), Project RA 870/7-1, and Collaborative Research Center SFB 876, A3. The authors acknowledge financial support by Deutsche Forschungsgemeinschaft and Technische Universität Dortmund within the funding programme Open Access Publishing.
Data Access
The R source code for the analyses of this article is publicly available at https://github.com/mllg/stability-bioinf.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Lang M., Kotthaus H., Marwedel P., Weihs C., Rahnenführer J., Bischl B. Automatic model selection for high-dimensional survival analysis. Journal of Statistical Computation and Simulation. 2015;85(1):62–76. doi: 10.1080/00949655.2014.929131. [DOI] [Google Scholar]
- 2.Kalousis A., Prados J., Hilario M. Stability of feature selection algorithms: a study on high-dimensional spaces. Knowledge and Information Systems. 2007;12(1):95–116. doi: 10.1007/s10115-006-0040-8. [DOI] [Google Scholar]
- 3.He Z., Yu W. Stable feature selection for biomarker discovery. Computational Biology and Chemistry. 2010;34(4):215–225. doi: 10.1016/j.compbiolchem.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Lausser L., Müssel C., Maucher M., Kestler H. A. Measuring and visualizing the stability of biomarker selection techniques. Computational Statistics. 2013;28(1):51–65. doi: 10.1007/s00180-011-0284-y. [DOI] [Google Scholar]
- 5.Nogueira S., Brown G. Machine Learning and Knowledge Discovery in Databases. Vol. 9852. Cham: Springer International Publishing; 2016. Measuring the stability of feature selection; pp. 442–457. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 6.Alelyani S., Zhao Z., Liu H. A dilemma in assessing stability of feature selection algorithms. Proceedings of the 2011 IEEE International Conference on High Performance Computing and Communications; September 2011; can. pp. 701–707. [DOI] [Google Scholar]
- 7.Wang H., Khoshgoftaar T. M., Wald R., Napolitano A. A novel dataset-similarity-aware approach for evaluating stability of software metric selection techniques. Proceedings of the 2012 IEEE 13th International Conference on Information Reuse and Integration, IRI 2012; August 2012; pp. 1–8. [DOI] [Google Scholar]
- 8.Meinshausen N., Bühlmann P. Stability selection. Journal of the Royal Statistical Society. Series B. Statistical Methodology. 2010;72(4):417–473. doi: 10.1111/j.1467-9868.2010.00740.x. [DOI] [Google Scholar]
- 9.Boulesteix A.-L., Slawski M. Stability and aggregation of ranked gene lists. Briefings in Bioinformatics. 2009;10(5):556–568. doi: 10.1093/bib/bbp034. [DOI] [PubMed] [Google Scholar]
- 10.Lee S., Rahnenführer J., Lang M., et al. Robust selection of cancer survival signatures from high-throughput genomic data using two-fold subsampling. PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0108818.e108818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awada W., Khoshgoftaar T. M., Dittman D., Wald R., Napolitano A. A review of the stability of feature selection techniques for bioinformatics data. Proceedings of the 2012 IEEE International Conference on Information Reuse and Integration; August 2012; usa. pp. 356–363. [DOI] [Google Scholar]
- 12.Abeel T., Helleputte T., Van de Peer Y., Dupont P., Saeys Y. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics. 2009;26(3):392–398. doi: 10.1093/bioinformatics/btp630. [DOI] [PubMed] [Google Scholar]
- 13.Davis C. A., Gerick F., Hintermair V., et al. Reliable gene signatures for microarray classification: assessment of stability and performance. Bioinformatics. 2006;22(19):2356–2363. doi: 10.1093/bioinformatics/btl400. [DOI] [PubMed] [Google Scholar]
- 14.Dessì N., Pascariello E., Pes B. A comparative analysis of biomarker selection techniques. BioMed Research International. 2013;2013:10. doi: 10.1155/2013/387673.387673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittman D., Khoshgoftaar T. M., Wald R., Wang H. Stability analysis of feature ranking techniques on biological datasets. Proceedings of the 2011 IEEE International Conference on Bioinformatics and Biomedicine; November 2011; pp. 252–256. [DOI] [Google Scholar]
- 16.Haury A., Gestraud P., Vert J. The influence of feature selection methods on accuracy, stability and interpretability of molecular signatures. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028210.e28210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H. W., Lawton C., Na Y. J., Yoon S. Robustness of chemometrics-based feature selection methods in early cancer detection and biomarker discovery. Statistical Applications in Genetics and Molecular Biology. 2013;12(2):207–223. doi: 10.1515/sagmb-2012-0067. [DOI] [PubMed] [Google Scholar]
- 18.Saeys Y., Abeel T., Van De Peer Y. Robust feature selection using ensemble feature selection techniques. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2008;5212(2):313–325. doi: 10.1007/978-3-540-87481-2_21. [DOI] [Google Scholar]
- 19.Schirra L.-R., Lausser L., Kestler H. A. Analysis of Large and Complex Data. Springer; 2016. [DOI] [Google Scholar]
- 20.P. Jaccard. Étude comparative de la distribution florale dans une portion des alpes et du jura. Bulletin de la Société Vaudoise des Sciences Naturelles. 1901;37:547–579. [Google Scholar]
- 21.Dice L. R. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- 22.Ochiai A. Zoogeographic studies on the soleoid fishes found in japan and its neighbouring regions. '' Bulletin of the Japanese Society for the Science of Fish. 1957;22(9):526–530. doi: 10.2331/suisan.22.526. [DOI] [Google Scholar]
- 23.Zucknick M., Richardson S., Stronach E. Comparing the characteristics of gene expression profiles derived by univariate and multivariate classification methods. Statistical Applications in Genetics and Molecular Biology. 2008;7(1):1–34. doi: 10.2202/1544-6115.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lustgarten J. L., Gopalakrishnan V., Visweswaran S. Measuring stability of feature selection in biomedical datasets. AMIA ... Annual Symposium proceedings/AMIA Symposium. AMIA Symposium. 2009;2009:406–410. [PMC free article] [PubMed] [Google Scholar]
- 25.Novovicová J., Somol P., Pudil P. A new measure of feature selection algorithms' stability. Proceedings of the 2009 IEEE International Conference on Data Mining Workshops, ICDMW 2009; December 2009; pp. 382–387. [DOI] [Google Scholar]
- 26.Somol P., Novovicová J. Evaluating stability and comparing output of feature selectors that optimize feature subset cardinality. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2010;32(11):1921–1939. doi: 10.1109/TPAMI.2010.34. [DOI] [PubMed] [Google Scholar]
- 27.Kuncheva L. I. A stability index for feature selection. Proceedings of the 25th IASTED International Conference on Artificial Intelligence and Applications (AIA '07); February 2007; pp. 390–395. [Google Scholar]
- 28.Sammut C., Webb G. I. Encyclopedia of Machine Learning. New York, NY, USA: Springer; 2011. [Google Scholar]
- 29.Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 30.Hofner B., Mayr A., Robinzonov N., Schmid M. Model-based boosting in R: a hands-on tutorial using the R Package mboost. Computational Statistics. 2014;29(1-2):3–35. doi: 10.1007/s00180-012-0382-5. [DOI] [Google Scholar]
- 31.Bühlmann P., Yu B. Boosting with the L2 loss. Journal of the American Statistical Association. 2003;98(462):324–339. doi: 10.1198/016214503000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan G.-X., Ho C.-H., Lin C.-J. Am improved GLMNET for L1-regularized logistic regression. Journal of Machine Learning Research (JMLR) 2012;13(1):1999–2030. [Google Scholar]
- 33.Izenman A. J. Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning. 2nd. New York, NY, USA: Springer; 2013. [Google Scholar]
- 34.Miettinen K. Nonlinear Multiobjective Optimization. 4th. Norwell, Mass, USA: Kluwer Academic Publishers; 2004. [Google Scholar]
- 35.Stiglic G., Kokol P. Stability of ranked gene lists in large microarray analysis studies. Journal of Biomedicine and Biotechnology. 2010;2010 doi: 10.1155/2010/616358.616358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanschoren J., van Rijn J. N., Bischl B., Torgo L. OpenML: networked science in machine learning. ACM SIGKDD Explorations Newsletter. 2013;15(2):49–60. [Google Scholar]
- 37.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core R., R Core Team R. Team, A Language and. Vienna: Environment for Statistical Computing. R Foundation for Statistical Computing; 2016. [Google Scholar]
- 39.Bischl B., Lang M., Kotthoff L., et al. mlr: machine learning in R. Journal of Machine Learning Research (JMLR) 2016;17(170):1–5. [Google Scholar]
- 40.Bischl B., Lang M., Mersmann O., Rahnenführer J., Weihs C. Batchjobs and batchexperiments: Abstraction mechanisms for using R in batch environments. Journal of Statistical Software. 2015;64(11):1–25. [Google Scholar]
- 41.Lang M. fmrmr: Fast mRMR. R package version 0.1. 2015 [Google Scholar]
- 42.Karatzoglou A., Hornik K., Smola A., Zeileis A. kernlab—an S4 package for kernel methods in R. Journal of Statistical Software. 2004;11(9):1–20. [Google Scholar]
- 43.Helleputte T., Gramme P. LiblineaR: Linear Predictive Models Based on the LIBLINEAR C/C++ Library. R package version 1.94-2. 2015 [Google Scholar]
- 44.Hothorn T., Bühlmann P., Kneib T., Schmid M., Hofner B., Bühlmann P. R package version 2.6-0. mboost: Model-Based Boosting; 2015. mboost: Model-Based Boosting; p. p. 2. [Google Scholar]
- 45.Wright M. N., Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. Journal of Statistical Software. 2017;77(1) doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 46.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of the study for finding desirable configurations for all stability measures.