Abstract
With the rise in whole slide scanner technology, large numbers of tissue slides are being scanned and represented and archived digitally. While digital pathology has substantial implications for telepathology, second opinions, and education there are also huge research opportunities in image computing with this new source of “big data”. It is well known that there is fundamental prognostic data embedded in pathology images. The ability to mine “sub-visual” image features from digital pathology slide images, features that may not be visually discernible by a pathologist, offers the opportunity for better quantitative modeling of disease appearance and hence possibly improved prediction of disease aggressiveness and patient outcome. However the compelling opportunities in precision medicine offered by big digital pathology data come with their own set of computational challenges. Image analysis and computer assisted detection and diagnosis tools previously developed in the context of radiographic images are woefully inadequate to deal with the data density in high resolution digitized whole slide images. Additionally there has been recent substantial interest in combining and fusing radiologic imaging and proteomics and genomics based measurements with features extracted from digital pathology images for better prognostic prediction of disease aggressiveness and patient outcome. Again there is a paucity of powerful tools for combining disease specific features that manifest across multiple different length scales.
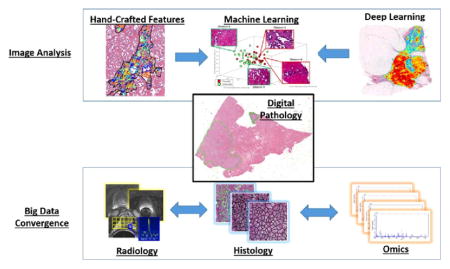
The purpose of this review is to discuss developments in computational image analysis tools for predictive modeling of digital pathology images from a detection, segmentation, feature extraction, and tissue classification perspective. We discuss the emergence of new handcrafted feature approaches for improved predictive modeling of tissue appearance and also review the emergence of deep learning schemes for both object detection and tissue classification. We also briefly review some of the state of the art in fusion of radiology and pathology images and also combining digital pathology derived image measurements with molecular “omics” features for better predictive modeling. The review ends with a brief discussion of some of the technical and computational challenges to be overcome and reflects on future opportunities for the quantitation of histopathology.
Keywords: Digital pathology, Deep learning, Radiology, Omics
Graphical Abstract
1. Introduction
Contrary to popular belief, the first forays into the use of digital image processing and computerized image analysis was not in face recognition or face detection, but rather for the analysis of cell and microscopy images. Over 50 years ago JM Prewitt and her colleagues wrote some of the very first papers on the use of computerized image analysis of cell images (Mendelsohn et al., 1965a, 1965b). Interestingly her most famous work, the Prewitt edge operator (Prewitt, 1970), was initially showcased in a 1965 paper (Mendelsohn et al., 1965a) on the morphological analysis of cells and chromosomes.
Over the last decade the advent and subsequent proliferation of whole slide digital slide scanners has resulted in a substantial amount of clinical and research interest in digital pathology, the process of digitization of tissue slides. Digitization of tissue glass slides facilitates telepathology, i.e. using telecommunication technology to facilitate the transfer of image-rich pathology data between distant locations for the purposes of research, diagnosis, and education. Telepathology can now be enabled more easily and seamlessly between multiple different remote sites and more critically connecting top academic centers with pathology labs in low resource settings. This is particularly convenient for solicitation of “second opinions” on challenging cases or the ability to do remote pathology consults without the need to physically ship slides around. Digital pathology also has the potential to help improve clinical workflows, reducing the need for storing glass slides on site and reducing the risk of physical slides getting broken or lost.
Additionally access to large digital repositories of tissue slides is a huge potential educational resource for medical students and pathology residents. Further in conjunction with smart analytics like content based image retrieval algorithms (Qi et al., 2014), students could be trained to identify and recognize pathology slides in a dynamic fashion.
Apart from education of medical students and residents, and clinical adoption, digital pathology has been transformative for computational imaging research. With digital slide archives such as the The Cancer Genome Atlas (TCGA), several thousands of slides are now freely available online and has resulted in a very large number of digital pathology image analysis related publications. A search for “digital pathology” and “image analysis” on Pubmed revealed over 1500 articles just in the last decade. While a number of these are application papers focusing on the use of computational image analysis tools to address specific targeted problems in digital pathology (e.g. quantifying specific biomarkers), a number of recent papers are focused on developing new algorithmic approaches, specifically for whole slide images.
The purpose of this review is to provide the reader with an update on the state of the art in terms of image analysis and machine learning tools being tailored specifically for digital pathology images. Specifically we seek to discuss how while early attempts at detection and segmentation of digital pathology images were rooted in traditional computer vision methods or tools hitherto developed for use in conjunction with radiographic images, there has been an evolution in the approaches in order to address the specific challenges associated with image analysis and classification of whole slide pathology images. Additionally we discuss recent advances in handcrafted and unsupervised feature analysis approaches for use in tissue classification, disease grading and in precision medicine. We also discuss new exciting directions involving multimodal imaging where digital pathology serves as a “bridge” for mapping disease extent from surgically excised tissue specimens on to radiologic in vivo imaging (Ward et al., 2012) and emerging research areas involving fusion of computer extracted tissue biomarkers with genomic and proteomic measurements for improved prediction of disease aggressiveness and patient outcome (Savage and Yuan, 2016). The review ends with a brief discussion of some of the regulatory and technical hurdles that need to be overcome prior to wide-spread dissemination and adoption of digital pathology, events which would no doubt further bolster active research in histopathology image computing.
2. Segmentation and detection of histologic primitives
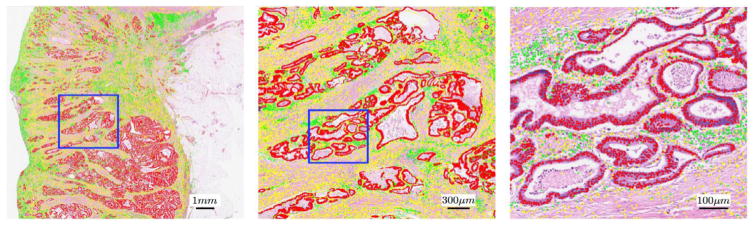
The recent advent of digital whole slide scanners has allowed for the development of quantitative histomorphometry (QH) analysis approaches which can now enable a detailed spatial interrogation (e.g. capturing nuclear orientation, texture, shape, architecture) of the entire tumor morphologic landscape and its most invasive elements from a standard hematoxylin and eosin (H&E) slide. However a pre-requisite to identifying these QH features is the need to detect and segment histologic primitives (e.g. nuclei, glands). Irshad et al. (2014) has compiled a resource in covering the extensive collection of nuclear segmentation papers for histologic analysis over the past 20 years. Fatakdawala et al. (2010) and Glotsos et al. (2004) used an active contour model to segment individual nuclei from H&E stained microscopic images. The approaches were based on the idea that the nuclei were differentially stained from the benign stroma and were associated with a prior shape. Xu et al. (2011) applied the idea of active contours to segment out individual glands from prostate cancer histopathology images. Sirinukunwattana et al. introduced a new stochastic model for glandular structures in histology images. The approach treats each glandular structure in an image as a polygon made of a random number of vertices, where the vertices represent approximate locations of epithelial nuclei. The approach was successfully employed for detection and extraction of glandular structures in histology images of normal colon tissue (Sirinukunwattana et al., 2015). Results pertaining to the approach are shown in the top row of Fig. 1.
Fig. 1.
Results of nuclear detection using the locally constrained deep learning approach proposed by Sirinukunwattana et al. (2016).
Work by Beck et al. (2011) has implicated stromal features in the prognosis of cancers. Consequently there is now interest in automated algorithms for determining epithelial and stromal tissue regions, in turn to help identify tumor infiltrating lymphocytes and also stromal specific features. Linder et al. (2012) employed a texture approach for segmentation of epithelium and stromal tissue partitions from tissue microarray (TMA) images. The authors showed that the use of local binary patterns (LBPs) could enable differentiation of stroma and epithelium with an area under the ROC curve of 0.995.
A more recent class of approaches are the so called “Deep Learning” based techniques. Deep learning paradigms represent end-to-end unsupervised feature generation methods that take advantage of large amounts of training data in conjunction with multi-layered neural network architectures. Histopathology images, given their data complexity and density are ideally suited for interrogation via deep learning approaches since they attempt to use deep architectures to learn complex features from data in an unsupervised manner. Additionally these approaches make less of a demand on the understanding the nuances of domain complexity.
Sirinukunwattana et al. recently showed the use of locality sensitive deep learning to automatically detect and classify individual nuclei in routine colon cancer histology images (Sirinukunwattana et al., 2016). Fig. 1 shows the results of nuclei detection and classification on a whole-slide image. Detected epithelial, inflammatory and fibroblast nuclei are represented as red, green, and yellow dots, respectively. The left, middle and right panels show the nuclear detection results overlaid on the image at 1, 5, and 20, respectively. The blue rectangle in the Fig. 1, left panel contains the region shown in the center panel, and the blue rectangle in the center panel contains the region shown in the right panel.
An area of substantial interest has been in the use of deep learning approaches for identifying and quantifying the number mitoses on cancer pathology images, a laborious and time consuming task for pathologists. In fact interest in this area has spawned a number of challenges for mitosis detection from routine H&E stained tissue images (Veta et al., 2015).
Deep learning approaches have however been criticized for their (a) dependency on large amounts of training data and (b) the lack of intuition associated with the deep learning generated image features. Wang et al. sought to address these challenges by presenting a convergent approach (Wang et al., 2014) to combine domain inspired features with deep learning features to detect mitoses. They showed that this integrated approach yielded superior detection accuracy compared to deep learning or hand-crafted feature based strategies alone.
3. Tissue classification, grading and precision medicine
For a number of diseases such as cancer, it has been long recognized that the underlying differences in the molecular expressions of the disease tend to manifest as tissue architecture and nuclear morphologic alterations.
Consequently there has been substantial interest in the digital pathology image analysis community to develop algorithms and feature approaches for automated tissue classification, disease grading and also developing histologic image based companion diagnostic tests for predicting disease outcome and precision medicine. Precision Medicine refers to the tailoring of medical treatment to the individual characteristics of each patient and a number of recent scientific papers (Lewis et al., 2014; Veta et al., 2012) appear to suggest that for a number of diseases, computer extracted image features from surgical and biopsy tissue specimens can help predict the level of disease aggressiveness and hence the escalation or more critically de-escalation of therapy. A critical component of these predictors is the image features mined from the tissue pathology images.
Work on quantitative feature modeling for tissue classification in the context of digital pathology can be classified into two general categories – handcrafted features and unsupervised feature based approaches. Broadly speaking, handcrafted features refer to those which can be connected to specific measurable attributes in the image and have some degree of interpretability. Unsupervised feature approaches such as deep learning based methods are less intuitive and rely on filter responses solicited from large numbers of training exemplars to characterize and model image appearance.
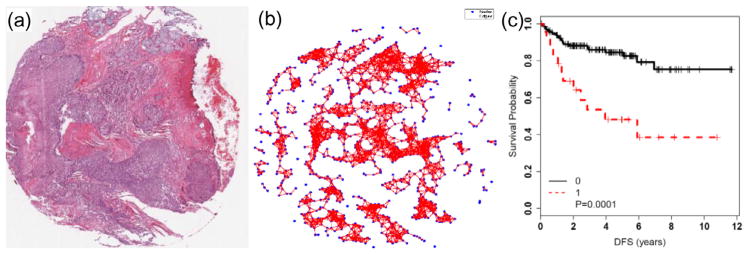
Handcrafted features can also in turn be thought to comprise domain agnostic and domain inspired features (Fig. 2). Domain agnostic features are those which though they may have intuitive meaning could nonetheless be applied for image characterization across a number of disease and tissue types. Examples include nuclear and gland shape and size, tissue texture and architecture. For instance Jafari-Khouzani and Soltanian-Zadeh (2003) used a series of wavelet and tissue texture features for automated Gleason grading of prostate pathology images. They showed that these texture features could allow for machine based separation of low and high Gleason grade prostate pathology images. Veta et al. (2012) showed that nuclear shape and texture features from tissue microarray images of male breast cancer were prognostic of survival. More recently there has been substantial interest in the use of graph based approaches (e.g. Voronoi and Delaunay tessellations, Minimum Spanning Trees) to characterize the spatial arrangement of nuclei and glands in tissue images and use features (e.g. statistics of edge lengths) from these graph representations for disease grading (Simon et al., 1998). Using a combination of nuclear architecture, shape and texture, Basavanhally et al. (2013) showed that these features could be used for accurate breast cancer grading. A variant of the global graph based approaches are the cell cluster graphs (CCG) which involved connecting clusters of proximal nodes or vertices with graph edges. These CCG features of local nuclear architecture were shown to be prognostic of disease progression in p16 + oropharyngeal cancers (Lewis et al., 2014).
Fig. 2.
Examples of domain agnostic and domain inspired handcrafted features for disease characterization and outcome prediction. An example of domain agnostic cell cluster graph features (b) to capture the spatial architecture of nuclei in p16 + oropharyngeal cancers (a). These CCG features were shown to predict progression (Lewis et al., 2014) in these cancers more accurately than T-stage and lymph node status (c).
Domain Inspired features unlike domain agnostic features represent a class of hand-crafted features that are typically specific to a particular domain or in some cases to a particular disease or organ site. An example of this class of feature is the co-occurring gland angularity feature presented by Lee et al. (2014) which involved computing the entropy of gland directions within local neighborhoods on tissue sections. The main finding of this paper was that glands tended to be more chaotically arranged in aggressive versus low to intermediate risk prostate cancer. Consequently the entropy associated with these “gland angularity features” (GAF) was found to be higher in aggressive and lower in indolent disease. For a cohort (n = 40) of grade and stage matched tumors, these gland tensor features were found to better predict biochemical recurrence in prostate cancer patients within 5 years of surgery, accurately predicting BCR over 75% of the time, while outperforming the Kattan nomogram, a popular tool for predicting BCR based off clinico-pathologic variables such as PSA, Gleason score, stage, etc.
In Yuan (2015) presented a new spatial modeling approach to identify new prognostic features reflecting the architectural appearance of tumor infiltrating lymphocytes in triple negative breast cancers. Automated image analysis approaches were used to identify intra-tumoral lymphocytes (ITLs), adjacent-tumor lymphocytes (ATLs) and distal-tumor lymphocytes (DTL). The ratio (ITLR) of ITLs to the total number of cancer cells was then calculated and found to be an independent prognostic predictor of disease-specific survival in two triple negative breast cancer cohorts.
More recently there has been interest in using deep learning based approaches for directly performing outcome prediction and disease grading. Bychkov et al. (2016) employed deep learning for outcome prediction in patients with colorectal cancer patients based off analysis of TMA images. CNN features extracted from the epithelial compartment only resulted in a prognostic discrimination comparable to that of visually determined histologic grade. Ertosun and Rubin (2015) employed a similar approach for automated grading of gliomas from digital pathology images.
Clearly there are strengths and weaknesses for both categories of features – handcrafted and unsupervised. Handcrafted features tend to provide more transparency and hence might be more intuitive to the end user – the pathologist or clinician as the case might be. On the other hand domain inspired features might be more challenging to develop since they involve more fundamentally understanding the nature of the disease and its manifestation within the tissue. The unsupervised feature generation based approach of deep learning strategies on the other hand means that they can be applied quickly and seamlessly to any domain or problem, but on the other hand suffer from the lack of feature interpretability. However there are efforts underway to be able to find ways to relate transformed feature spaces (such as might be obtained via deep learning) to handcrafted attributes (Ginsburg et al., 2016). As proposed by Wang et al. (2014), perhaps finding ways to converge two families of feature modeling techniques might provide the ideal blend of generalizability and interpretability for disease characterization and modeling in digital pathology.
4. Digital pathology as a bridge between radiology and “omics”
It is clear that molecular changes in gene expression solicit a structural and vascular change in phenotype that is in turn observable on the imaging modality under consideration. For instance, tumor morphology in standard H&E tissue specimens reflects the sum of all molecular pathways in tumor cells. By the same token radiographic imaging modalities such as MRI and CT are ultimately capturing structural and functional attributes reflective of the biological pathways and cellular morphology characterizing the disease. Historically the concept and importance of radiologypathology fusion has been around and recognized. Certainly for prostate cancer imaging, crude attempts at radiology-pathology fusion were attempted in the early 1990s with some limited success in terms of discovering radiographic correlates of histopathologic features of the disease (Schnall et al., 1991).
Recently there has been a recognition of the importance of carefully and spatially aligning in vivo radiographic imaging and ex vivo histology in order to spatially map the extent of pathology onto the corresponding imaging (Ward et al., 2012). This radiology-pathology co-registration could also enable identification of radiologic image markers correlated with tissue histomorphometric changes and thereby better disease characterization. In fact including tissue stained biomarkers as part of this radiologypathology co-registration, could enable spatially resolved radiogenomics studies, or the identification of radiologic imaging markers correlated with expressions of specific molecular and genetic biomarkers (Lenkinski et al., 2008). Digital pathology thus could serve as the “bridge” or conduit that could enable the discovery of radiographic imaging biomarkers associated with molecular pathways implicated in disease severity or progression.
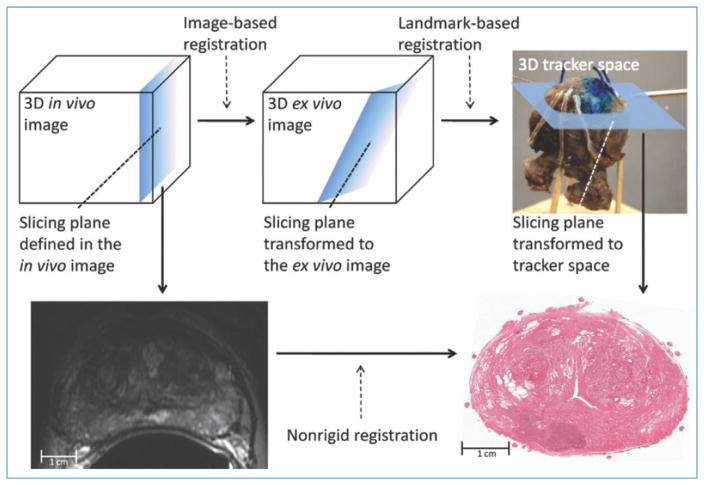
A critical aspect of this radiology-pathology fusion is the ability to carefully and spatially co-register ex vivo pathology specimens with radiographic imaging. This is non-trivial for a number of reasons, not least because of the way the tissue sections tend to created and deformations induced during the process. Another issue is carefully establishing slice correspondences between the radiologic and pathologic data. One of the approaches to overcome the aforementioned issues is via 3D histologic reconstruction of the individual 2D digitized slides. This then allows for a 3d-3d co-registration between the radiologic imaging and ex vivo pathology. Another approach described by Ward et al. (2012) involving registration of in vivo prostate MRI scans to digital histopathologic images by using image-guided specimen slicing based on strand-shaped fiducial markers relating specimen imaging to histopathologic examination. Non-rigid registration of the MRI and the histopathology yielded a mean target registration error of 1.1 mm. The approach is illustrated in Fig. 3.
Fig. 3.
Image-guided specimen slicing process. The first step is to select a desired slicing plane orientation and location from the 3D in vivo image. By using image-based registration, this plane is transformed to the space of the ex vivo specimen image. Corresponding fiducial markers are localized on the ex vivo image (by using software) and on the physical specimen (by using a tracked stylus), and a landmark transform aligning these fiducial markers transforms the slicing plane into the space of the tracker. The tracked stylus is used to direct the insertion of three slicing plane–defining pins into the specimen, which align the specimen in a slotted forceps for slicing. This permits the establishment of a correspondence between stained histologic slices and in vivo imaging planes, which are registered by using a non-rigid registration. Reproduced from (Ward et al., 2012).
Another area where digital pathology is serving as a bridge between different length scales is in the recent interest to combine histomorphometry with molecular “omics” measurements for better disease characterization. Savage and Yuan (2016) recently presented FusionGP, a new tool for selecting informative features from heterogeneous data types and predicting treatment response and prognosis. Specifically they showed the ability of FusionGP in a cohort of 119 estrogen receptor (ER) negative and 345 ER positive breast cancers to predict two important clinical outcomes: death and chemoinsensitivity by combining gene expression, copy number alteration and digital pathology image data.
5. Future directions and opportunities
The digitization of tissue glass slides is clearly opening up exciting opportunities as well as challenges to the world of computational imaging scientists. It is clear that while computational imaging can clearly play a role in better quantitative characterization of disease and precision medicine, there still remain a number of substantial technical and computational challenges that need to be overcome before computer assisted image analysis of digital pathology can become part of the routine clinical diagnostic workflow.
On the technical side, one of the main challenges in the computational interpretation of digital slide images has to do with color variations in the tissue induced by differences in slide preparation, staining, and even whole slide scanners. Clearly decision support algorithms that aim to work on digital pathology images will have to contend with and be resilient to these variations. A second technical challenge has to with the fact that most whole slide digital scanners are only able to generate 2D planar images of the slides. Pathologists however routinely take advantage of depth information which is available on most microscopes. This dept or z-axis information is useful for a number of tasks such as in confirming the presence of mitotic figures. However some whole slide scanner manufacturers are already beginning to recognize the importance of accommodating the z-stack and we can anticipate 3d whole slide scanners soon. The availability of a new dimension to accompany the dense planar data will no doubt further put pressure on the algorithmic scientists to develop more intelligent and efficient approaches for detecting, segmenting, analyzing, and interrogating 3d stacks of digitized slide images. This issue of computational complexity will become further exacerbated with the spread and availability of multi-spectral imaging cameras for investigation of multiple different tissue analytes, where each tissue section could be imaged at multiple different wavelengths and hence comprise hundreds of accompanying images. Approaches like deep learning which attempt to perform unsupervised feature analysis and discovery will clearly need to be operating at much higher levels of computational efficiency and in conjunction with high performance computing and GPU clusters (Jeong et al., 2010) to deal with the ongoing data deluge.
Despite the aforementioned challenges, the opportunities opened up by computational imaging of digital pathology are tantalizing. In spite of the reluctance thus far by the regulatory agencies to grant approval to whole slide scanned images for use for primary diagnosis, it is clear that the use of computer aided analysis with digital pathology will be part of clinical decision making in the near future. Apart from substantially aiding the pathologists in decision making, the use of computational imaging tools could enable the creation of digital imaging based companion diagnostic assays that could allow for improved disease risk characterization (Lewis et al., 2014; Veta et al., 2012). Unlike expensive molecular based assays that involve destroying the tissue and invariably capture genomic or proteomic measurements from a small part of the tumor, these digital imaging based companion diagnostic tests could be offered for a fraction of the price, could enable characterization of disease heterogeneity across the entire landscape of the tissue section, and would not need physical shipping of the tissue samples.
For the biomedical image computing, machine learning, and bioinformatics scientists, the aforementioned challenges will present new and exciting opportunities for developing new feature analysis and machine learning opportunities. Clearly though, the image computing community will need to work closely with the pathology community and potentially whole slide imaging and microscopy vendors to be able to develop new and innovative solutions to many of the critical image analysis challenges in digital pathology.
One very interesting image computing research area that digital pathology opens up is the ability to combine traditional handcrafted feature approaches with deep learning methodologies, thereby taking advantage of domain knowledge while also enabling the classifier to discover new features. Another exciting research avenue will be in the development of new data fusion algorithms for combining radiologic, histologic, and molecular measurements for improved disease characterization.
Computational imaging advances for digital pathology will finally begin to make pathology more quantitative, a field that has thus far significantly lagged behind radiology in this regard. By all indications, this transformation from qualitative to quantitative pathology is in the not too distant future.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374, 1R01CA202752, R21CA179327; R21CA195152, K01ES026841, the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the DOD Prostate Cancer Idea Development Award (PCRP W81XWH-15-1-0558); the Case Comprehensive Cancer Center Pilot Grant, VelaSano Grant from the Cleveland Clinic, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Basavanhally A, Ganesan S, Feldman M, Shih N, Mies C, Tomaszewski J, Madabhushi A. Multi-field-of-view framework for distinguishing tumor grade in ER + breast cancer from entire histopathology slides. IEEE Trans Biomed Eng. 2013;60:2089–2099. doi: 10.1109/TBME.2013.2245129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- Bychkov D, Turkki R, Haglund C, Linder N, Lundin J. Deep learning for tissue microarray image-based outcome prediction in patients with colorectal cancer. In: Gurcan MN, Madabhushi A, editors. Proceedings of the SPIE9791, Medical Imaging 2016. Digital Pathology; San Diego, California, United States. 2016. p. 979115. [DOI] [Google Scholar]
- Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc. 2015;2015:1899–1908. [PMC free article] [PubMed] [Google Scholar]
- Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, Tomaszewski JE, Madabhushi A. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:1676–1689. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- Ginsburg SB, Lee G, Ali S, Madabhushi A. Feature importance in nonlinear embeddings (FINE): applications in digital pathology. IEEE Trans Med Imaging. 2016;35:76–88. doi: 10.1109/TMI.2015.2456188. [DOI] [PubMed] [Google Scholar]
- Glotsos D, Spyridonos P, Cavouras D, Ravazoula P, Dadioti PA, Nikiforidis G. Automated segmentation of routinely hematoxylin-eosin-stained microscopic images by combining support vector machine clustering and active contour models. Anal Quant Cytol Histol. 2004;26:331–340. [PubMed] [Google Scholar]
- Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: a review–current status and future potential. IEEE Rev Biomed Eng. 2014;7:97–114. doi: 10.1109/RBME.2013.2295804. [DOI] [PubMed] [Google Scholar]
- Jafari-Khouzani K, Soltanian-Zadeh H. Multiwavelet grading of pathological images of prostate. IEEE Trans Biomed Eng. 2003;50:697–704. doi: 10.1109/TBME.2003.812194. [DOI] [PubMed] [Google Scholar]
- Prewitt JMS. Picture Processing and Psychopictorics. Academic Press; New York: 1970. Object enhancement and extraction; pp. 75–149. [Google Scholar]
- Jeong WK, Schneider J, Turney SG, Faulkner-Jones BE, Meyer D, Westermann R, Reid RC, Lichtman J, Pfister H. Interactive histology of largescale biomedical image stacks. IEEE Trans Vis Comput Graph. 2010;16:1386–1395. doi: 10.1109/TVCG.2010.168. [DOI] [PubMed] [Google Scholar]
- Lee G, Sparks R, Ali S, Shih NNC, Feldman MD, Spangler E, Rebbeck T, Tomaszewski JE, Madabhushi A. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients. PloS One. 2014;9:e97954. doi: 10.1371/journal.pone.0097954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkinski RE, Bloch BN, Liu F, Frangioni JV, Perner S, Rubin MA, Genega EM, Rofsky NM, Gaston SM. An illustration of the potential for mapping MRI/MRS parameters with genetic over-expression profiles in human prostate cancer. Magma N Y N. 2008;21:411–421. doi: 10.1007/s10334-008-0133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JSJ, Ali S, Luo J, Thorstad WL, Madabhushi A. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2014;38:128–137. doi: 10.1097/PAS.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder N, Konsti J, Turkki R, Rahtu E, Lundin M, Nordling S, Haglund C, Ahonen T, Pietikäinen M, Lundin J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn Pathol. 2012;7:22. doi: 10.1186/1746-1596-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn ML, Kolman WA, Perry B, Prewitt JM. Morphological analysis of cells and chromosomes by digital computer. Methods Inf Med. 1965a;4:163–167. [PubMed] [Google Scholar]
- Mendelsohn ML, Kolman WA, Perry B, Prewitt JM. Computer analysis of cell images. Postgrad Med. 1965b;38:567–573. doi: 10.1080/00325481.1965.11695692. [DOI] [PubMed] [Google Scholar]
- Qi X, Wang D, Rodero I, Diaz-Montes J, Gensure RH, Xing F, Zhong H, Goodell L, Parashar M, Foran DJ, Yang L. Content-based histopathology image retrieval using CometCloud. BMC Bioinformatics. 2014;15:287. doi: 10.1186/1471-2105-15-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage RS, Yuan Y. Predicting chemoinsensitivity in breast cancer with ’omics/digital pathology data fusion. R Soc Open Sci. 2016;3:140501. doi: 10.1098/rsos.140501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall MD, Imai Y, Tomaszewski J, Pollack HM, Lenkinski RE, Kressel HY. Prostate cancer: local staging with endorectal surface coil MR imaging. Radiology. 1991;178:797–802. doi: 10.1148/radiology.178.3.1994421. [DOI] [PubMed] [Google Scholar]
- Simon I, Pound CR, Partin AW, Clemens JQ, Christens-Barry WA. Automated image analysis system for detecting boundaries of live prostate cancer cells. Cytometry. 1998;31:287–294. doi: 10.1002/(sici)1097-0320(19980401)31:4<287::aid-cyto8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Sirinukunwattana K, Raza S, Tsang Y-W, Snead D, Cree I, Rajpoot N. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016 doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- Sirinukunwattana K, Snead DRJ, Rajpoot NM. A stochastic polygons model for glandular structures in colon histology images. IEEE Trans Med Imaging. 2015;34:2366–2378. doi: 10.1109/TMI.2015.2433900. [DOI] [PubMed] [Google Scholar]
- Veta M, Kornegoor R, Huisman A, Verschuur-Maes AHJ, Viergever MA, Pluim JPW, van Diest PJ. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol Off J U S Can Acad Pathol Inc. 2012;25:1559–1565. doi: 10.1038/modpathol.2012.126. [DOI] [PubMed] [Google Scholar]
- Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, Gonzalez F, Larsen ABL, Vestergaard JS, Dahl AB, Ciresan DC, Schmidhuber J, Giusti A, Gambardella LM, Tek FB, Walter T, Wang CW, Kondo S, Matuszewski BJ, Precioso F, Snell V, Kittler J, de Campos TE, Khan AM, Rajpoot NM, Arkoumani E, Lacle MM, Viergever MA, Pluim JPW. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, Tomaszewski J, Gonzalez F, Madabhushi A. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging Bellingham Wash. 2014;1:34003. doi: 10.1117/1.JMI.1.3.034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AD, Crukley C, McKenzie CA, Montreuil J, Gibson E, Romagnoli C, Gomez JA, Moussa M, Chin J, Bauman G, Fenster A. Prostate: registration of digital histopathologic images to in vivo MR images acquired by using endorectal receive coil. Radiology. 2012;263:856–864. doi: 10.1148/radiol.12102294. [DOI] [PubMed] [Google Scholar]
- Xu J, Janowczyk A, Chandran S, Madabhushi A. A high-throughput active contour scheme for segmentation of histopathological imagery. Med Image Anal. 2011;15:851–862. doi: 10.1016/j.media.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J R Soc Interface R Soc. 2015:12. doi: 10.1098/rsif.2014.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]