Abstract
Choline (Cho) is the precursor of the osmoprotectant glycine betaine and is itself an essential nutrient for humans. Metabolic engineering of Cho biosynthesis in plants could therefore enhance both their resistance to osmotic stresses (drought and salinity) and their nutritional value. The key enzyme of the plant Cho-synthesis pathway is phosphoethanolamine N-methyltransferase, which catalyzes all three of the methylations required to convert phosphoethanolamine to phosphocholine. We show here that overexpressing this enzyme in transgenic tobacco increased the levels of phosphocholine by 5-fold and free Cho by 50-fold without affecting phosphatidylcholine content or growth. Moreover, the expanded Cho pool led to a 30-fold increase in synthesis of glycine betaine via an engineered glycine betaine pathway. Supplying the transgenics with the Cho precursor ethanolamine (EA) further enhanced Cho levels even though the supplied EA was extensively catabolized. These latter results establish that there is further scope for improving Cho synthesis by engineering an increased endogenous supply of EA and suggest that this could be achieved by enhancing EA synthesis and/or by suppressing its degradation.
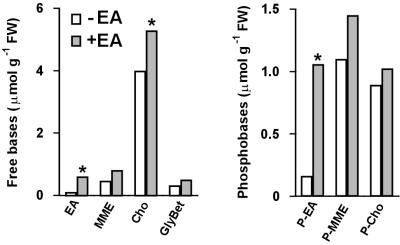
Choline (Cho) is a vital metabolite in plants and other eukaryotes because it is needed to synthesize the membrane phospholipid phosphatidylcholine (1, 2). In addition, certain plants such as spinach have chloroplast enzymes that catalyze the two-step oxidation of Cho to glycine betaine (GlyBet; Fig. 1; refs. 1 and 3). GlyBet is important because it has strong osmoprotectant properties and confers tolerance to salinity, drought, and other stresses (1, 3). Many bacteria also have enzymes that oxidize Cho to GlyBet (3). Plant or bacterial genes for Cho oxidation have therefore been used to engineer GlyBet synthesis in tobacco, Arabidopsis thaliana, and other plants that lack it, and this has increased their stress tolerance (3, 4). However, the levels of GlyBet thus far attained by engineering are low, and the increments in stress tolerance are commensurately small (4). It has been shown that the low levels of GlyBet in the engineered plants are due in large part to an inadequate capacity for Cho synthesis (4–6). This constraint on GlyBet synthesis provides one reason to enhance Cho biosynthesis in plants. There is also a second reason, which is nutritional. Cho has recently been classified as an essential nutrient for humans, and the Institute of Medicine of the United States National Academy of Sciences has accordingly set adequate intake values for Cho in the diet (7, 8). Dietary Cho is of course needed to synthesize phosphatidylcholine, but it is also required for cholinergic neurotransmission, methyl metabolism, and other functions (9). Cho has a particularly critical role in brain development (7, 8).
Figure 1.
Pathways of Cho and GlyBet synthesis in plants. Note that not all plants produce GlyBet. Bold arrows show the three methylation reactions that are catalyzed by PEAMT. The conversion of phospho (P-) bases to phosphatidyl (Ptd-) bases occurs via cytidine 5′-diphosphate (CDP) derivatives, which have been omitted for simplicity. MME and DME, mono- and dimethylethanolamine; Bet Ald, betaine aldehyde.
The biosynthesis of Cho in plants (Fig. 1) differs from that in other eukaryotes, where the only route is stepwise methylation of the phospholipid phosphatidylethanolamine (4, 9). In plants, the first and committing step is methylation of phosphoethanolamine, catalyzed by the cytosolic enzyme phosphoethanolamine N-methyltransferase (PEAMT; EC 2.1.1.103; refs. 10–12). PEAMT also mediates the next two methylations, producing phosphocholine (11, 12), which is either directly dephosphorylated to free Cho, as in spinach (13), or incorporated into phosphatidylcholine and then metabolized to Cho, as in tobacco (14). The final two methylations of Cho synthesis also can occur at the phospholipid level, but the importance of this route varies with species, and in spinach and tobacco it accounts for ≤15% of the total (13, 14). Whatever the later biosynthetic steps, all metabolic flux to Cho goes through PEAMT, and various lines of evidence indicate that this enzyme exerts major control over flux through the whole pathway (6, 12, 15, 16). Most notably, PEAMT is sensitive to feedback inhibition by phosphocholine (11, 12) and is repressed by adding Cho to the medium (15). Also, PEAMT is induced by salt stress, which enhances the rate of GlyBet synthesis and hence the demand for Cho (12, 16). PEAMT is thus the prime target for manipulation to enhance Cho synthesis in plants.
Accordingly, we transformed tobacco plants with a spinach PEAMT cDNA under the control of a strong constitutive promoter. These plants were already expressing transgenes encoding chloroplast enzymes for GlyBet synthesis, choline monooxygenase (CMO) and betaine aldehyde dehydrogenase (BADH). This strategy enabled us to evaluate effects on GlyBet as well as Cho production.
Materials and Methods
Construction of the PEAMT Plant Expression Vector.
The PEAMT expression cassette was constructed in pFMV, which contains the figwort mosaic virus 34S promoter and the nos terminator (17). The PEAMT coding sequence was amplified by PCR, using Pfu polymerase (Stratagene) with the forward primer 5′-AGCTTCTAGATTTTTACAACAATTACCAACAACAACAAACAACAAACAACATTACAATTACTATTTACAATTACAAAAATGGCCGCTTCAGCTATGG-3′, the reverse primer 5′-ACGTGGATCCTCACATTTTCTTGGCAATG-3′, and pREP3-PEAMT (12) as template. The forward primer includes the tobacco mosaic virus Ω sequence (18) and alters the sequence context of the start codon as described (19). The reaction product was ligated to pFMV as an XbaI/BamHI fragment to yield pFMV-PEAMT. The binary vector was derived from pMON51745, which carries genes for spectinomycin resistance in bacteria and glyphosate resistance in plants. pMON51745 was cut with PacI/BstXI to remove the reporter cassette, end-polished with T4 DNA polymerase, ligated to BglII adaptors (12), and circularized to give pMON51745B. The pFMV-PEAMT expression cassette was excised as a NotI fragment and ligated to pMON51745B. Sequence analysis verified construct integrity at each step. The PEAMT construct and the empty pMON51745B vector were mobilized into Agrobacterium tumefaciens strain ABI for plant transformation.
Construction of Plants Expressing a GlyBet Synthesis Pathway and PEAMT.
Tobacco plants expressing CMO and BADH were obtained by crossing lines homozygous for the ΩCMOnos (19) and beet BADH (20) transgenes, each of which is linked to a kanamycin-resistance marker. Enzyme assays confirmed that the F1 progeny expressed both genes. The PEAMT construct and the empty vector were introduced into F1 plants by leaf-disk transformation (21). Transformants were selected and regenerated on medium containing 100 μg ml−1 kanamycin and 25 μM glyphosate.
Plant Growth Conditions.
Plants were cultured at 25°C in 12-h days in magenta boxes on Murashige and Skoog medium containing sucrose and hormones as described (20). Where specified, media were supplemented with 5 mM ethanolamine (EA) that had been neutralized with HCl. For one experiment, plants were transferred to a light soil mixture and grown in a greenhouse; irrigation was with Peters nutrient solution.
PEAMT Extraction and Assay.
Soluble proteins were extracted from the uppermost three to five leaves of cultured plants and desalted as described (13). PEAMT activity was assayed radiometrically (12) at 30°C. Protein was determined by using the Bradford (22) assay.
Ab Production and Immunoblotting.
Recombinant spinach PEAMT with a C-terminal histidine tag was produced by using the pET28b vector (Novagen), purified by Ni2+ affinity chromatography, and used to raise rabbit Abs. Immunoblotting was as described (19); the anti-PEAMT serum was diluted 1:1000; no crossreaction with tobacco PEAMT was detectable.
Metabolite Analysis.
Free bases, phosphobases, and GlyBet were isolated as described (6) from 0.5-g fresh weight (FW) samples of leaf material or from whole plants. GlyBet was converted to its n-butyl ester and quantified by fast atom bombardment MS (23). EA, mono-, and dimethylethanolamine, and their phospho-esters were quantified from the intensity of ninhydrin- (EA, monomethylethanolamine) or Dragendorff- (dimethylethanolamine) reactive TLC zones relative to standards (6). Cho and phosphocholine (after acid hydrolysis) were determined enzymatically by using Cho oxidase (6). Phosphatidyl bases were extracted, acid-hydrolyzed, and determined as the corresponding free bases (14). All metabolite levels reported are corrected for recovery, based on recoveries obtained for standard compounds. EA remaining in culture media at the end of the supplementation experiment was extracted from the agar with hot water, purified by ion exchange chromatography by using AG-1 (OH−) and BioRex 70 (H+) resins (Bio-Rad; ref. 6), and determined as above.
Results
PEAMT Overexpression Enhances Cho Levels.
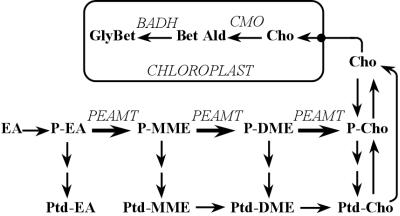
The spinach PEAMT coding sequence (12), preceded by the Ω translational enhancer (18) and the figwort mosaic virus 34S promoter (17), was cloned into an Agrobacterium binary vector with a glyphosate-resistance marker. This construct was used to transform tobacco plants that were coexpressing spinach CMO (19) and beet BADH (20) and so contained a transgenic GlyBet pathway in their chloroplasts. Glyphosate-resistant transformants were identified that had levels of spinach PEAMT protein close to those in spinach itself (Fig. 2A). These PEAMT+ transgenics contained up to 50-fold more free Cho than plants transformed with vector alone (Fig. 2B). Consistent with the high PEAMT protein levels, PEAMT activity was elevated at least 40-fold (Table 1). The PEAMT transgene did not affect growth; at 4 weeks after subculture the FW (mean ± SE) of PEAMT+ plants was 3.6 ± 0.6 g vs. 3.6 ± 0.3 g for empty-vector controls.
Figure 2.
PEAMT expression and Cho levels in leaves of tobacco primary transformants grown in culture. (A) Immunoblot analysis of recombinant spinach PEAMT protein in leaf extracts of eight individual PEAMT+ transformants (tracks 1–8) and an empty-vector control (V); salinized spinach (SS) is included for comparison. Tracks contained 50 μg of protein. (B) Free Cho levels in the same plants.
Table 1.
PEAMT activity and levels of Cho and its metabolites in leaves of transgenic tobacco plants expressing spinach PEAMT
| Transgenic line | Growth conditions | PEAMT activity, pkat mg−1 protein | Cho moieties, μmol
g−1FW
|
||||
|---|---|---|---|---|---|---|---|
| Cho | P-Cho | Ptd-Cho | GlyBet | Total | |||
| Vector | Culture | <0.1 | 0.11 | 0.11 | 1.56 | 0.02 | 1.80 |
| 1 | Culture | 5.00 | 4.78 | 0.63 | 1.42 | 0.45 | 7.28 |
| 4 | Culture | 4.63 | 5.03 | 0.94 | 1.62 | 0.41 | 8.00 |
| 5 | Culture | 3.41 | 5.27 | 0.35 | 1.38 | 0.62 | 7.62 |
| Vector (YL) | Greenhouse | — | 0.13 | 0.08 | — | 0.04 | — |
| 1 (YL) | Greenhouse | — | 5.21 | 0.23 | — | 1.81 | — |
| 1 (OL) | Greenhouse | — | 5.75 | 0.19 | — | 1.16 | — |
Three PEAMT+ lines and an empty-vector control were cultured in vitro. Line 1 and the empty-vector control were also grown in the greenhouse for 10 weeks, after which a half-expanded leaf (YL) and the third fully expanded leaf (OL) were analyzed. All data are values for individual plants. P-Cho, phosphocholine; Ptd-Cho, phosphatidylcholine.
Effects of PEAMT Overexpression on Levels of Cho Metabolites.
PEAMT+ transformants had greatly elevated phosphocholine levels, but their phosphatidylcholine levels were normal (Table 1). Consistent with their larger free Cho pools, GlyBet accumulation was enhanced up to 30-fold in PEAMT+ plants. This enhancement of GlyBet synthesis fits predictions from modeling of the transgenic GlyBet synthesis pathway (24), as we discuss later. When the levels of individual Cho metabolites are summed (Table 1, last column), it can be seen that PEAMT+ plants contained 4 times more Cho moieties than controls. Because neither Cho moieties (15) nor GlyBet (1, 6) are metabolically degraded by plants, it follows that metabolic flux through PEAMT to Cho must have increased 4-fold.
The effects of PEAMT overexpression were not confined to plants cultured in vitro. When a representative PEAMT+ line was grown in soil in the greenhouse, the free Cho levels in expanding and mature leaves were comparable to those in cultured plants, and GlyBet levels were higher (Table 1); only the phosphocholine levels were lower. A lower level of phosphocholine in greenhouse-grown tobacco relative to cultured plants was noted in previous work (6).
PEAMT Overexpression Depletes EA and Phosphoethanolamine Pools.
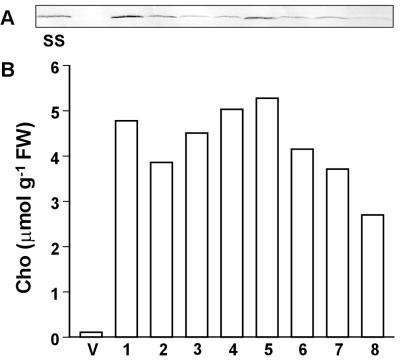
The levels of free Cho accumulated by PEAMT+ transgenics are well above normal ranges for plant tissues, where the ceiling is ≈1 μmol g−1 FW (25–27). However, these free Cho levels are below those that can accumulate in plant tissues supplied with Cho, as are the phosphocholine levels (15, 28). This situation suggests that Cho synthesis in the transgenics might be constrained by steps lying upstream from PEAMT. Analysis of the pool sizes of Cho precursors supported the possibility of an upstream constraint (Fig. 3). PEAMT+ plants had smaller pools of both EA and phosphoethanolamine but a larger pool of phosphomonomethylethanolamine, the first product of PEAMT action. These data suggest that the internal supply of EA becomes limiting in PEAMT+ plants.
Figure 3.
Evidence that overexpression of PEAMT depletes the endogenous pools of EA and phosphoethanolamine in transgenic tobacco. Levels of EA, phosphoethanolamine (P-EA), and phosphomonomethylethanolamine (P-MME) were determined in leaves of transgenic line 1 expressing spinach PEAMT (PEAMT+) and an empty-vector control line (V). Plants were grown for 4 weeks in culture. Data are means and SE for three or four replicate plants.
Metabolism of Supplied EA in PEAMT+ Plants.
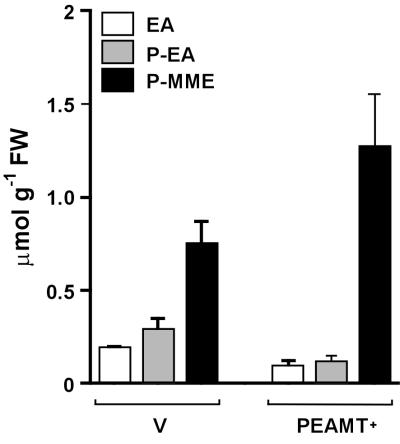
To investigate whether EA is indeed limiting, we cultured PEAMT+ plants on media with or without 5 mM EA, and measured the levels of EA metabolites in leaves (Fig. 4). Supplying EA increased the endogenous pools of free EA and phosphoethanolamine by ≈7-fold and significantly increased the flux to Cho and GlyBet. The increases in Cho and GlyBet were smaller than might at first sight be expected from the large increase in phosphoethanolamine, the substrate for PEAMT. This result can be ascribed, at least in part, to feedback inhibition of PEAMT by phosphocholine (11, 12). Because phosphocholine is located solely in the cytoplasm of tobacco leaves (24), its in vivo concentration in PEAMT+ plants (Fig. 4) would be ≈40 mM, which is enough to inhibit PEAMT activity by >95% (12). This inhibition is not simply competitive with respect to phosphoethanolamine and thus cannot be completely reversed by higher phosphoethanolamine levels (11, 12). It is also possible that part of the EA that enters tobacco leaf tissue from the outside, as opposed to endogenously made EA, is diverted into a pool of phosphoethanolamine to which PEAMT has poor access (14).
Figure 4.
The effect of EA supplementation on the levels of free bases and phosphobases in leaves of transgenic tobacco. Plants expressing spinach PEAMT (transgenic line 1) were grown for 4 weeks on medium with or without 5 mM EA, and the levels of free bases and phosphobases were determined in the uppermost leaves. Phosphodimethylethanolamine levels were below the detection limit (0.5 μmol g−1 FW). Data are means of four replicates; asterisks denote differences between treatments that are significant at P ≤ 0.05 by ANOVA.
Importantly, the total increase in EA moieties (i.e., in EA and its metabolites; see Fig. 1) in the leaves of EA-fed plants was only 4.6 μmol g−1 FW, which is far less than the increases observed when 5 mM mono- or dimethylethanolamine is supplied (21.7 and 12.9 μmol g−1 FW, respectively; ref. 6). This situation might be because EA is less readily absorbed or more readily catabolized than its partially methylated derivatives. To test these possibilities, we carried out an EA feeding experiment in which EA metabolites were measured in entire plants, and EA disappearance from the medium was quantified. This experiment enabled us to construct a balance sheet showing the fate of the supplementary EA (Table 2). Much more EA was removed from the medium (100 μmol per plant) than was present as extra EA moieties in the harvested plants (29 μmol per plant). Only about half the extra EA moieties in the plants (14 μmol per plant) were methylated. Thus, 71% of the absorbed EA was catabolized during the 4-week culture period, whereas only 14% entered the Cho synthesis pathway.
Table 2.
Balance sheet showing the fates of EA supplied to transgenic tobacco plants expressing spinach PEAMT
| Metabolites | −EA, μmol per plant | +EA, μmol per plant | Difference,* μmol per plant |
|---|---|---|---|
| EA | 0.7 | 14.5 | +13.8 |
| Monomethylethanolamine | 3.3 | 2.4 | −0.9 |
| Cho | 20.6 | 34.1 | +13.5 |
| Phosphoethanolamine | 1.6 | 2.6 | +1.0 |
| Phosphomonomethylethanolamine | 12.1 | 9.6 | −2.5 |
| Phosphocholine | 1.5 | 2.7 | +1.2 |
| Phosphatidylethanolamine | 2.6 | 3.4 | +0.8 |
| Phosphatidylmonomethylethanolamine | 0.7 | 1.3 | +0.6 |
| Phosphatidylcholine | 4.2 | 5.3 | +1.1 |
| GlyBet | 1.0 | 1.6 | +0.6 |
| Totals | 48.3 | 77.5 | +29.2 |
| EA lost from medium† | 100 | ||
Individual plants were cultured for 4 weeks on 50 ml of medium with or without 5 mM EA; the EA dose was thus 250 μmol per plant. Each plant then was harvested and assayed for EA metabolites, and the quantity of EA left in the medium was determined. Data are means for duplicate plants. Dimethylethanolamine and its esters were undetectable.
Metabolite level in the +EA treatment minus that in the −EA treatment.
EA loss from the medium after incubation for 4 weeks with no plant present was insignificant.
Discussion
We have applied rational metabolic engineering principles (29) to enhance Cho synthesis in plants. The levels of free Cho we achieved are about 5 times the natural maximum levels reported in leaves and other fleshy plant tissues, and 50 times those in normal tobacco. The Cho-accumulating transgenics appeared to grow normally, and had normal phosphatidylcholine contents. The latter finding is consistent with the view that phosphatidylcholine synthesis is governed downstream of Cho and phosphocholine formation, by the rate of the phosphocholine cytidylyltransferase reaction (30). As predicted by metabolic modeling (19, 24), the expanded Cho pool drives a higher flux through the transgenic GlyBet synthesis pathway. That this flux is not even higher can be explained by additional constraints on the operation of the transgenic pathway in tobacco, particularly a low capacity to import Cho into the chloroplast from its site of synthesis in the cytosol (19, 24). It should be noted that this constraint is missing from plants engineered with a cytosolic Cho → GlyBet pathway by using bacterial Cho-oxidizing enzymes (3, 5, 19), so that overexpressing PEAMT in such plants could lead to higher GlyBet levels than those we attained. Because the GlyBet levels we achieved are comparable to those already demonstrated by several groups to enhance stress resistance in tobacco and other plants (5, 25, 31), we did not reduplicate this work with our transgenics.
In human nutrition, phosphocholine is at least equivalent to Cho and may be the preferred form of Cho for infants because it is the dominant Cho-containing compound in human milk (32). It is therefore significant that phosphocholine as well as free Cho accumulated in PEAMT+ plants. The total Cho plus phosphocholine level in these plants is ≈7 mM (assuming 80% water content), which is five times higher than the total of Cho-containing compounds in human milk (32). This level is promising from a dietary standpoint because a single 200-ml serving of vegetable or fruit juice containing 7 mM Cho compounds would meet at least half the adequate intake value for a preteen child and one-third for an adult (8).
Moreover, our results suggest that higher Cho contents might be achieved by further engineering. One approach would be to introduce a truncated PEAMT (12) that is less sensitive to inhibition by phosphocholine, as proposed (12). However, it is doubtful that this maneuver in itself would be optimal, for analysis of the pools of Cho synthesis intermediates and the effect of supplying EA indicate that PEAMT+ plants suffer from an inadequate internal EA supply. The EA-feeding experiments further suggest that this supply depends on the rate of EA degradation as well as synthesis. Both these processes could in principle be engineered to make more EA available for Cho production.
EA synthesis in plants is thought to proceed mainly by decarboxylation of free serine (33), not phosphatidylserine as in other organisms (2). Because serine decarboxylases were recently cloned from plants (ref. 34, and D. Rontein, I. Nishida, G. Tashiro, D. Voelker, and A.D.H., unpublished data), it may prove straightforward to enhance the capacity for EA synthesis. Reducing the capacity for EA catabolism is more problematic because little is known about how plants degrade EA. An oxidative route by means of glycolaldehyde and glycolate has been proposed (35). Alternatively, plants may have phosphoethanolamine phospho-lyase, a pyridoxal phosphate enzyme known from mammals and bacteria that cleaves phosphoethanolamine to phosphate, ammonia, and acetaldehyde (36, 37). The large accumulation of phosphoethanolamine in EA-fed plants could drive a high flux through such a route, rendering phosphoethanolamine unavailable for methylation.
Finally, it should be noted that we achieved a dramatic increase in the level of Cho by overexpressing a single gene, and moreover one whose product is subject to feedback inhibition. Simple manipulations of primary metabolism are rarely so effective, either in plants or other organisms (29, 38).
Acknowledgments
We thank Harry Klee (University of Florida, Gainesville) for pFMV and Karen Fincher of Monsanto (St. Louis) for pMON51745 and A. tumefaciens strain ABI. This research was supported by the Florida Agricultural Experiment Station, by a grant from the United States Department of Agriculture National Research Initiative Competitive Grants Program, and by an endowment from the C. V. Griffin, Sr., Foundation, and was approved for publication as Journal Series No. R08116.
Abbreviations
- BADH
betaine aldehyde dehydrogenase
- Cho
choline
- CMO
choline monooxygenase
- EA
ethanolamine
- FW
fresh weight
- GlyBet
glycine betaine
- PEAMT
phosphoethanolamine N-methyltransferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rhodes D, Hanson A D. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:357–384. [Google Scholar]
- 2.Kent C. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto S, Murata N. J Exp Bot. 2000;51:81–88. [PubMed] [Google Scholar]
- 4.Nuccio M L, Rhodes D, McNeil S D, Hanson A D. Curr Opin Plant Biol. 1999;2:128–134. doi: 10.1016/s1369-5266(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Hirji R, Adam L, Rozwadowski K L, Hammerlindl J K, Keller W A, Selvaraj G. Plant Physiol. 2000;122:747–756. doi: 10.1104/pp.122.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuccio M L, Russell B L, Nolte K D, Rathinasabapathi B, Gage D A, Hanson A D. Plant J. 1998;16:487–496. doi: 10.1046/j.1365-313x.1998.00316.x. [DOI] [PubMed] [Google Scholar]
- 7.Blusztajn J K. Science. 1998;281:794–795. doi: 10.1126/science.281.5378.794. [DOI] [PubMed] [Google Scholar]
- 8.Zeisel S H. Nutrition. 2000;16:669–671. doi: 10.1016/s0899-9007(00)00349-x. [DOI] [PubMed] [Google Scholar]
- 9.Zeisel S H, Blusztajn J K. Annu Rev Nutr. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- 10.Datko A H, Mudd S H. Plant Physiol. 1988;88:1338–1348. doi: 10.1104/pp.88.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith D D, Summers P S, Weretilnyk E A. Physiol Plant. 2000;108:286–294. [Google Scholar]
- 12.Nuccio M L, Ziemak M J, Henry S A, Weretilnyk E A, Hanson A D. J Biol Chem. 2000;275:14095–14101. doi: 10.1074/jbc.275.19.14095. [DOI] [PubMed] [Google Scholar]
- 13.Summers P S, Weretilnyk E A. Plant Physiol. 1993;103:1269–1276. doi: 10.1104/pp.103.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNeil S D, Nuccio M L, Rhodes D, Shachar-Hill Y, Hanson A D. Plant Physiol. 2000;123:371–380. doi: 10.1104/pp.123.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mudd, S. H. & Datko, A. H. (1989) 90, 296–395. [DOI] [PMC free article] [PubMed]
- 16.Weretilnyk E A, Smith D D, Wilch G, Summers P S. Plant Physiol. 1995;109:1085–1091. doi: 10.1104/pp.109.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger M, Daubert S, Goodman R M. Plant Mol Biol. 1990;14:433–443. doi: 10.1007/BF00028779. [DOI] [PubMed] [Google Scholar]
- 18.Dowson-Day M J, Ashurst J L, Mathias S F, Watts J W, Wilson T M, Dixon R A. Plant Mol Biol. 1993;23:97–109. doi: 10.1007/BF00021423. [DOI] [PubMed] [Google Scholar]
- 19.Nuccio M L, McNeil S D, Ziemak M J, Hanson A D. Metab Eng. 2000;2:300–311. doi: 10.1006/mben.2000.0158. [DOI] [PubMed] [Google Scholar]
- 20.Rathinasabapathi B, McCue K F, Gage D A, Hanson A D. Planta. 1994;193:155–162. doi: 10.1007/BF00192524. [DOI] [PubMed] [Google Scholar]
- 21.Horsch R B, Fry J E, Hoffmann N L, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 22.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes D, Rich P J, Myers A C, Reuter C C, Jamieson G C. Plant Physiol. 1987;84:781–788. doi: 10.1104/pp.84.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil S D, Rhodes D, Russell B L, Nuccio M L, Shachar-Hill Y, Hanson A D. Plant Physiol. 2000;124:153–162. doi: 10.1104/pp.124.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi H, Alia, Mustardy L, Deshnium P, Ida M, Murata N. Plant J. 1997;12:133–142. doi: 10.1046/j.1365-313x.1997.12010133.x. [DOI] [PubMed] [Google Scholar]
- 26.Lerma C, Rich P J, Ju G C, Yang W J, Hanson A D, Rhodes D. Plant Physiol. 1991;95:1113–1119. doi: 10.1104/pp.95.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson A D, Rhodes D. Plant Physiol. 1983;71:692–700. doi: 10.1104/pp.71.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bligny R, Foray M-F, Roby R, Douce R. J Biol Chem. 1989;264:4888–4895. [PubMed] [Google Scholar]
- 29.Stephanopoulos G. Metab Eng. 1999;1:1–11. doi: 10.1006/mben.1998.0101. [DOI] [PubMed] [Google Scholar]
- 30.Kinney A J, Clarkson D T, Loughman B C. Biochem J. 1987;242:755–759. doi: 10.1042/bj2420755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T. Plant J. 1997;11:1115–1120. doi: 10.1046/j.1365-313x.1997.11051115.x. [DOI] [PubMed] [Google Scholar]
- 32.Holmes-McNary M Q, Cheng W L, Mar M H, Fussell S, Zeisel S H. Am J Clin Nutr. 1996;64:572–576. doi: 10.1093/ajcn/64.4.572. [DOI] [PubMed] [Google Scholar]
- 33.Mudd S H, Datko A H. Plant Physiol. 1989;91:587–597. doi: 10.1104/pp.91.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida I, Tashiro G, Sugiura M, Watanabe A. In: Advances in Plant Lipid Research. Sanchez J, Cerda-Olmedo E, Martinez-Force E, editors. Seville, Spain: University of Seville; 1998. pp. 255–258. [Google Scholar]
- 35.Miedema E, Richardson K E. Plant Physiol. 1966;41:1026–1030. doi: 10.1104/pp.41.6.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleshood H L, Pitot H C. J Biol Chem. 1970;245:4414–4420. [PubMed] [Google Scholar]
- 37.Jones A, Faulkner A, Turner J M. Biochem J. 1973;134:959–968. doi: 10.1042/bj1340959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinney A J. Curr Opin Plant Biol. 1998;1:173–178. doi: 10.1016/s1369-5266(98)80021-6. [DOI] [PubMed] [Google Scholar]