Abstract
Integrative analysis of whole-genome/exome-sequencing data has been challenging, especially for the non-programming research community, as it requires simultaneously managing a large number of computational tools. Even computational biologists find it unexpectedly difficult to reproduce results from others or optimize their strategies in an end-to-end workflow. We introduce Germline Mutation Scoring Tool fOr Next-generation sEquencing data (GeMSTONE), a cloud-based variant prioritization tool with high-level customization and a comprehensive collection of bioinformatics tools and data libraries (http://gemstone.yulab.org/). GeMSTONE generates and readily accepts a shareable ‘recipe’ file for each run to either replicate previous results or analyze new data with identical parameters and provides a centralized workflow for prioritizing germline mutations in human disease within a streamlined workflow rather than a pool of program executions.
INTRODUCTION
Next-generation sequencing (NGS) has significantly reduced the cost of obtaining genomic data for increasingly large sample sizes (1), facilitating discovery of causal genes and mutation candidates for various disorders (2), and providing sizable genetic variant datasets (3). As a result, the process of filtering, annotating and prioritizing variants from large-scale studies has grown in complexity and computational burden. It has become increasingly difficult to organize, maintain and standardize the variant analysis workflows, increasing the time and monetary investment for less computationally oriented biologists and labs. Some integrative frameworks (4–7) have been developed to enhance the reproducibility and accessibility of NGS studies. This same initiative inspired the framework for GeMSTONE: recording all analysis metadata for reproducible computational experiments, specifically focusing on germline mutation prioritization in human disease.
Although other platforms bring together different bioinformatics tools and allow users to schedule their analyzes online, none of them are built with an emphasis on streamlined single-run scheduling and automatic fetching of the necessary supplementary public data. Platforms like Galaxy (4), for instance, allow the user to combine many different tools from an impressive catalog, but require the user to reformat their data depending on the particular input format of the database or tool that they want to add to their analysis. A major design goal in the development of GeMSTONE is the ability to maximize customization for studies in a streamlined workflow rather than a pool of program executions. Within the GeMSTONE interface, databases required by the user-selected tools are pre-loaded and the user-input data will be automatically reformatted to fit query requirements. Therefore, adding an extra layer of analysis to any workflow requires minimal effort.
There is a large research community focusing on genetic variation study relating to human disease (8–18). This community often performs their analysis in-house rather than using any of the currently available tools for variant analysis. GeMSTONE facilitates the process of integrating and assessing evidence for causal inferences while automating the whole workflow in a reproducible way. Through its design GeMSTONE fills a significant gap in the online analysis landscape. GeMSTONE provides centralized workflows: embedding key features of variant prioritization for DNA sequencing data, focused on but not limited to germline mutations, with a collection of current bioinformatics tools and data libraries in a highly-customizable and reproducible manner. In short, we created GeMSTONE to organize, schedule, document and reproduce our variant analysis workflows from a single interface.
We show that the GeMSTONE workflow is consistent with consensus guidelines for interpreting sequence variants in human disease (19,20) (Supplementary Table S1). A demo study is fully described and explained as it is designed, scheduled and analyzed through the chained GeMSTONE functionalities (http://gemstone.yulab.org/manual.html); we also demonstrate its feasibility and efficiency in a proof-of-concept case by recapitulating results of a published variant analysis (9).
MATERIALS AND METHODS
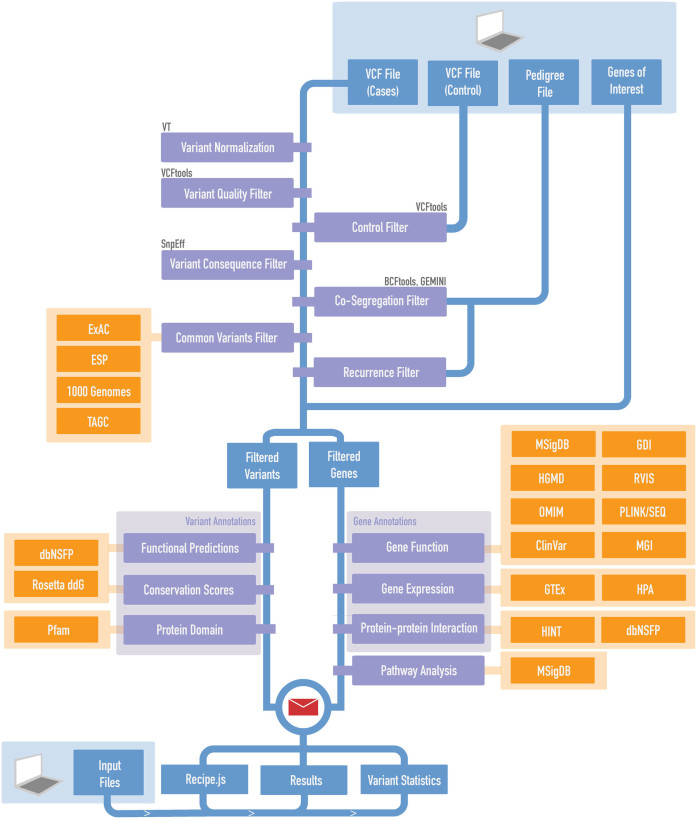
GeMSTONE serves as an online variant prioritization framework that leverages seven popular bioinformatics suites [VT (21), VCFtools (22), BCFtools (23), SnpEff (24), GEMINI (25), dbNSFP (26) and PLINK/SEQ (27)] in connection to 46 meta-information and prediction resources (Figure 1; Supplementary Table S2) to provide a smooth, customizable workflow for variant analysis.
Figure 1.
GeMSTONE pipeline overview. The schematic represents the GeMSTONE's central analysis pipeline. The fundamental backbone filter cascade can be seen in blue, prioritizing rare and putatively damaging variants and genes. Different libraries are grouped in orange, used in annotation or filtering steps throughout the workflow as indicated.
Users of the GeMSTONE web portal can customize their analyzes of genomic data from Variant Call Format (VCF) files by using tools from a range of different classes (Figure 1). These include (i) variant normalization for unified representation of genetic variants using VT, (ii) variant/genotype quality filters on matrices encoded in the VCF file such as QUAL (Phred-scaled quality score), GQ (genotype quality), DP (read depth) and filter status using VCFtools, (iii) variant type filters on variant consequence and transcript biotype based on SnpEff annotations, (iv) common variant filter on allele frequency in the general population [ExAC (28), 1000 Genomes (29), ESP6500 (30) and TAGC (31)], (v) variant function filters on predicted damaging effects [18 methods (e.g. Polyphen-2 (32), SIFT (33), CADD (34)) complied in dbNSFP (Supplementary Table S2), Rosetta ddG (35)] and protein domains [Pfam (36)], and (vi) comprehensive annotations (and filters) on gene and gene product attributes [Gene Ontology (37)], biological pathways [KEGG (38), BioCarta (39) and Reactome (40) complied in MSigDB (41)], human disease association [HGMD (42), ClinVar (43), OMIM (44)] and mouse model knockout phenotypes [MGI (45)], gene-based scores on accumulated mutational damage [GDI (46)] and genic intolerance [RVIS (47)], gene expression [GTEx (48), HPA (49)], protein–protein interaction network [IntAct (50), BioGRID (51) and ConcesusPathDB (52) complied in dbNSFP, and HINT (53)], and (vii) pathway enrichment analysis using a fisher exact test. Users may also choose to include supplementary files, such as a pedigree (PED) file for co-segregation analysis, a list of genes for personalized annotation, or a second VCF file with a control cohort for genetic association tests [BURDEN (27), Calpha (54), vt (55) and SKAT (56) implemented in PLINK/SEQ]. All these options come together to provide a holistic filtering, annotation and prioritization pipeline (Figure 1).
The customized pipeline is then scheduled for processing on a protected server, alleviating the user's burden to update software, parse data libraries, store large derivative files and dedicate processing time. The web server and database server are running as virtual machines (VMs) on shared physical infrastructure. Both the database and web host VMs can be expanded or moved to an upgraded physical machine or granted more resources in their current depending on demand, making the hardware setup easily scalable to more traffic. The average turnaround time is about 11 min for a 1MB VCF input file containing ∼13,800 variants under default settings, of which querying up to 18 in silico predictions takes a static 8 min searching through 76GB dbNSFP database on all chromosomes. Although the processing time will vary depending on the choice of options and the number of concurrent users, GeMSTONE in general can handle a single ∼500M VCF input per run within 1 day. Once the job is finished, the user can log into the GeMSTONE portal to interact with the completed workflow by selectively downloading step-by-step snapshots of their workflow, interactively visualizing their variant statistics and downloading their recipe (JSON) file, which can be uploaded or shared to replicate or modify the same workflow.
An essential design to reinforce GeMSTONE's reproducibility function and to ensure the sustainability of our web tool is our rigorous versioning system. We keep in our system static versions of all the external resources, where all the tools and datasets that we use for GeMSTONE are loaded onto our server so that it does not go to any external program or server when running. Thus we are able to ensure backward compatibility as we add updated versions of software or new tools. GeMSTONE records the versions of each tool and database used in a job in the recipe file and if users submit a recipe whose workflow uses older software or datasets, they will be prompted on the fly asking whether they want to use the legacy version or the latest version of the resources. GeMSTONE also records the versions in a human-readable summary file for easy access and reference.
One important function for germline mutation prioritization in human disease is GeMSTONE's co-segregation analysis, which provides six common inheritance models (autosomal dominant, autosomal recessive, recessive compound heterozygous (via GEMINI (25)), X-linked dominant, X-linked recessive and Y-linked dominant) based on the user-defined pedigree structure in PED file. GeMSTONE screens sample genotypes (using BCFtools (23)) in each family and seeks for variants that are co-segregating with disease status under selected mode of inheritance. Additionally, a recurrence filter constrains the degree to which co-segregation events are allowed across multiple families and the prevalence of the variants in sporadic samples. We found this option to be seldom implemented by previous web tools yet often recommended by American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) (20). The benefits of this analysis are many-fold: (i) increasing segregation data in families or (ii) high mutation frequency affecting multiple sporadic cases suggests stronger evidence for pathogenicity; (iii) whereas a upper limit of such recurrence can help eliminate potential false positives in large samples. This process of user-driven development by which GeMSTONE morphs to the community's needs is the key behind GeMSTONE's ability to grow as a knowledge bank with a robust and updated set of functionalities. Small but necessary prioritizing steps like these, now explicitly documented in the GeMSTONE summary and recipe files, can become an active component of study replication.
Another supporting evidence for disease association comes from in silico predictions of variant functional effects. Predictions from different algorithms are considered as a single piece of evidence in sequence interpretation in part due to the underlying similarities in the basis in these software suites (19,20). GeMSTONE's variant functional prediction step allows the user to choose up to 19 different in silico predictors (Supplementary Table S2) with customizable thresholds. More dedicatedly, a ‘global deleteriousness filter’ allows users to set a threshold on the number of selected predictors needed for a variant to pass the filter. This set of filters is useful in that it allows users to adjust the stringency of each algorithm while balancing and investigating any inconsistency among different predictions. The availability of these filters and annotations also provide an environment in which users can choose predictive metrics solely based on their relative merit rather than the programming investment that it would take to install, query and customize them for a study.
Most options within the GeMSTONE workflow can serve dual purposes, acting as either filters or annotations. For the ‘global deleteriousness filter’ mentioned above, the count of deleterious predictions and their individual scores will be annotated next to each variant, providing information that can be used for variant prioritization without being part of any filter. We also provide the option to combine information across libraries, for example, we allow for known disease gene annotation on candidates to be supplemented with their interaction partners as reported in other databases, asking whether those interactors were previously implicated in the disease of interest. This distribution and coverage of tools (Figure 1) have never been collected and connected in a centralized workflow before.
By maintaining an updated set of bioinformatics tools for variant analysis, GeMSTONE decreases the barrier to entry, for less computationally oriented research groups and establishes a central bioinformatics hub for researchers who study sequence variants implicated in severe familial diseases as well as rare, large-effect risk variants in complex disease. The options offered by the web interface also serve as a way for users to explore and learn about new tools and data sources while providing developers with an overview of the current variant analysis landscape to fill any gaps in the current tool-space. New tools can be easily added to GeMSTONE and presented to the community through the web interface, removing platform-specific barriers.
RESULTS
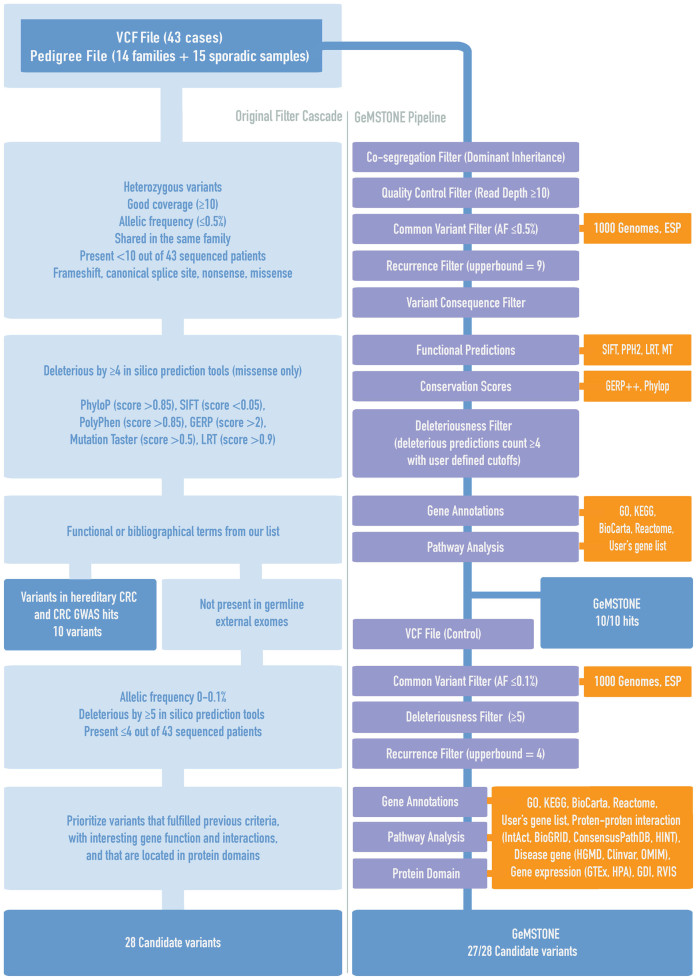
As an example of a GeMSTONE use case, we replicated a published analysis of rare pathogenic variants in new predisposition genes for familial colorectal cancer (CRC) (9). A side-by-side demonstration of the study's workflow and GeMSTONE's reimplementation using the same dataset and prioritization criteria is shown in Figure 2. The original analyzes were conducted in two sequences of prioritization, progressively looking for predisposing mutations with stronger evidence for causality to CRC as they underwent increasingly stringent criteria (lower allele frequency in general populations; rarer presence among the affected samples; more deleterious molecular impact by in silico predictions; more interesting biological functions of the genes and their protein product, e.g. domains and interactions) (9). While formerly requiring in-house scripting for co-segregation analysis, in silico analysis and a series of gene function annotations querying and parsing several databases, the entirety of each sequence of prioritization pipeline can be performed with a single run through our interactive, lightweight web form using GeMSTONE.
Figure 2.
Recapitulation of a published colorectal cancer (CRC) study. As a proof-of-concept case study, GeMSTONE recapitulated every step in the original Colorectal-cancer prioritization workflow1, rescuing 27 out of 28 candidate variants from the ∼30,000 variants in the raw whole exome sequencing dataset and hitting all hereditary CRC and CRC GWAS variants.
Perhaps the most convenient feature within GeMSTONE is its recipe file generator. The recipe file from any given run can be shared and readily uploaded to our site to modify any part of the filtering and annotation pipeline for more stringent prioritization in a follow-up run. Once uploaded, the recipe file (JSON) will populate the web form dynamically, giving the user the ability to modify the run using the same interface that created it. In our CRC case, we lowered the upper-bound of allele frequency filter from 0.5% to 0.1% [in 1000 Genomes (29) and ESP6500 (30)] and recurrence filter from 9 to 4, requiring variants to be present in ≤4 individuals in our dataset. Next, we increased the lower-bound of deleteriousness filter from 4 to 5 without changing the user-defined deleterious thresholds of any single predictor [PhyloP (33) score >0.85, SIFT (57) score <0.05, PolyPhen-2 (32) score >0.85, GERP++ (58) score >2, Mutation Taster (59) score >0.5 and LRT (60) score >0.9]. Finally, we added variant and gene annotations with interesting gene function, interactions and locations in protein domains. This workflow leverages a variety of public databases, including Gene Ontology (37), KEGG (38), Reactome (40), HINT (53), Pfam (36) and HGMD (42), as well as a complementary list of cancer terms collected by the authors. This modified workflow was automatically recorded in a JSON recipe file and packaged with corresponding results and intermediate output files. Through the above two automated runs, GeMSTONE recapitulated every step of the original prioritization workflow. A total of 27 out of 28 candidate variants were identified (the missing variant was filtered out due to slightly higher allele frequency in a sub-population database from 1000 Genomes), as well as all hereditary CRC and CRC Genome-wide Association Study (GWAS) variants (9) (Figure 2).
DISCUSSION
GeMSTONE provides a code-free portal for variant filtering, annotation and prioritization, which not only helps standardize genetic variation analyzes (Supplementary Table S1) but also offers the means to replicate and share computational protocols easily. From a user's perspective, GeMSTONE is a reliable one-stop shop for variant analysis where they can find a collection of tools spanning a broad range of applications through an intuitive, unified user interface subsuming all general-purpose workflows from comparable toolkits (Figure 3).
Figure 3.
Heatmap comparison of GeMSTONE and other variant prioritization tools. This heatmap compares with other tools that have similar objectives on the aspects of (A) raw data inputs and prioritization, (B) knowledge-based annotation from external data resources and libraries, (C) inheritance models for co-segregation analysis and (D) strategy of reproducibility. Each row represents a different tool, while each column represents a specific feature. Dark blue indicates that a tool has similar capacity for a specific function while light blue indicates that a tool has a similar feature but with less powerful functionality than GeMSTONE (see Discussion).
Although currently most of other variant prioritization tools accept VCF and pedigree files as inputs and can perform routine filtering on quality control and variant consequence (Figure 3A), GeMSTONE stands out as a more powerful tool by including annotations at the variant, gene, pathway and network level (Figure 3B) and co-segregation analysis using different inheritance models for potential germline mutation prioritization (Figure 3C). We consider certain features in GeMSTONE to be ‘more powerful’ in the aspect of comprehensiveness or/and flexibility: GeMSTONE often provides more comprehensive options for filtering and annotation linking to external resources than others and most of the GeMSTONE options flexibly allow for annotation or filtering, or both. See detailed reasons in Supplementary Table S3.
A keystone of GeMSTONE is the recipe file (Figure 3D), which records all workflow parameters in a single file that can be shared and uploaded onto the site to reproduce a previous run. The recipe file can be used to (i) replicate results by rerunning the same workflow on the same dataset, (ii) process new data with a known workflow or (iii) modify parameters in a known workflow to evaluate study design. This approach has the potential to bring more transparency and openness to the bioinformatics community by enhancing the reproducibility of large-scale genomic studies.
CONCLUSIONS
GeMSTONE allows for accessible, collaborative, replicable and holistic analysis of genetic variants. First, it seamlessly knits together filters and annotations through different tools with either stringent, study-specific parameters or general best-practice settings. Second, it eliminates the time and space burdens associated with modern variant analysis tools, saving users dozens of gigabytes of potential disk space per run for the same workflow on a medium-sized dataset. Third, it significantly lowers the barrier to entry for traditional biologists by eliminating the installation and scripting sinkholes that may dissuade researchers from pursuing large-scale analysis or trying new tools. Fourth, it provides a readable, shareable log—both programmatic and human—to allow other researchers to understand and replicate study results given the same starting data. Finally, GeMSTONE encourages the growth of the genomics research community by maintaining and updating a bank of best-practice bioinformatics methods and tools. We expect our GeMSTONE will greatly aid in automating the (re)analysis of genome-wide genetic variation data and enhance the reproducibility of large-scale genomic studies.
DECLARATIONS
Availability of data and material
Exome sequence data for 43 CRC patients were provided by Esteban-Jurado et al. (9) through private communication.
Ethics approval
Ethics approval was not needed for this study.
Supplementary Material
ACKNOWLEDGEMENTS
IDIBAPS: we are sincerely grateful to the patients and their families for their participation. We are thankful to the Centre Nacional d’Anàlisi Genòmica and the Biobank of Hospital Clínic–IDIBAPS, Barcelona, for technical help. The work was carried out (in part) at the Esther Koplowitz Centre, Barcelona.
BRC: we are thankful to the Cornell University Biotechnology Resource Center for providing facilities' resources and services, and we would like to thank James VanEe for his invaluable support throughout the project.
Authors’ contributions: X.W. and H.Y. conceived of the GeMSTONE concept and oversaw all aspects of the study. S.C. designed and implemented the GeMSTONE pipeline with supervision and input from H.Y. and X.W. J.F.B. designed and implemented the GeMSTONE web interface. C.E.J., S.F.E. and S.C.B. provided CRC study datasets and the original study design. J.F.B., S.C. and H.Y. wrote the manuscript. All authors reviewed and approved the final manuscript as submitted.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [R01 GM097358, R01 GM104424, R01 GM108716]; National Cancer Institute [R01 CA167824]; Eunice Kennedy Shriver National Institute of Child Health and Human Development [R01 HD082568]; National Human Genome Research Institute [UM1 HG009393]; Simons Foundation Autism Research Initiative [367561 to H.Y.]; Fondo de Investigación Sanitaria/FEDER [14/00173]; Fundación Científica de la Asociación Española contra el Cáncer [GCB13131592CAST]; COST Action BM1206, Beca Grupo de Trabajo ‘Oncología’ AEG (Asociación Española de Gastroenterología); Agència de Gestió d’Ajuts Universitaris i de Recerca [Generalitat de Catalunya, 2014SGR255]; CIBERehd by the Instituto de Salud Carlos III (to C.E.J., S.F.E). Funding for open access charge: National Institutes of Health; Simons Foundation Autism Research Initiative [367561 to H.Y.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010; 11:31–46. [DOI] [PubMed] [Google Scholar]
- 2. Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E.. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 2013; 14:681–691. [DOI] [PubMed] [Google Scholar]
- 3. Nekrutenko A., Taylor J.. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet. 2012; 13:667–672. [DOI] [PubMed] [Google Scholar]
- 4. Goecks J., Nekrutenko A., Taylor J., Galaxy T.. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010; 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P.. GenePattern 2.0. Nat. Genet. 2006; 38:500–501. [DOI] [PubMed] [Google Scholar]
- 6. Halbritter F., Vaidya H.J., Tomlinson S.R.. GeneProf: analysis of high-throughput sequencing experiments. Nat. Methods. 2011; 9:7–8. [DOI] [PubMed] [Google Scholar]
- 7. Lushbough C., Bergman M.K., Lawrence C.J., Jennewein D., Brendel V.. BioExtract server–an integrated workflow-enabling system to access and analyze heterogeneous, distributed biomolecular data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010; 7:12–24. [DOI] [PubMed] [Google Scholar]
- 8. Farlow J.L., Robak L.A., Hetrick K., Bowling K., Boerwinkle E., Coban-Akdemir Z.H., Gambin T., Gibbs R.A., Gu S., Jain P. et al. . Whole-exome sequencing in familial Parkinson disease. JAMA Neurol. 2016; 73:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esteban-Jurado C., Vila-Casadesus M., Garre P., Lozano J.J., Pristoupilova A., Beltran S., Munoz J., Ocana T., Balaguer F., Lopez-Ceron M. et al. . Whole-exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genet. Med. 2015; 17:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey J.N., Patterson C., de Nijs L., Duron R.M., Nguyen V.H., Tanaka M., Medina M.T., Jara-Prado A., Martinez-Juarez I.E., Ochoa A. et al. . EFHC1 variants in juvenile myoclonic epilepsy: reanalysis according to NHGRI and ACMG guidelines for assigning disease causality. Genet. Med. 2017; 19:144–156. [DOI] [PubMed] [Google Scholar]
- 11. Bellido F., Pineda M., Aiza G., Valdes-Mas R., Navarro M., Puente D.A., Pons T., Gonzalez S., Iglesias S., Darder E. et al. . POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 2016; 18:325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackay D.S., Bennett T.M., Culican S.M., Shiels A.. Exome sequencing identifies novel and recurrent mutations in GJA8 and CRYGD associated with inherited cataract. Hum. Genomics. 2014; 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Medeiros A.M., Alves A.C., Bourbon M.. Mutational analysis of a cohort with clinical diagnosis of familial hypercholesterolemia: considerations for genetic diagnosis improvement. Genet. Med. 2016; 18:316–324. [DOI] [PubMed] [Google Scholar]
- 14. Cox S.N., Pesce F., El-Sayed Moustafa J.S., Sallustio F., Serino G., Kkoufou C., Giampetruzzi A., Ancona N., Falchi M., Schena F.P. et al. . Multiple rare genetic variants co-segregating with familial IgA nephropathy all act within a single immune-related network. J. Intern. Med. 2017; 281:189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Radovica-Spalvina I., Latkovskis G., Silamikelis I., Fridmanis D., Elbere I., Ventins K., Ozola G., Erglis A., Klovins J.. Next-generation-sequencing-based identification of familial hypercholesterolemia-related mutations in subjects with increased LDL-C levels in a latvian population. BMC Med. Genet. 2015; 16:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mackay D.S., Bennett T.M., Shiels A.. Exome sequencing identifies a missense variant in EFEMP1 co-segregating in a family with autosomal dominant primary open-angle glaucoma. PLoS One. 2015; 10:e0132529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Einarsdottir E., Svensson I., Darki F., Peyrard-Janvid M., Lindvall J.M., Ameur A., Jacobsson C., Klingberg T., Kere J., Matsson H.. Mutation in CEP63 co-segregating with developmental dyslexia in a swedish family. Hum. Genet. 2015; 134:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T. et al. . Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015; 385:1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. et al. . Guidelines for investigating causality of sequence variants in human disease. Nature. 2014; 508:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. et al. . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015; 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan A., Abecasis G.R., Kang H.M.. Unified representation of genetic variants. Bioinformatics. 2015; 31:2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. . The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paila U., Chapman B.A., Kirchner R., Quinlan A.R.. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013; 9:e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Jian X., Boerwinkle E.. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011; 32:894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. PLINK/SEQ 2014; https://atgu.mgh.harvard.edu/plinkseq/.
- 28. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Tukiainen T.. Analysis of protein-coding genetic variationin 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A.. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J. et al. . Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013; 493:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carmi S., Hui K.Y., Kochav E., Liu X., Xue J., Grady F., Guha S., Upadhyay K., Ben-Avraham D., Mukherjee S. et al. . Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates jewish and european origins. Nat. Commun. 2014; 5:4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R.. A method and server for predicting damaging missense mutations. Nat. Methods. 2010; 7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rohl C.A., Strauss C.E., Misura K.M., Baker D.. Protein structure prediction using Rosetta. Methods Enzymol. 2004; 383:66–93. [DOI] [PubMed] [Google Scholar]
- 36. Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. et al. . Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. . Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M.. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014; 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nishimura D. BioCarta. Biotech. Softw. Internet Rep. 2004; 2:117–120. [Google Scholar]
- 40. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al. . The reactome pathway knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N.. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014; 133:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A.. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015; 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blake J.A., Eppig J.T., Kadin J.A., Richardson J.E., Smith C.L., Bult C.J. the Mouse Genome Database, G. . Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017; 45:D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Velez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G. et al. . The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13615–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B.. The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet. 2015; 11:e1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. . Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
- 50. Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A. et al. . IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004; 32:D452–D455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O’Donnell L. et al. . The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015; 43:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kamburov A., Wierling C., Lehrach H., Herwig R.. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009; 37:D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das J., Yu H.. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 2012; 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J.. Testing for an unusual distribution of rare variants. PLoS Genet. 2011; 7:e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R.. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010; 86:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X.. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011; 89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar P., Henikoff S., Ng P.C.. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009; 4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 58. Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S.. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010; 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D.. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010; 7:575–576. [DOI] [PubMed] [Google Scholar]
- 60. Chun S., Fay J.C.. Identification of deleterious mutations within three human genomes. Genome Res. 2009; 19:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.