Abstract
Background
Informaticians at any institution that are developing clinical research support infrastructure are tasked with populating research databases with data extracted and transformed from their institution’s operational databases, such as electronic health records (EHRs). These data must be properly extracted from these source systems, transformed into a standard data structure, and then loaded into the data warehouse while maintaining the integrity of these data. We validated the correctness of the extract, load, and transform (ETL) process of the extracted data of West Virginia Clinical and Translational Science Institute’s Integrated Data Repository, a clinical data warehouse that includes data extracted from two EHR systems.
Methods
Four hundred ninety-eight observations were randomly selected from the integrated data repository and compared with the two source EHR systems.
Results
Of the 498 observations, there were 479 concordant and 19 discordant observations. The discordant observations fell into three general categories: a) design decision differences between the IDR and source EHRs, b) timing differences, and c) user interface settings. After resolving apparent discordances, our integrated data repository was found to be 100% accurate relative to its source EHR systems.
Conclusion
Any institution that uses a clinical data warehouse that is developed based on extraction processes from operational databases, such as EHRs, employs some form of an ETL process. As secondary use of EHR data begins to transform the research landscape, the importance of the basic validation of the extracted EHR data cannot be underestimated and should start with the validation of the extraction process itself.
Keywords: extract transform load, electronic health record, correctness, clinical data warehouse, informatics
1. Introduction
The widespread adoption of Electronic Health Records (EHR) offers great potential for clinical translational research through reuse of the data. As federal funding agencies heavily incentivize this reuse of EHR data, the conduct of clinical research will be greatly affected. A major caveat however is that EHR systems were not designed to be used for research. While it may be debated whether “data shall only be used for the purpose for which they were collected” (1) or whether that data simply needs to meet the criteria of “fitness for use,” (2) EHR data were collected to support healthcare clinical decision making and not for research purposes. Unless its data are carefully validated for such repurposing, the integrity of the research results generated from it may be questionable at best.
A critical step in ensuring the validity of research is making sure the data are ‘correct.’ Correctness is one of the five dimensions of data quality put forth by Weiskopf and Weng in assessing the fitness of EHR data for its reuse for research. Their meta-analysis evaluated how 60 studies assessed correctness in the reuse of EHR data. For example, the definition of correctness suggested by Hogan and Wagner is summarized as the “proportion of data elements present that are correct.” Weiskopf and Weng found that the most common method used for assessing correctness was comparison of EHR data to some gold standard. (3)
The purpose of this study was to validate the correctness of the West Virginia Clinical and Translational Science Institute (WVCTSI)’s Integrated Data Repository (IDR) data elements. In this study we evaluated the IDR using the EHR as the gold standard in order to validate the correctness of the extract, transform and load (ETL) process used in migrating the data from the EHR sources to the IDR target. To do this, we used a two-step process in which we randomly selected data from a subset of patients and compared them to the EHR databases from which they were extracted.
2. Materials and Methods
The WVCTSI IDR is a comprehensive clinical data warehouse, first deployed in June 2012. Currently, it contains observations on approximately 2 million patients, that is, information such as lab tests, medications, diagnoses and procedures as well as demographic data including but not limited to patient age, race and gender. The IDR contains over 250 million observations, captured from records of both inpatient and outpatient visits. The IDR uses the widely-employed database model of the i2b2 (Informatics for Integrating Biology and the Bedside) platform to store data. The i2b2 platform was designed by Partners Health System in conjunction with Harvard University faculty as part of an NIH-supported effort to develop a scalable informatics framework for translational research. This framework has been adopted by many major research institutions (4) and has become a standard tool used to support cohort discovery, clinical trial recruitment and hypothesis generation.
The IDR currently includes data from two sources, West Virginia University Healthcare’s (WVUH)’s EpicCare and Medsite systems. WVUH is a multi-hospital entity, with over sixty affiliated physician practices and clinics, whose largest facility is Ruby Memorial Hospital, a 531 bed tertiary care hospital and Level One Trauma Center. The EpicCare application, from Epic Systems Corporation, provides WVU Healthcare with a full suite of integrated financial and clinical applications. The EpicCare application was implemented in 2008. Prior to that time, WVUH used the Medsite application as its EHR. Medsite was developed in-house and was in full scale usage by staff and clinicians from late 1998 until the Epic EHR implementation (March 2008). Medsite captured and integrated data from WVU Healthcare’s inpatient and outpatient registration systems as well as ancillary systems such as laboratory, radiology and cardiology.
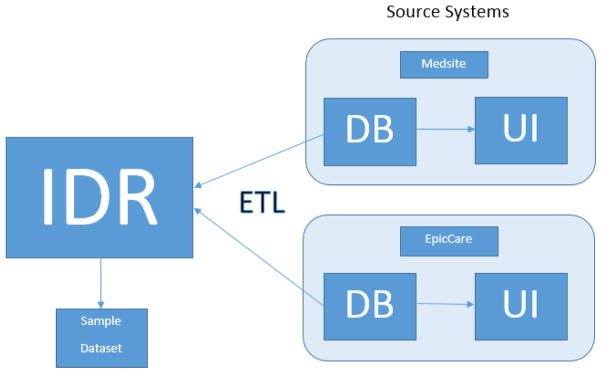
The WVCTSI’s IDR was developed by an extract, transform and load (ETL) process [Figure 1]. In this ETL process, data was first extracted from the source systems’ databases (in this case, the Medsite and EpicCare applications); second, the extracted data was then transformed to make it accommodate the requirements of the IDR; and, third, once transformed the data was then loaded into the IDR’s database. The ETL process was designed and developed entirely by the WVCTSI’s Biomedical Informatics staff, including MJD, who were provided access to WVUH’s Medsite and EpicCare data via direct Oracle database to Oracle database links. The ETL software was developed using Oracle’s PL/SQL programming language and its Integrated Development Environment tool, SQL Developer. The initial ETL development began in early 2011 with Medsite’s data and was completed with the first extract, transform and load of EpicCare data in mid-2012. The on-going ETL process is designed to run quarterly against WVUH’s EpicCare database.
Figure 1. Overview of ETL Process and Sample Dataset Extraction.
IDR data extracted, transformed, and loaded (ETL) from source systems (Medsite and EpicCare). Sample dataset generated from IDR for comparison with source systems. Comparison done by Health Information Management staff member who used the source system’s user interfaces (UI) to validate data contained within their respective databases (DB).
Occasions for error occur in all three steps of the ETL process. For example, during the extraction phase, a field may be extracted incorrectly such as a secondary diagnosis being inadvertently selected as a primary diagnosis. In the transform phase, many opportunities for error exist, as the ETL software makes the source systems’ data “fit” the needs of the IDR’s display and reporting requirements. Observational data, such as laboratory results, have to be categorized so that they can be used within the ontologies or structured hierarchies of standardized terminologies. So, if the identifying terms for laboratory results are locally developed, they may need to be translated into a standardized terminology such as LOINC (Logical Observation Identifiers Names and Codes). Finally, in the load phase of the ETL process in which the extracted and transformed data is placed in the IDR’s data structures, errors can occur that are, in effect, the mirror image of those that might happen in the extract phase; for example, a primary diagnosis is placed in a field defined as reserved for the secondary diagnosis.
Our goal was to match data obtained for the IDR to a “gold standard” in order to evaluate correctness. For our purposes, the gold standard was the data contained in the EpicCare and Medsite applications. As stated above, wanting different types of observations, we chose five commonly searched types: laboratory results, medication, diagnosis, procedure, and race. We assumed at minimum 95% correctness, thus collecting 500 observations would yield a margin of error of ±2%, at five observations from 100 patients. The method employed was to extract the random observations for evaluation by WVU’s Department of Health Information Management (HIM)’s data integrity coordinator (JLA), who is responsible within HIM for reviewing, researching and resolving medical records data validity and correctness issues. First, patients were randomly selected using ORACLE 11g’s “sample” function with a specified percentage to get the desired number of patients. Next, ORACLE 11g’s dbms_random package’s “value” function was used to select five observations (one each from laboratory results, CPT procedure codes, race, ICD-9-CM diagnoses, and medications) per patient. Initially, we randomly selected five unique patients and then five observations per patient to determine quickly if any major issues existed with our method of selecting patients and observations within patients, the IDR extracted data, or the HIM verification process. After the initial 25 observations were evaluated and issues resolved as outlined below, we then selected 100 patients with five observations per patient. Once these data were extracted, the data were sent to the HIM data integrity coordinator for verification who then determined the correctness of the data by comparing the randomly selected IDR observations to data she observed via the EpicCare or Medsite user interfaces. If differences were observed, they were resolved on a case-by-case basis to determine where the discrepancies occurred.
3. Results
As a first step in the verification process, we submitted five patients with five observations each for a total of 25 observations. This process identified several naming issues, e.g. generic names vs. brand names and lab result codes vs. expanded names. Once these naming issues were resolved in favor of the EpicCare and Medsite naming conventions, we proceeded to the main validation phase.
For the main phase, we randomly selected 100 patients with five observations each. Two of these patients had only four observations. As this reduction of observations only has very minimal effect on margin of error, no additional patients were selected.
Thus, 498 observations were submitted to the HIM data integrity coordinator for validation via the user interfaces to the EpicCare or Medsite applications. On review by the coordinator, there were 479 matches and 19 initially unmatched observations. These initial unmatched observations were reviewed further to determine the reason for the discrepancies. The discrepancies were of three types: (1) design decision differences, (2) timing issues and (3) reviewer user interface settings. The design decision discrepancy type accounted for a majority (16) of the differences and were the result of either ETL features that excluded certain data or EHR user interface features that caused certain data to not be displayed. The timing issue accounted for one discrepancy and is somewhat inevitable given the nature of the ETL process in relation to the operational, up-to-date EHR. Reviewer user interface settings accounted for the remaining two discrepancies and are often initially “invisible” as they are a by-product of the EHR’s security and privacy configurations. Once these discrepancies were resolved, there was no discordance between the sample dataset and the EpicCare and Medsite source systems. See Table 1 for how these discrepancies were resolved.
Table 1.
Resolution of initially unmatched observations
| Observation Type | n | Resolution | IDR Correct? | Discrepancy Type |
|---|---|---|---|---|
| Lab Result | 5 | EHR used lab IDs while IDR merged these to patient’s MRN | Yes | Design Decision |
| Race | 1 | Field was empty in EHR at time of ETL but updated before validation | Yes | Timing Issue |
| Medication | 8 | Route of admin deliberately not captured by IDR but displayed in EHR | Yes | Design Decision |
| Diagnosis | 1 | EHR listed diagnosis twice for same date; IDR considers this just one observation as it occurs on the same date | Yes | Design Decision |
| Lab Result | 1 | IDR observation order date (near midnight) confused with EHR collection date (of the following day) | Yes | Design Decision |
| Diagnosis | 1 | IDR observation found in EHR after user’s account settings modified | Yes | Reviewer Setting |
| Medication | 1 | IDR observation found in EHR after user’s account settings modified | Yes | Reviewer Setting |
| Lab Result | 1 | IDR observation order date (near midnight) confused with EHR collection date (of the following day) | Yes | Design Decision |
EHR=electronic health record ID=identification IDR=Integrated Data Repository MRN=medical record number ELT=extract load transform
4. Discussion
The Electronic Health Record of today has its origins in the hospital systems of the 1960s, designed to serve billing and accounting purposes.(5, 6) Later, in the 1970s and 80s, systems oriented toward clinical decision-making, such as radiology results reporting, were developed as generally stand-alone applications. (5, 6) Then in the late 1990s, as previously stand-alone applications began to be integrated into larger offerings, software vendors started to market the notion of the complete “electronic healthcare record” (EHR) as a solution to healthcare’s integration needs.(5, 6) These were billed as comprehensive applications that included everything from patient registration through order communications to billing and accounting, all under one software roof, so to speak. As EHRs became more common, it became increasingly clear that huge amounts of electronic data were available for purposes other than clinical and financial decision making and support. These data were a byproduct of operational systems, but could also provide researchers with clinical and demographic data on a scale previously unimaginable. However, it is important to remember that the typical EHR’s primary purpose remains to provide billing, reimbursement, and clinical care support. This wealth of data is segmented off in transaction-based systems that are used for operational purposes. Consequently this data must be extracted, transformed, and loaded in order for it to be potentially useful for investigators.
To our knowledge, outside of natural language processing studies, we are the first to report on a validation study of the data extracted, transformed and loaded from a healthcare institution’s operational database into a large-scale, health and clinical data warehouse designed for research; Table 2 illustrates the PubMed search strategies used and their results. Per Logan (2001), our results suggest confidence in the correctness of the IDR’s data, i.e. that the integrity of the EHR data was maintained during the IDR’s ETL process. (7) It was not our purpose to examine the accuracy or correctness of the data in the EHR itself in this effort, but whether data extracted from the source systems actually matched the data in the IDR. Multiple papers in the past several years examined this issue of correctness of EHR data. (3, 8, 9) However, as typical clinical data warehouses such as the IDR extract data from multiple sources, it is important to ensure that there is no breakdown in the integrity of the data during the ETL process since each instance of data migration introduces the possibility of extraction and transcription errors.
Table 2.
PubMed Search Strategy for Clinical Operational Database to Clinical Data Warehouse Extract, Transform, Load (ETL) Validation Studies
| PubMed Search Criteria | Results | Examination |
|---|---|---|
| ETL [All Fields] AND extract [All Fields] AND transform [All Fields] and load [All Fields] | 11 articles | None of the articles evaluated the validation of the ETL process from a healthcare institution’s operational database (EHR) to a clinical data warehouse |
| clinical [All Fields] AND research [All Fields] AND database [All Fields] AND accuracy [All Fields] AND validation [All Fields] | 231 articles | With the exception of natural language processing studies, none of the articles evaluated the validation of the ETL process from a healthcare institution’s operational database (EHR) to a clinical data warehouse |
Ensuring the integrity of this ETL process was the first step in our validation studies of the clinical warehouse data for research. It should be noted that while we were able to validate the integrity of the ETL process, our initial comparison of the ETL against the EHR did find apparent discrepancies that were later determined to be either a result of design decisions made by the ETL process itself or the designers of the EHR’s user interface or timing issues between the time of the ETL process and the operational EHR or the EHR’s user interface privacy and security settings. Such apparent discrepancies can cause some confusion and delays during the validation process as they are sorted out and resolved.
4.1 Strengths and Limitations
This study is our phase 0 of a multi-phase validation study of our clinical warehouse. Though future phases will include validation of disease phenotype identification developed for EHRs, our focus in this current study has been on validating the ETL process which populates that data warehouse. There has been little research in validating the ETL-processed data used for secondary research. By using a random sample of patients and observations, we have minimized potential selection bias. By having a validator (from HIM) who is external to WVCTSI’s Biomedical Informatics, there is limited potential for observer bias.
The assumption that the EHR is a gold standard can be seen as limitation. However, our phase 0 did not depend on the actual correctness of the source systems’ data, just that the IDR was correct relative to those source systems. The accuracy of the source systems’ data relative to the patient is beyond the scope of this study but will be examined in future studies. Finally, while the IDR data is accurate with regard to its source data, choices made during the ETL process may affect researcher query results. Informatics professionals need a thorough understanding of source data and the ETL process in order to provide researchers with the most accurate data sets.
4.2 Conclusion
Any institution that uses a clinical data warehouse that is developed based on extraction processes from operational databases, such as EHRs, employs some form of an ETL process. It is important to validate this process. Our study validated the correctness of the WVCTSI IDR data extracted via an ETL process from the EpicCare and Medsite source systems. After resolving apparent discordances, the WVCTSI IDR was found to be 100% correct relative to the source systems. This result will insure confidence in our, and others’, subsequent studies using the WVCTSI IDR. The push to use EHR data for secondary analysis is immense in both research funding and efficiency. As secondary use of EHR data begins to transform the research landscape, the importance of the basic validation of the extracted EHR data cannot be underestimated and should start with the validation of the extraction process itself. Such validation should be part of an iterative quality control process for all clinical data warehouses.
Summary Table.
What was already known on the topic (2–4 points)
Secondary use of EHR data is transforming the research landscape
EHR data were collected to support healthcare clinical decision making, not for research purposes
Correctness is a dimension of data quality that determines the fitness of EHR data for its repurposing in research
What this study added to our knowledge (2–4 points)
A simple reproducible outline of validating the ETL process is presented
While data from clinical data warehouses may be accurate with regard to its source data, choices made during the ETL process may affect researcher query results.
Acknowledgments
The authors would like to thank Charles Mullett, MD and Abhishek Vishnu, MD, PhD for their helpful feedback on this manuscript. This work was supported in part by National Institutes of Health grant U54GM1049.
Footnotes
Authors’ contributions
MJD conceived the study, collected and researched the data, and wrote the manuscript. MGA conceived the study and wrote the manuscript. JLA collected and researched the data, and contributed to the discussion. DML conceived the study, performed the sample size calculation, and wrote the paper. BNC conceived and directed the overall study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van der Lei J. Use and abuse of computer-stored medical records. Methods of information in medicine. 1991;30(2):79–80. Epub 1991/04/01. [PubMed] [Google Scholar]
- 2.Juran JM, Gryna FM. Juran’s Quality Control Handbook. New York: McGraw-Hill; 1988. [Google Scholar]
- 3.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. Journal of the American Medical Informatics Association : JAMIA. 2013;20(1):144–51. doi: 10.1136/amiajnl-2011-000681. Epub 2012/06/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.i2b2. Informatics for Integrating Biology and the Bedside. 2016 cited 2016 May 31, 2016. Available from: https://www.i2b2.org/work/i2b2_installations.html.
- 5.Grandia L. Healthcare Information Systems: A Look at the Past, Present, and Future. cited 2016 May 25. Available from: https://www.healthcatalyst.com/healthcare-information-systems-past-present-future.
- 6.Collen MF, RAM . The Early History Information Systems for Inpatient Care in the United States. In: Collen MF, MJB, editors. A History of Medical Informatics in the United States. New York: Springer; 2015. [Google Scholar]
- 7.Logan JR, Gorman PN, Middleton B. Measuring the quality of medical records: a method for comparing completeness and correctness of clinical encounter data. Proceedings / AMIA Annual Symposium AMIA Symposium; 2001; pp. 408–12. Epub 2002/02/05. [PMC free article] [PubMed] [Google Scholar]
- 8.Lo Re V, 3rd, Haynes K, Forde KA, Localio AR, Schinnar R, Lewis JD. Validity of The Health Improvement Network (THIN) for epidemiologic studies of hepatitis C virus infection. Pharmacoepidemiology and drug safety. 2009;18(9):807–14. doi: 10.1002/pds.1784. Epub 2009/06/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seminara NM, Abuabara K, Shin DB, Langan SM, Kimmel SE, Margolis D, et al. Validity of The Health Improvement Network (THIN) for the study of psoriasis. The British journal of dermatology. 2011;164(3):602–9. doi: 10.1111/j.1365-2133.2010.10134.x. Epub 2010/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]