Abstract
We have used magnetic resonance to map the interaction surface of an integral membrane protein for its regulatory target, an integral membrane enzyme. Phospholamban (PLN) regulates cardiac contractility via its modulation of sarco(endo)plasmic reticulum calcium ATPase (SERCA) activity. Impairment of this regulatory process causes heart failure. To map the molecular details of the PLN/SERCA interaction, we have functionally reconstituted SERCA with labeled PLN in dodecylphosphocholine micelles for high-resolution NMR spectroscopy and in both micelles and lipid bilayers for EPR spectroscopy. Differential perturbations in NMR linewidths and chemical shifts, measured as a function of position in the PLN sequence, provide a vivid picture of extensive SERCA contacts in both cytoplasmic and transmembrane domains of PLN and provide structural insight into previously reported functional mutagenesis data. NMR and EPR data show clear and complementary evidence for a dynamic (μs-to-ms) equilibrium between two conformational states in the cytoplasmic domain of PLN. These results support the hypothesis that SERCA attracts the cytoplasmic domain of PLN away from the lipid surface, shifting the preexisting equilibrium of PLN conformers toward a structure that is poised to interact with the regulatory target. EPR shows that this conformational switch behaves similarly in micelles and lipid membranes. Based on structural and dynamics data, we propose a model in which PLN undergoes allosteric activation upon encountering SERCA.
Keywords: NMR spectroscopy, protein-protein interaction
Sarco(endo)plasmic reticulum calcium ATPase (SERCA) designates a family of P-type calcium pumps embedded in the sarco(endo)plasmic reticulum. In muscle, SERCA is a 110-kDa enzyme, containing 10 transmembrane helices and three distinct cytoplasmic domains (1), which restores cytosolic calcium to submicromolar levels via ATP hydrolysis, resulting in relaxation. In cardiac muscle, phospholamban (PLN) regulates SERCA, inhibiting the enzyme at submicromolar Ca2+ (2). After β-adrenergic stimulation, PLN is phosphorylated by protein kinase A, reversing SERCA inhibition. SERCA1a, the fast-twitch skeletal muscle isoform, is functionally identical to SERCA2a, the cardiac isoform, both in the presence and absence of PLN (3). Because SERCA1a is a better-characterized enzyme and is readily purified in 100-mg quantities, the present study used this isoform.
PLN is a 52-aa, single-pass transmembrane protein that undergoes a dynamic equilibrium between a monomeric inhibitory form and a pentameric storage form (4). PLN has three structural domains, as determined by NMR in dodecylphosphocholine (DPC) micelles (5). The N-terminal cytosolic helix (domain Ia: residues 2-16) is amphipathic, with the hydrophobic face pointing toward the lipid surface (5, 6). A flexible loop (residues 17-21) connects this helix to the C-terminal helix (residues 22-50), introducing an average angle of ≈80° between the two helices (5, 6). The C-terminal helix ends with a hydrophobic membrane-embedded sequence.
NMR relaxation studies further dissected PLN into four domains, in terms of ns dynamics: the N-terminal cytosolic helix (domain Ia), the connecting loop, and two segments within the C-terminal helix (domain Ib, part of the cytoplasmic domain, and domain II, the transmembrane domain) (7). In addition, motions on the μs-to-ms time scale occur within the cytoplasmic domain Ia, the flexible loop, and a few residues within transmembrane domain II. The NMR dynamics data in DPC micelles are supported by EPR data in lipid bilayers, which reveal that the cytoplasmic helix is characterized by a slow conformational equilibrium (≥μs) between two states, one of which displays considerably more ns backbone dynamics (8). Both fast (ps-ns) and slow (μs-ms) motions are proposed to play fundamental roles in mediating biologically relevant conformational transitions and protein-protein interactions (9-11).
Recently, by using biochemical and mutagenesis data, two computational studies have produced models for the PLN/SERCA complex (12, 13). In one model (12), PLN docks between transmembrane helices M4 and M6 of SERCA, whereas the second model (13) placed PLN in a groove formed by transmembrane helices M2, M4, M6, and M9. Although both models propose that the SERCA-bound form of PLN is more extended than that of free PLN in lipid, their predictions differ about (i) the extent of PLN′s cytoplasmic helix unwinding, and (ii) specific residues in both helical domains of PLN contacting the enzyme. To test and refine these models, high-resolution structural data from a functional PLN/SERCA complex must be obtained.
Because PLN is a prime target for therapeutic intervention in human heart disease (14), our objective was to identify the PLN residues mediating its interaction with SERCA. We used a fully functional monomeric mutant of PLN, termed AFA-PLN, in which the three transmembrane cysteines were replaced with Ala, Phe, and Ala. Building on our recently determined high-resolution structure and dynamics characterization of AFA-PLN in micelles (5, 7), we used solution NMR to define its binding epitope, with the dual objective of (i) establishing the structural basis for the mechanism of calcium regulation in muscle and (ii) providing a better template for rational drug design.
To map the SERCA-binding epitope of AFA-PLN, we measured chemical shift perturbations and differential line broadenings (15, 16) of 15N-labeled AFA-PLN upon addition of SERCA. We studied a functional complex between two integral membrane proteins probed by solution NMR spectroscopy. Our investigation demonstrates that it is possible to use solution NMR to characterize, at the atomic level, interactions between integral membrane proteins, which are pivotal to the regulation of many cellular events. To complement the NMR data and extend the study from detergent micelles to lipid membranes, we performed EPR experiments by using the 2,2,6,6,-tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid (TOAC) spin label.
Materials and Methods
Protein Preparations. SERCA was purified from rabbit skeletal muscle (17), using 5 mM DPC instead of 1% C12E8 in the column elution buffer. Recombinant uniformly and selectively 15N-labeled AFA-PLN was produced as reported (18). Syntheses of AFA-PLN (used for activity assays) and its TOAC spin-labeled derivatives were carried out by using fluorenylmethoxycarbonyl chemistry (8, 19).
Activity Assays. A mixture of 0 or 0.9 μM AFA-PLN, 0.1 μM SERCA, and varying concentrations of DPC (300, 600, or 900 μM) were incubated on ice for 20 min, then diluted 1:10 into the standard assay mix (20). The final sample composition was 0 or 0.09 μM AFA-PLN, 0.01 μM SERCA, 30, 60, or 90 μM DPC, 0.2% C12E8, 4.5 mM MgCl2, 2 mM Mops (pH 7.0), 2% glycerol, 25 μM DTT, 6 μM ADP, 6 μM PBS, 18 mM imidazole, 88 mM KCl, 0.44 mM EGTA, 2.1 mM ATP, 0.16 mM NADH, 0.44 mM phosphoenolpyruvate, 169 units of lactic dehydrogenase, 169 units of PK, and 0.012-0.534 mM CaCl2. NADH consumption was monitored by absorbance at 340 nm by using a Spectromax plate reader at 37°C. To confirm the reversibility of SERCA inhibition by PLN, 1% C12E8 was added (21).
NMR Sample Preparation and Spectroscopy. Uniformly 15N-labeled AFA-PLN was dissolved at 0.9 mM in 20 mM PBS (pH 6.0), 300 mM DPC, containing 10% D2O (total volume 300 μl). SERCA, initially at a concentration of 9.1 μM in 5 mM DPC, 1 mM CaCl2, 1 mM MgCl2, 20 mM Mops (pH 7.0), 20% glycerol, 0.25 mM DTT, 4 mM ADP, and ±5 mM EGTA (total volume 20 ml), was concentrated at 4°C with an Amicon stirred cell by using a YM10 membrane to a concentration of 1.12 mM (150 μl). Subsequently, SERCA was titrated into the AFA-PLN sample at molar ratios of 0.20, 0.40, and 0.59. Higher SERCA concentrations caused sample precipitation. NMR spectra were acquired on a Varian Inova spectrometer operating at 600 MHz at 37°C, using an inverse detection triple-resonance and triple-axis gradient probe. The heteronuclear single quantum correlation (HSQC) pulse program was equipped with pulse field gradients for both coherence selection and sensitivity enhancement (22). The final matrix size was 1,024 × 512 real points after zero filling in both dimensions and Fourier transformation. Data were processed with nmrpipe (23) and nmrview (24). Molecular graphics were prepared by using molmol and pymol (25).
EPR Sample Preparation and Spectroscopy. AFA-PLN was synthesized with the TOAC spin label at positions 11 and 46 (8). This spin label is rigidly coupled to the α-carbon and thus reports directly the dynamics of the peptide backbone (8), providing information about protein dynamics that is comparable to that of 15N-NMR. Spin-labeled PLN was reconstituted into DPC micelles (NMR conditions) or lipid bilayers (1,2-dioleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-sn-glycero-3-phospho-ethanolamine, 4:1, 200 lipids per PLN), with or without SERCA. Samples in DPC micelles were prepared as for NMR spectroscopy, except that the final PLN concentration was ≈100 μM. Spin-labeled AFA-PLN was reconstituted in lipid bilayers in the presence and absence of SERCA (26) and was found to have normal inhibitory activity (8). EPR absorption derivative spectra were acquired at X band (9.4 GHz) with a Bruker E500 EPR spectrometer (11); spectra were analyzed to resolve distinct conformational components (8, 26).
Results
SERCA Activity Under NMR Conditions. We measured SERCA's ATPase activity and its regulation by AFA-PLN under NMR conditions in DPC micelles. The calcium dependence of SERCA activity and its reversible inhibition by AFA-PLN are illustrated in Fig. 1. Over a wide range of DPC concentrations, up to a molar ratio of 9,000:1 (DPC/SERCA), the ATPase activity was ≈23 units, which is comparable to typical values for SERCA in muscle membranes (27). The dependence of SERCA activity on Ca2+ concentration ([Ca2+]) exhibits the typical sigmoidal curve (2) (Fig. 1, black line). At a molar ratio of 9:1, AFA-PLN inhibits SERCA activity markedly, particularly at saturating [Ca2+] (≈40% inhibition at pCa 4) (Fig. 1, red line). In contrast, in lipid membranes obtained from muscle or cell culture (2, 28) or reconstitution (18, 29), inhibition is most clearly observed at submicromolar [Ca2+]. Upon addition of 1% C12E8, we observed full recovery of SERCA function (Fig. 1, blue line) (21), showing that inhibition by AFA-PLN is fully reversible under NMR conditions. Because only a minimal loss (≤20%) of SERCA activity was observed upon incubation at 37°C for 5 h, all NMR titration experiments were conducted within a 3-h time frame.
Fig. 1.
Ca-dependent ATPase activity of SERCA (0.1 μM) in DPC (0.3 mM), in the absence (black line) and presence (red line) of 0.9 μM AFA-PLN. One percent C12E8 (blue line) reverses this inhibition.
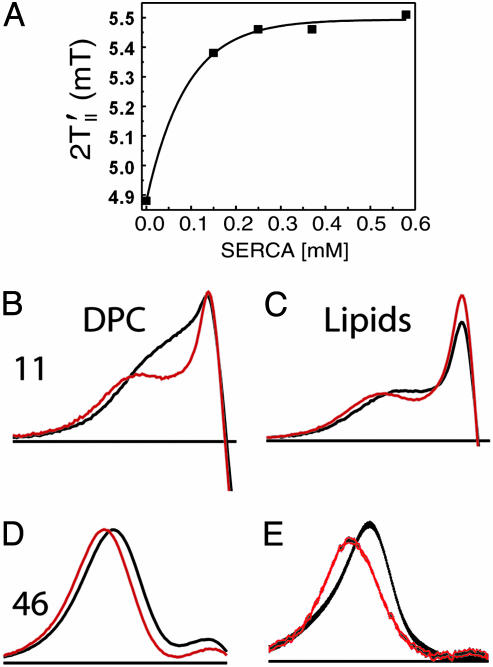
Kd Measurement by EPR. The affinity of AFA-PLN for SERCA in DPC micelles was assessed by measuring the dissociation constant (Kd) by using EPR. The higher sensitivity of EPR compared with NMR allowed us to use lower protein concentrations and determine Kd more accurately. AFA-PLN with the TOAC spin label at position 11 (8) was reconstituted with SERCA in DPC micelles. Upon addition of SERCA, spin-label dynamics was restricted, as monitored by the splitting between the outer extrema of the EPR spectrum. The dependence of this splitting on SERCA concentration was analyzed (30) to determine the Kd (Fig. 2). Under these conditions, Kd was 60 ± 12 μM, a value significantly greater (weaker binding) than the value reported (Kd ≤ 20 μM) for specific PLN-SERCA binding in a lipid bilayer (30).
Fig. 2.
EPR spectra of TOAC-labeled PLN at positions 11 and 46 in the presence and absence of SERCA. (A) Binding isotherm for PLN/SERCA complex formation in DPC micelles, from the increase of the EPR splitting (between the outer peaks of the spectrum) for spin-labeled PLN (100 μM A11-TOAC-AFA-PLN) as a function of [SERCA] (Kd = 60 ± 12 μM). (B-E) EPR absorption derivative spectra (low-field region, horizontal axis 2.5 mT) of AFA-PLN with the TOAC spin label at position 11 (B and C) or 46 (D and E) in the absence (black line) and presence (red line) of 2 mol SERCA per mole PLN. (B and D) NMR conditions in DPC. (C and E) NMR conditions in dioleoylphosphatidylcholine/dioleoylphosphatidylethanolamine lipid bilayers. Spectra are normalized to the double integral, so that they correspond to the same number of spins.
Identification of the AFA-PLN Binding Interface. The activity assays and Kd in DPC yielded sample conditions suitable for probing the AFA-PLN/SERCA complex formation by NMR differential line-broadening analysis (15, 16). Control titrations were performed with the SERCA purification buffer to rule out changes caused by dilution (31). To ensure reproducibility, titrations were repeated in triplicate with independent preparations of each protein. The full assignment of the PLN 1H-15N-HSQC spectrum is based on our previous work (5). When needed, the assignment of splitting and overlapping resonances was confirmed by experiments on selectively labeled samples.
Fig. 3 shows the superposition of 1H-15N-HSQC spectra of uniformly 15N-labeled AFA-PLN in DPC micelles titrated with unlabeled SERCA. The addition of SERCA caused a global reduction of signal intensities, showing that AFA-PLN binds to SERCA, which increases the overall correlation time. A closer inspection shows differential line broadening, which reveals a dynamic interaction between these two proteins on an intermediate/slow time scale (μs-ms) with respect to NMR chemical exchange (15). With intermediate exchange, NMR lines are significantly broadened, whereas under slow exchange, each spin resonates at a separate frequency. Because the PLN/SERCA complex is too large to be detected directly by NMR (>130 kDa), the appearance of new peaks in the HSQC spectrum (Fig. 3) upon SERCA addition was attributed to an intermediate PLN species in equilibrium between the original free and SERCA-bound forms (Fig. 3). Exchange between the unobservable bound state and the free species influences the observed peak position of the free species, facilitating identification of the AFA-PLN binding interface (15, 16). Broadening is caused by changes in chemical shift, conformational changes or direct contact with SERCA, or immobilization (15). If chemical shift changes tend to dominate the broadening, in their absence immobilization is the primary source of decreased intensity (15).
Fig. 3.
Chemical shift mapping of SERCA binding to AFA-PLN. A wide range of differential line broadenings are observed between the 1H/15N HSQC spectra of AFA-PLN in the absence (black) and presence (red) of SERCA. Boxes highlight examples of resonances belonging to groups 1-5 (see text).
The normalized peak intensities of AFA-PLN at a molar ratio of 0.59:1 (SERCA/AFA-PLN) (Fig. 4) are mapped onto the AFA-PLN structure in Fig. 5. Depending on their behavior in the presence of SERCA, the residues were clustered into five groups. Group 1 (Fig. 5, yellow) contains residues with reduced intensities, such as M1, Y6, and the majority of the transmembrane residues (from Q23 through L52), with the exclusion of A36 and F41. Group 2 (Fig. 5, violet) includes E2, K3, Q5, T8, and R9, whose original intensities are completely abolished. Group 3 (Fig. 5, orange) includes residues L7, R13, S16, and I18, whose original peaks disappear, with a new peak observable at a slightly different frequency, and Q22 and A24, which show reduction of the original resonances and subtle splitting. Group 4 (Fig. 5, pink) includes S10, I12, and T17, in which the original peak moves to a distinct new frequency, and A11, R14, A15, A36, and F41, for which the original peak is reduced in intensity and one new frequency appears. Finally, group 5 (Fig. 5, pink) includes only residue E19, for which the original peak is reduced and two new frequencies appear. Because of overlapping resonances, R25 and V4 were not included in the analysis.
Fig. 4.
Observed changes in peak intensity (Upper) and chemical shift (Lower) at SERCA/AFA-PLN = 0.59 (molar ratio), normalized to control buffer titrations. In Lower, black bars correspond to the original peaks and red bars represent newly appearing (splitting) peaks. Missing and overlapping residues (residues 4, 10, 25, 27, and 39), indicated by *, were eliminated from the analysis. Secondary structure domains of PLN are indicated at the top.
Fig. 5.
NMR mapping of PLN hot spots in the PLN/SERCA complex. (A) 1H/15N HSQC spectra as a function of SERCA concentration, for representative AFA-PLN residues in each of the five groups. Gray peaks, control buffer titrations. Colored peaks, after addition of SERCA. Group 1 is yellow; group 2 is violet; groups 3, 4, and 5 are pink (significant chemical shift perturbations) or orange (subtle splitting). (B) Mapping of interacting residues onto the AFA-PLN solution NMR structure (5). PLN domains are indicated. (C) Mapping of the interacting residues onto the computational model of the PLN/SERCA complex (13). Horizontal lines indicate the boundary of the hydrophobic core of the lipid bilayer.
Group 1 includes residues from domain Ib and domain II and a few in domain Ia (Fig. 3). The overall decrease in intensity probably is caused by immobilization of these residues upon complex formation, in proportion to the fraction of PLN molecules in the bound state (15). Given a Kd of 60 μM, the average decrease in signal intensity should correspond to ≈50% of PLN molecules in the free state, which is in good agreement with the average resonance intensities of 44 ± 8% and 45 ± 5% for domains Ib and II, respectively. Residues clustered in group 2 were exclusively located in domain Ia, including E2, K3, Q5, T8, and R9. The complete disappearance of peaks is most likely caused by the dominating effect of chemical shift changes upon complex formation (15).
SERCA Induces a Switch Between AFA-PLN Conformations. Concomitant with the reduction of all peak intensities (Fig. 4), several of them split into two resolved peaks in slow exchange (Fig. 3, group 4). These residues are located primarily in domain Ia and the loop region, with a few residues found in domains Ib and II (L7, S10, A11, I12, R13, R14, A15, S16, T17, I18, E19, Q22, A24, A36, and F41). E19 showed two new peaks in slow exchange (Fig. 3, group 5). The perturbations in chemical shift values caused by SERCA binding (plotted in Fig. 4 for each AFA-PLN residue) reveal minimal variations for residues without splitting (average of 0.017 ± 0.009 ppm), but the majority of residues converting into a new position gave significant chemical shift perturbations (average of 0.065 ± 0.034 ppm). These residues were placed in group 4. The remaining residues, defined as group 3 (L7, R13, S16, I18, Q22, and A24), did not show a marked chemical shift change, yet showed subtle conversion to a new frequency (Fig. 3). Fig. 5 shows examples of the different groups in expanded scale (A) and the mapping of interacting residues onto the AFA-PLN NMR structure (B) and onto one of the proposed SERCA-PLN models (C). For the most part, each group covers a contiguous surface area, with the splitting residues located primarily within domains Ia, loop, and Ib, with the exception of Y6 and M1, whose signals are reduced and do not show the presence of two conformers. These residues in the model of Fig. 5 face the opposite site of the cytoplasmic domain of the enzyme. Two residues of domain II (A36 and F41) also show significant splitting (Fig. 5B), implying that the conformational switching also involves domain II.
Differential behavior of resonances in different domains often has been observed in soluble enzymes (32), where the doubling of specific resonances in the presence of the binding partner indicates slow conformational switching (32). SERCA induces a conformational change in AFA-PLN, most likely priming it for interaction and giving rise to the interconverting free species observable in solution.
Conformational Switch of AFA-PLN Observed in Lipid Membranes by EPR. To compare the behavior of the SERCA/PLN complex in DPC micelles to more biologically relevant lipid membranes, we performed EPR experiments under both conditions. AFA-PLN was synthesized with the TOAC spin label at positions 11 and 46 (8) and reconstituted into DPC micelles (NMR conditions) or lipid bilayers (dioleoylphosphatidylcholine/dioleoylphosphatidylethanolamine, 4:1, 200 lipids per PLN). Regulatory function was retained in both micelles (Fig. 1) and bilayers (8). The TOAC spin label's rigid connection with the peptide backbone makes it possible to monitor backbone dynamics accurately in a manner very similar to 15N NMR, and EPR has sufficient sensitivity to do this in the membrane (Fig. 2 C and E) as well as in solution (Fig. 2 B and D). Most importantly, EPR detects directly not only the free form of PLN (as in NMR), but also the SERCA-bound form, which cannot be detected directly by NMR. The EPR spectrum of the spin label at position 46 (Fig. 2 D and E) in the transmembrane domain shows a single component with highly restricted ns dynamics (order parameter S >0.8). In contrast, the spin label at 11 (Fig. 2 B and C) in the cytoplasmic domain shows two resolved components: a broad one, indicating restriction comparable to that at position 46, and a narrow component, corresponding to large-amplitude ns motions (S <0.3). Previous EPR studies have shown that the restricted component, corresponds to an ordered helix that is stabilized by interaction with the membrane surface, whereas the dynamic component corresponds to a more dynamic structure that has lost contact with the surface (8, 26). SERCA increases the population of this dynamically disordered conformation in both DPC (from 4% to 12%) and lipid (from 11% to 19%, indicated by increased intensity of the narrow peak in Fig. 2 B and C), while decreasing the mobility of the restricted component (indicated by leftward shift of the broad peak in Fig. 2 B and C). We conclude that in both DPC solution and lipid bilayers (i) position 11 is much more dynamically disordered than 46, in agreement with NMR dynamics data (7), (ii) position 11 is much more affected by SERCA, confirming the present NMR results, (iii) there are two resolved components at 11, but only one at 46, and (iv) at 11 (but not at 46) SERCA increases the mole fraction of the more dynamic of the two components and increases the resolution between the components. Both in lipids and DPC EPR reveals that even in the absence of SERCA there are two conformational states intrinsic to AFA-PLN, which are particularly evident in the cytoplasmic domain. SERCA induces a shift toward the more dynamically disordered of the two states (Fig. 2). Thus, the EPR results in both micelles and bilayers confirm and clarify the key NMR conclusions.
Discussion
The boundaries of structural biology have expanded with the application of solution NMR to determine the structures of small membrane-bound peptides as well as large, multidomain membrane proteins (33-39). Here, we report the functional interaction between two membrane proteins probed by solution NMR in detergent micelles. Our approach reveals the direct physical interactions between PLN and SERCA from the perspective of the inhibitor, PLN. Under NMR conditions, SERCA is fully functional, binds PLN with a Kd of 60 μM, and is reversibly inhibited by it. The Ca dependence of this functional interaction in DPC, inhibition at micromolar Ca2+ and above, is different from that observed in lipids, where inhibition is observed only at submicromolar Ca2+. Thus the SERCA-PLN complex is altered by the DPC environment. However, EPR shows that the structural interactions of both PLN′s transmembrane and cytoplasmic domains with SERCA are similar in DPC and lipids (Fig. 2), supporting the conclusion that the NMR data report accurately the functional inhibitory interaction of PLN with SERCA.
NMR reveals a dynamic mechanism by which PLN controls SERCA, and EPR indicates that these results also hold in lipids. These findings indicate a dynamic equilibrium within and between these two proteins (40). The current view, based on the functional effects of site-directed mutagenesis, proposes that both domains of PLN participate in SERCA regulation, with the C-terminal transmembrane domain primarily responsible for inhibition, and the N-terminal cytoplasmic domain primarily responsible for the relief of inhibition (28, 41). Some of the PLN “hot spots” identified by mutagenesis coincide with residues observed to disappear or double upon interaction with SERCA (E2, K3, L7, R9, A11, I12, R13, R14, A15, S16, T17, and I18). However, our studies showed additional residues, not detected in previous biochemical assays, to be perturbed in the presence of SERCA (Q5, T8, S10, E19, Q22, A24, A36, and F41). It is likely that residues showing the greatest intensity perturbation (E2, K3, Q5, T8, and R9; Fig. 5) are those involved in PLN conformational changes with the largest SERCA-induced chemical shift variations. When these sites are mapped onto a recently proposed model of the SERCA/PLB complex (13) (Fig. 5C), the most affected residues cluster close to K3, which was identified previously as a crucial residue in the complex. The only two residues of domain Ia that do not show conformational exchange (M1 and Y6) face the opposite side of SERCA's cytoplasmic domain. E19, which “hops” between three different conformations, may play a unique role in the dynamics of this interaction. Finally, the two residues of the transmembrane region that show conformational exchange, A36 and F41, are adjacent to the transmembrane helices of SERCA that are responsible for calcium translocation (42). Additional dynamics in that region recently was detected for both the AFA mutant in micelles and WT-PLN in lipid environments (7, 43). We conclude that our mapping of the hot spots generally supports previous molecular modeling (13).
The existence of splitting residues (L7, S10, A11, I12, R13, R14, A15, S16, T17, I18, E19, Q22, A24, A36, and F41; Fig. 5) confirms that PLN switches between at least two conformations when it binds to SERCA. Most of these residues are located in the cytoplasmic helix, with a few found within the loop region and the two dynamic domains of the hydrophobic helix. E19, which lies within a more dynamic region of the hinge, is the only residue that exhibits its original peak plus two new peaks upon SERCA titration. Although our average NMR structure suggests that the hinge is between T17 and Q22 (5), mutational analysis (7, 28, 44) and dynamics data from NMR (7, 28, 44) and EPR (8) suggest that this dynamic region can become extended to a much broader region, from A11 to Q29. This flexible region allows the cytoplasmic domain of PLN to undergo a large concerted motion (26), facilitating the formation of a viable complex with SERCA. In addition, the appearance of splitting residues within the transmembrane helix implies long-range coupling between this centrally located hinge and the two helices of PLN.
Both NMR dynamics (7) and EPR (8) reveal the interconversion of AFA-PLN in its free state between two or more conformations. SERCA perturbs this preexisting equilibrium of PLN conformers, slowing the exchange and shifting the equilibrium distribution. The lifetime of exchange between the two PLN conformations, averaged over all 15 residues with resonance doubling, is 54 ± 27 ms, exceeding the inverse of their chemical shift differences. The quantification of these two PLN species in the NMR spectra, based on the endpoint of SERCA titration, gave a distribution of 7 ± 2% in the original free state and 37 ± 4% in the intermediate form. The remaining 56% of the intensity corresponds to the undetectable bound form.
EPR complements NMR in two important ways. First, EPR has sufficient sensitivity to obtain information about PLN conformational dynamics directly in “immobilized” states, in lipid bilayers as well as in DPC micelles, and in the SERCA-bound form as well as in the dissociated form. Second, EPR's high-resolution sensitivity to ns dynamics allows it to resolve two distinct conformational states in the cytoplasmic domain even in the absence of SERCA (Fig. 2, black lines, both in DPC and lipid), where the 1H/15N HSQC spectrum resolves only one state. A previous EPR study showed that these two states correspond to a highly ordered conformation, probably helical, and a dynamically disordered conformation (8). Addition of SERCA (Fig. 2, red line) enhances the more dynamically disordered of the two populations, confirming and explaining the key NMR result.
The observed SERCA-dependent conformational switching in the PLN structure suggests an allosteric activation mechanism (45-48) (Fig. 6). In this model, PLN in its free form exists in two (or more) conformations, with a thermodynamic preference for the inactive or T state over the active or R state. Both NMR (5, 6) and EPR (8, 26) of monomeric free AFA-PLN confirm this preexisting equilibrium (shown explicitly by EPR in Fig. 2) and show that the predominant state (T) is bent, with the highly ordered amphipathic cytoplasmic helix facing the lipid environment. Both NMR and EPR show that interaction with SERCA shifts this preexisting equilibrium toward the R state, in which the cytoplasmic domain is more dynamically disordered and extended and has higher affinity for SERCA. The role of the binding partner can be played either by the regulator, as in the classical model (40), or the regulatory target, as in the present study (41-43). The conformational switching induced by SERCA involves the PLN structure as a whole, propagating throughout the protein backbone (see splitting in residues A36 and F41), and mediating the reported functional coupling between these domains (49).
Fig. 6.
Allosteric model of PLN interaction with SERCA. PLN monomer interconverts between the bent form (T state) and the less stable (more dynamically disordered) extended form (R state). The T state inhibits SERCA because of its interaction in the transmembrane domain. Only in the R state, in which the cytoplasmic domains interact, is SERCA-bound PLN capable of reversing SERCA inhibition, because of PLN phosphorylation or mutation. SERCA has a greater affinity for the R form and thus shifts the equilibrium toward this regulatory intermediate.
Based on chemical crosslinking and mutagenesis data, it has been proposed that PLN′s cytoplasmic domain must assume an extended conformation when interacting with SERCA (12, 13) (Fig. 5C). This idea suggests that PLN must switch from its bent L shape to a more elongated conformation, with the amphipathic cytoplasmic helix undergoing more dramatic change (Figs. 5C and 6). This hypothesis is strongly supported by previous EPR data, showing that a spin label at position 11 in PLN interacts much less with the membrane surface upon interaction with SERCA (26). The present NMR data show that the majority of residues having a significant perturbation in chemical shift upon interaction with SERCA line the hydrophobic side of the amphipathic cytoplasmic helix (Figs. 3 and 5). Thus for functional association to occur, the cytoplasmic domain of PLN must reconcile between adhering to the lipids and extending itself to contact the cytoplasmic portion of SERCA (Fig. 6).
In summary, we have used complementary NMR and EPR techniques to map the PLN residues that interact with SERCA and show that this interaction affects all PLN domains, perturbing the preexisting structural equilibrium of PLN. This conformational switch exemplifies an allosteric activation mechanism of protein-protein interaction. The cytoplasmic helix of AFA-PLN undergoes the most dramatic changes in conformation, but significant alterations in the loop and the hydrophobic helix confirm coupling throughout the entire protein. Our mapping is limited to the perturbation of signals in the protein backbone; further high-resolution studies are needed to clarify the role of PLN side chains in the functional interaction and elucidate the conformation of PLN when bound to SERCA. The methods developed in the present study should prove applicable for probing the interactions and functional dynamics of a wide range of membrane protein complexes.
Acknowledgments
We thank C. Toyoshima and D. MacLennan for coordinates of the PLN/SERCA model; R. Di Fonzo, H. Matsuo, and K. Walters for helpful comments; and D. Live and B. Ostrowski for NMR technical assistance. This work was supported by National Institutes of Health Grant GM64742 and American Heart Association Grant 0160465Z (to G.V.), National Institutes of Health GM27906 (to D.D.T.), and the Minnesota Medical Foundation (C.K.). NMR instrumentation was provided with funds from National Science Foundation Grant BIR-961477 and the University of Minnesota Medical School.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PLN, phospholamban; SERCA, sarco(endo)plasmic reticulum calcium ATPase; DPC, dodecylphosphocholine; TOAC, 2,2,6,6,-tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid; HSQC, heteronuclear single quantum correlation.
References
- 1.Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. (2000) Nature 405, 647-655. [DOI] [PubMed] [Google Scholar]
- 2.Simmerman, H. K. & Jones, L. R. (1998) Physiol. Rev. 78, 921-947. [DOI] [PubMed] [Google Scholar]
- 3.Lytton, J., Westlin, M., Burk, S. E., Shull, G. E. & MacLennan, D. H. (1992) J. Biol. Chem. 267, 14483-14489. [PubMed] [Google Scholar]
- 4.Cornea, R. L., Jones, L. R., Autry, J. M. & Thomas, D. D. (1997) Biochemistry 36, 2960-2967. [DOI] [PubMed] [Google Scholar]
- 5.Zamoon, J., Mascioni, A., Thomas, D. D. & Veglia, G. (2003) Biophys. J. 85, 2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mascioni, A., Karim, C., Zamoon, J., Thomas, D. D. & Veglia, G. (2002) J. Am. Chem. Soc. 124, 9392-9393. [DOI] [PubMed] [Google Scholar]
- 7.Metcalfe, E. E., Zamoon, J., Thomas, D. D. & Veglia, G. (2004) Biophys. J. 87, 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karim, C. B., Kirby, T. L., Paterlini, M. G., Zhang, Z., Nitu, F., Nesmelov, Y. & Thomas, D. D. (2004) Proc. Natl. Acad. Sci. USA, 101, 14437-14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanagh, J. & Venters, R. A. (2001) Nat. Struct. Biol. 8, 912-914. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson, A. & Kieffer, B. (2004) Progr. Nuclear Magn. Reson. Spectrosc. 44, 141-187. [Google Scholar]
- 11.Wand, A. J. (2001) Science 293, 1395. [DOI] [PubMed] [Google Scholar]
- 12.Hutter, M. C., Krebs, J., Meiler, J., Griesinger, C., Carafoli, E. & Helms, V. (2002) Chembiochemistry 3, 1200-1208. [DOI] [PubMed] [Google Scholar]
- 13.Toyoshima, C., Asahi, M., Sugita, Y., Khanna, R., Tsuda, T. & MacLennan, D. H. (2003) Proc. Natl. Acad. Sci. USA 100, 467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt, J. P., Kamisago, M., Asahi, M., Li, G. H., Ahmad, F., Mende, U., Kranias, E. G., MacLennan, D. H., Seidman, J. G. & Seidman, C. E. (2003) Science 299, 1410-1413. [DOI] [PubMed] [Google Scholar]
- 15.Matsuo, H., Walters, K. J., Teruya, K., Tanaka, T., Gassner, G. T., Lippard, S. J., Kyogoku, Y. & Wagner, G. (1999) J. Am. Chem. Soc. 121, 9903-9904. [Google Scholar]
- 16.Panchal, S. C., Kaiser, D. A., Torres, E., Pollard, T. D. & Rosen, M. K. (2003) Nat. Struct. Biol. 10, 591-598. [DOI] [PubMed] [Google Scholar]
- 17.Stokes, D. L. & Green, N. M. (1990) Biophys. J. 57, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buck, B., Zamoon, J., Kirby, T. L., DeSilva, T. M., Karim, C., Thomas, D. & Veglia, G. (2003) Protein Expression Purif. 30, 253-261. [DOI] [PubMed] [Google Scholar]
- 19.Lockwood, N. A., Tu, R. S., Zhang, Z., Tirrell, M. V., Thomas, D. D. & Karim, C. B. (2003) Biopolymers 69, 283-292. [DOI] [PubMed] [Google Scholar]
- 20.Madden, T. D., Chapman, D. & Quinn, P. J. (1979) Nature 279, 538-541. [DOI] [PubMed] [Google Scholar]
- 21.Reddy, L. G., Cornea, R. L., Winters, D. L., McKenna, E. & Thomas, D. D. (2003) Biochemistry 42, 4585-4592. [DOI] [PubMed] [Google Scholar]
- 22.Kay, L. E., Keifer, P. & Saarinen, T. (1992) J. Am. Chem. Soc. 114, 10663-10665. [Google Scholar]
- 23.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 603-614. [DOI] [PubMed] [Google Scholar]
- 25.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 29-32. [DOI] [PubMed] [Google Scholar]
- 26.Kirby, T. L., Karim, C. B. & Thomas, D. D. (2004) Biochemistry 43, 5842-5852. [DOI] [PubMed] [Google Scholar]
- 27.Shivanna, B. D. & Rowe, E. S. (1997) Biochem. J. 325, 533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLennan, D. H., Kimura, Y. & Toyofuku, T. (1998) Ann. N.Y. Acad. Sci. 853, 31-42. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki, T., Inui, M., Kimura, Y., Kuzuya, T. & Tada, M. (1992) J. Biol. Chem. 267, 1674-1679. [PubMed] [Google Scholar]
- 30.Mueller, B., Karim, C. B., Negrashov, I. V., Kutchai, H. & Thomas, D. D. (2004) Biochemistry 43, 8754-8765. [DOI] [PubMed] [Google Scholar]
- 31.McDonnell, P. A., Shon, K., Kim, Y. & Opella, S. J. (1993) J. Mol. Biol. 233, 447-463. [DOI] [PubMed] [Google Scholar]
- 32.Stevens, S. Y., Sanker, S., Kent, C. & Zuiderweg, E. R. (2001) Nat. Struct. Biol. 8, 947-952. [DOI] [PubMed] [Google Scholar]
- 33.Mascioni, A., Karim, C., Barany, G., Thomas, D. D. & Veglia, G. (2002) Biochemistry 41, 475-482. [DOI] [PubMed] [Google Scholar]
- 34.Mascioni, A., Porcelli, F., Ilangovan, U., Ramamoorthy, A. & Veglia, G. (2003) Biopolymers 69, 29-41. [DOI] [PubMed] [Google Scholar]
- 35.Porcelli, F., Buck, B., Lee, D. K., Hallock, K. J., Ramamoorthy, A. & Veglia, G. (2004) J. Biol. Chem. 279, 45815-45823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, C., Marassi, F. M., Jones, D. H., Straus, S. K., Bour, S., Strebel, K., Schubert, U., Oblatt-Montal, M., Montal, M. & Opella, S. J. (2002) Protein Sci. 11, 546-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora, A., Abildgaard, F., Bushweller, J. H. & Tamm, L. K. (2001) Nat. Struct. Biol. 8, 334-338. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez, C., Hilty, C., Wider, G., Guntert, P. & Wuthrich, K. (2004) J. Mol. Biol. 336, 1211-1221. [DOI] [PubMed] [Google Scholar]
- 39.Hwang, P. M., Bishop, R. E. & Kay, L. E. (2004) Proc. Natl. Acad. Sci. USA 101, 9618-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, D. D., Reddy, L. G., Karim, C. B., Li, M., Cornea, R., Autry, J. M., Jones, L. R. & Stamm, J. (1998) Ann. N.Y. Acad. Sci. 853, 186-194. [DOI] [PubMed] [Google Scholar]
- 41.Cornea, R. L., Autry, J. M., Chen, Z. & Jones, L. R. (2000) J. Biol. Chem. 275, 41487-41494. [DOI] [PubMed] [Google Scholar]
- 42.Toyoshima, C., Nomura, H. & Tsuda, T. (2004) Nature 432, 361-368. [DOI] [PubMed] [Google Scholar]
- 43.Tiburu, E. K., Karp, E. S., Dave, P. C., Damodaran, K. & Lorigan, G. A. (2004) Biochemistry 43, 13899-13909. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, A. G., Zhai, J., Carr, A. N., Gerst, M. J., Lorenz, J. N., Pollesello, P., Annila, A., Hoit, B. D. & Kranias, E. G. (2002) Cardiovasc. Res. 56, 248-259. [DOI] [PubMed] [Google Scholar]
- 45.Monod, J., Wyman, J. & Changeux, J. P. (1965) J. Mol. Biol. 12, 88-118. [DOI] [PubMed] [Google Scholar]
- 46.Volkman, B. F., Lipson, D., Wemmer, D. E. & Kern, D. (2001) Science 291, 2429-2433. [DOI] [PubMed] [Google Scholar]
- 47.Kern, D. & Zuiderweg, E. R. (2003) Curr. Opin. Struct. Biol. 13, 748-757. [DOI] [PubMed] [Google Scholar]
- 48.Buck, M., Xu, W. & Rosen, M. K. (2004) J. Mol. Biol. 338, 271-285. [DOI] [PubMed] [Google Scholar]
- 49.Kimura, Y., Asahi, M., Kurzydlowski, K., Tada, M. & MacLennan, D. H. (1998) J. Biol. Chem. 273, 14238-14241. [DOI] [PubMed] [Google Scholar]