Abstract
Assessment of a tumor’s molecular makeup using biofluid samples, known as liquid biopsy, is a prominent research topic in precision medicine for cancer, due to its noninvasive property allowing repeat sampling for monitoring molecular changes of tumors over time. Circulating exosomes recently have been recognized as promising tumor surrogates because they deliver enriched biomarkers, such as proteins, RNAs, and DNA. However, purification and characterization of these exosomes are technically challenging. Microfluidic lab-on-a-chip technology effectively addresses these challenges owing to its inherent advantages in integration and automation of multiple functional modules, enhancing sensing performance, and expediting analysis processes. In this article, we review the state-of-the-art development of microfluidic technologies for exosome isolation and molecular characterization with emphasis on their applications toward liquid biopsy–based analysis of cancer. Finally, we share our perspectives on current challenges and future directions of microfluidic exosome analysis.
Keywords: microfluidic lab-on-a-chip technology, exosomes, liquid biopsy, cancer, precision medicine
Introduction
Tissue biopsy is often required for cancer diagnosis and prognosis. However, tissue biopsy is highly invasive for most primary tumors and metastatic diseases,1,2 especially brain cancer, lung cancer, and ovarian cancer, which require difficult surgeries.3 The obtained tissue quality and quantity highly determine the diagnostic precision at the molecular level, including mutation characterization. Tumor tissues are heterogeneous and evolve over time.2,4 Sampling of entire tissue with dynamic representatives is not possible. Therefore, assessment of the molecular makeup of tumors from a biofluid sample is of great research interest.2 Although technology challenging, noninvasive blood-based liquid biopsy allows repeat monitoring for clinical oncologists to gain a broader molecular understanding of tumors without the need for a tissue biopsy.5,6 Indeed, several blood-based biomarker tests that have been developed have been around for decades and are still debatable, including prostate-specific antigen (PSA) screening for prostate cancer, carcinoembryonic antigen (CEA) for colorectal cancer, CA19-9 for pancreatic cancer, and CA125 for ovarian cancer, due to the lack of reliability and specificity.7,8 Recent technological developments being applied to liquid biopsies are capable of reproducibly detecting mutations at very low allelic frequencies.9 However, lack of confidence in blood-based biomarkers still prevents widespread utilization of liquid biopsy for cancers.
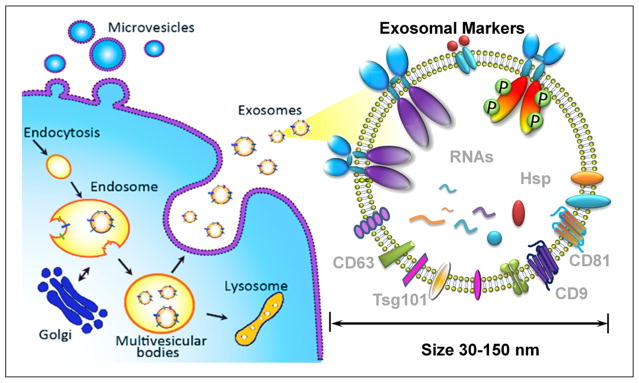
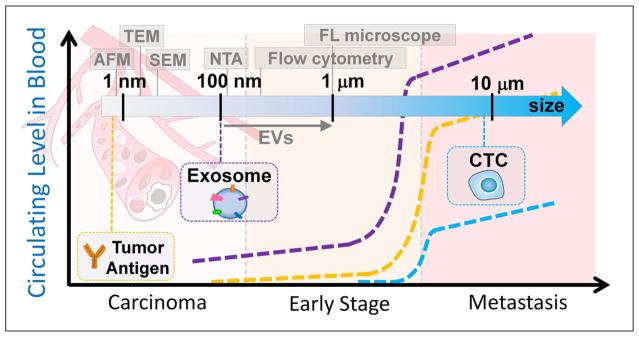
Recent research of extracellular vesicles (EVs) and exosomes has revealed that early-stage tumors constitutively release vesicles carrying various tumor markers.10–12 Exosomes, which are membrane vesicles of endocytic origin (30–150 nm diameter, Fig. 1), are systemically detectable in the blood of various cancer patients and have been shown to correlate well with tumor progression, immune response suppression, angiogenesis, and metastasis.13–15 Exosomes are stable carriers of enriched genetic material and proteins from their cell of origin, thereby holding great promise for identifying early-stage tumors.16,17 Compared to well-studied circulating tumor cells (1–10 circulating tumor cells [CTCs]/mL of blood), exosome release from tumor cells is an active process with concentrations of ≥109 vesicles/mL in blood.11,18 As illustrated in Figure 2, exosomes sensitively reflect tumor status; therefore, substantial investigations have focused on the essential physiologic and pathophysiologic functions of circulating exosomes as a surrogate for tumor liquid biopsy.14,19–22
Figure 1.
Exosome biogenesis, properties, and molecular composition.
Figure 2.
Circulating tumor antigens, exosomes, and CTCs associated with tumor status (not to scale). Scale bar shows the detection resolution of various benchtop instruments.
However, isolation and molecular analysis of such diverse nanoscale exosome vesicles for clinical utilization are technically challenging.21,23–26 Size overlap complicates exosome definition due to the presence of other membrane-derived subcellular structures, such as apoptotic vesicles, exosome-like vesicles, membrane particles, and ectosomes.27,28 Current exosome purification methods, including ultrafiltration and sucrose gradient ultracentrifugation, are tedious and time-consuming (>10 h) and cannot completely discriminate exosomes from other EVs.29–31 Although conventional filtration isolates microvesicles (MVs) with a uniform size of less than 150 nm, forced filtration and shearing force may cause membrane fusion and loss of integrity.32 Molecular analysis of isolated MVs is primarily performed using Western blot, enzyme-linked immunosorbent assay (ELISA), or mass spectrometry, which require lengthy processes and concentrated exosome samples.33 Nanoparticle tracking analysis (NTA) and flow cytometry have demonstrated limited reliability for detecting particles smaller than 200 nm.33,34 Transmission electron microscopy (TEM) and atomic force microscopy (AFM) have been utilized often to investigate exosome morphology and size distribution (Fig. 2),35 but microscopic protocols do not inherently exhibit high-throughput and rapid measurement. Microfluidic lab-on-a-chip technology has recently been spotlighted as a promising approach for exosome isolation and molecular analysis, owing to its low-volume consumption,36 high capability of functional module integration,37 quick analysis time, and high sensitivity,38 as well as a sample-to-answer format.39–41 Several microfluidic approaches, such as isolation, quantification, and molecular profiling, have been previously developed for exosome study.42 We review the state-of-the-art microfluidic technologies for isolating exosomes and their applications for exosome-based liquid biopsy analysis of cancer. Other emerging microfluidic approaches developed for fabrication and production of therapeutic exosomes43 are not within the scope of this article.
Microfluidic Exosome Isolation
Since 2012, research efforts have increased dramatically to develop microfluidic platforms in order to isolate exosomes. Compared to benchtop methods, microfluidic technology offers fast isolation speed, high yield and efficiency, automation, and functional integration for streamlined exosome molecular analysis.42 Vesicle size (30–150 nm) and surface markers (immunoaffinity) are mandatory in order to identify a specific population of circulating MVs that primarily consists of exosomes.10 Therefore, current reported microfluidic platforms can be classified as immunoaffinity-based exosome isolation and size-based exosome isolation.
Immunoaffinity-based exosome isolation can be implemented into microfluidic devices by manipulating affinity particles/magnetic beads, or modifying microchannel surfaces with antibodies. In 2010, Chen et al. reported the first microfluidic exosome isolation platform, which used an anti-CD63 functionalized surface for immunocapture of exosomes from human sera.44 Kanwar et al. developed a similar platform called ExoChip, which utilized a surface-functionalized (anti-CD63) circular microchamber to capture exosomes, followed by fluorescent carbocyanine dye (DiO) staining for quantitation.45 Although various microstructures were configured to enhance mixing and capture efficiency, capture capacity was still limited by available surface area and antibody immobilization density. Therefore, spherical particles or immunomagnetic beads were introduced into microfluidic devices to enhance capture capacity. We recently reported two microfluidic platforms46,47 for large-scale exosome isolation and molecular profiling of both surface and intravesicular markers by manipulating immunomagnetic capture beads in a microfluidic, multistage circuit. Dudani et al. introduced 20 μm polystyrene beads conjugated with biotinylated anti-human CD63 into a microfluidic device that utilized inertial lift forces at a finite Reynolds number, in order to position microparticles and exchange solutions for rapid purification.48 This approach provided high-flow-rate isolation of exosomes, which greatly increased throughput and processing volume compared to other methods. However, premixing and incubation of capture beads with samples were needed.
Although immunoaffinity-based isolation generates specific exosome populations and reflects molecular expression levels, size-based isolation has advantages of size uniformity without sample bias. Wang et al. fabricated a microfluidic device consisting of an array of porous silicon nanowire-on-micropillar.49 The inter-nanowire spacing was tuned within a range of 30–200 nm to create a high density of interstitial sites, which allowed physical trap of exosomes. Davies et al. in situ prepared nanoporous membranes in a microfluidic filtration system to isolate vesicles from whole blood with tunable size cutoff (<500 nm).50 However, these physical trapping approaches were restricted by exosome saturation limit and recovery rate. A continuous flow design, without capacity limitation, is ideal for on-chip high-throughput processing and integration of downstream sample preparation. Lee et al. developed an acoustic nano-filter chip that can fractionate exosomes (diameter < 200 nm) from cell culture media and blood in a continuous flow manner.51 Santana et al. built a microfluidic obstacle array based on the principle of deterministic lateral displacement and fractionated MVs with size cutoff of 250 nm.52 Currently, isolation with greater size resolution below 200 nm still needs to be improved.
Table 1 summarizes exosome isolation performance from various microfluidic platforms in terms of yield, capacity, and efficiency. As shown in the table, microfluidic technology as an advanced approach substantially increased exosome isolation speed and efficiency compared to a classic benchtop ultracentrifugation approach. In addition, it has been reported that diverse subpopulations of vesicles secreted by various intracellular mechanisms were present in exosome preparations obtained by differential ultracentrifugation.31 Although size is the most acceptable criterion for exosome identification (smaller than 150 nm), it is not a strict feature of exosomes. Several current preparations invariably contain varying proportions of other membranous vesicles that co-purified with exosomes, such as shed MVs and apoptotic blebs. Tauro et al. observed that EpCAM immunoaffinity-captured exosomes contain at least double the amount of enriched exosome markers and exosome-associated proteins, compared to centrifugation and density-based separations, although these MVs all range from 40 to 100 nm.53 Therefore, it is crucial in the future to design novel microfluidic methods that combine size and marker features for purifying and characterizing vesicle types and allowing precise analysis of respective exosome functions.
Table 1.
Exosome Isolation Performance of Various Microfluidic Platforms.
| Techniques | Sample | Isolation Speed | Yield | Isolation Capacity | Reference | |
|---|---|---|---|---|---|---|
| Microfluidic technology | Anti-CD63 functionalized surface with herringbone groves | Serum | 16 μL/min in 25 min | 42%–94% | 400 μL | 43 |
| Array of surface-functionalized circular microchambers | Serum | 8 μL/min in60 min | 15–18 μg of total proteins | 400 μL | 44 | |
| Online mixing in serpentine channel with immunomagnetic beads | Plasma | 10 μL/min in100 min | 42%–97.3% | Continuous flow 10 mL | 45, 46 | |
| Precaptured polystyrene beads with inertial solution exchange | Cell culture media | 140 μL/min | n/a | Continuous flow | 47 | |
| Acoustic nanofilter chip | Cell culture media | n/a | >80% | Continuous flow 50 μL | 48 | |
| Obstacle arrays in deterministic lateral displacement | Cell culture media | n/a | 39% | 170 μL | 49 | |
| Array of porous silicon nanowire-on-micropillar | Standard liposome solution | 10 μL/min in 10 min | 45%–60% | Saturation 100 μL | 50 | |
| Filtration with photopolymerized nanoporous membranes | Blood | 1 μL/min | 3% | Saturation 100 μL | 51 | |
| Ultra-centrifugation | Plasma | ≥600 min | 5%–25% | 12 mL | 30, 52 |
Microfluidic Exosome Sensing toward Liquid Biopsy
Liquid biopsy analysis of tumors has beneficially impacted clinical care, especially regarding treatment decision guidance; exosomes are significant contributors to such progress.54 Several advanced microfluidic exosome measurement and sensing technologies have been developed to characterize exosome physical, biological, and molecular properties. Akagi et al. developed a microfluidic immunoelectrophoresis approach to detect bound antibodies to exosome surface markers without need of fluorescent labeling.55 Immunobinding of exosomes with antibodies has resulted in zeta potential changes, thus exhibiting different migration mobilities under electrophoresis depending on marker expression levels. Equipped with a laser dark-field microscope, this platform can measure size, zeta potential, and surface markers from exosomes.55 In contrast to flow cytometry, this method does not require fluorescent labeling for detecting bound antibodies. Additionally, the shift in the zeta potential is not associated with the size of vesicles, as long as the density of bound antibodies is not changed. However, the sensitivity or resolution of the zeta potential change in terms of exosome particle number has not been discussed, and the equipment setup is relatively expensive and complicate. Vaidyanathan et al. introduced a tunable alternating current electrohydrodynamic flow (nanoshearing) in a microfluidic channel that enhances the specificity of capture for sensitive detection of exosomes.56 The multiplexed device allows simultaneous detection of multiple exosome markers on-chip using colorimetric readout visible to the naked eye. The detection method is simple and straightforward. Quantitative detection of markers HER2, PSA, and CD9 was demonstrated for breast cancer diagnosis. More efforts have been made to extract intravesicular exosomal biomarkers (microRNA) for seeking interconnections with cancer disease study and diagnostic potential. Taller et al. substantially improved exosome lysing and the RNA detection process in ~1.5 h by integrating a surface acoustic wave with an ion exchange nanomembrane sensor.57 Note that conventional benchtop methods require at least ~24 h of processing time. Wei et al. employed a nonuniform electrical field to disrupt exosomes and release harbored exosomal RNA/protein biomarkers for on-site monitoring through immune recognition.58,59 The hCD63-GFP expressing exosomes from lung cancer cell line H460, which was stably transfected with hCD63-GFP, were found in vivo to transport to saliva, in addition to serum.
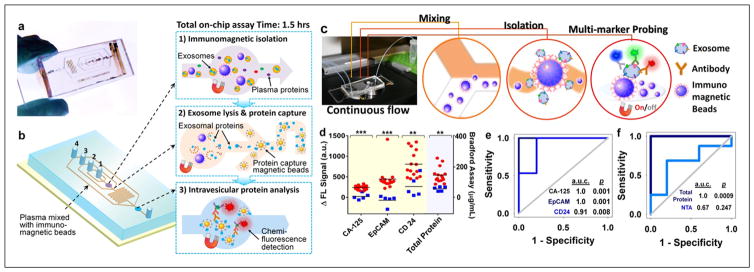
In order to profile both surface and intravesicular exosomal markers, we have developed a cascading chip to integrate exosome isolation, enrichment, chemical lysis, protein immunoprecipitation, and sandwich immunoassay assisted by chemifluorescence detection47 (Fig. 3a,b). Analysis of both type 1 insulin growth factor receptor (IGF-1R) and its intravesicular phosphorylation status from non-small-cell lung cancer patient plasma-derived exosomes was demonstrated. Significant overexpression of exosomal IGF-1R was observed in lung cancer patients compared to healthy individuals. In addition, we employed an ExoSearch chip for blood-based diagnosis of ovarian cancer using multiplexed measurement of three exosomal tumor markers (CA125, EpCAM, CD24) from the same population of CD9 positive exosomes46 (Fig. 3c). Results showed significant diagnostic power (a.u.c. = 1.0, p = 0.001) through a training set of ovarian cancer patient plasma (Fig. 3d,e). In contrast, the diagnostic accuracy of using exosomal particle concentrations measured by NTA was poor, with an a.u.c. of only 0.67 (Fig. 3f), due to the relative large uncertainty in size and concentration measured by NTA. Note that counting exosomes alone has been found insufficient for cancer diagnosis. The results from receiver operating characteristic (ROC) analysis suggested that ExoSearch chip enables sensitive multiplexed exosomal marker detection for blood-based diagnosis of ovarian cancer with significant predictive power. In order to further improve the exosome detection sensitivity and boost the limit for early detection of cancer, we recently introduced a novel graphene oxide/polydopamine (GO/PDA) nanointerface.60 The GO-induced formation of 3D nanoporous PDA surface coating enabled ultrasensitive on-chip ELISA assay of ovarian cancer plasma-derived exosomes at 50 exosomes/μL.
Figure 3.
Microfluidic analysis of exosomal protein markers for liquid biopsy of cancer. (a) Polydimethylsiloxane chip containing a microchannel network for cascading exosome analysis. (b) Integration of streamlined on-chip immunomagnetic isolation of exosomes, chemical lysis, and intravesicular protein analysis. (c) Setup of ExoSearch chip for continuous mixing, isolation, enrichment, and multimarker probing of circulating exosomes. (d) Expression level and ROC analysis (e,f) of three tumor markers (CA125, p < 10−4; EpCAM, p = 0.0009; CD24, p = 0.003) from blood plasma–derived exosomes (nOvCa = 15, nhealthy = 5) using ExoSearch chip, compared to standard Bradford assay and NTA analysis of ultracentrifugation-purified exosomes from matched human subjects. Ovarian cancer patients are represented by red dots, and healthy controls are represented by blue dots. (Figures are adapted with permission from the Royal Society of Chemistry.)
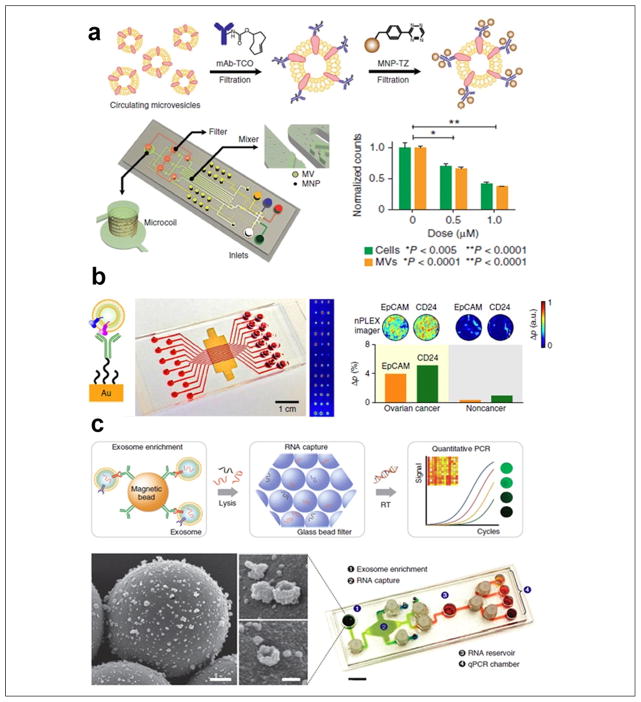
Dr. Hakho Lee’s research group pioneered a study of diagnostic and prognostic roles of circulating exosomes using microfluidic sensing technology. In 2012, Shao et al. devised a microfluidic nuclear magnetic resonance system (μNMR) in which MVs bound with marker-specific magnetic nanoparticles resulted in faster decay of the NMR signal depending on protein expression levels (Fig. 4a). The significance of using blood exosomes in diagnosis and monitoring treatment responses for patients with glioblastoma was demonstrated by probing nine exosomal markers.61 Efficacy of drug treatment, including temozolomide (TMZ) and geldanamycin, has been evaluated using μNMR, as MV numbers decreased in proportion to drug concentration. This platform has better detection sensitivity than standard ELISA and flow cytometry analyses, and further enhancement of sensitivity is under development. Rho et al. refined the exosome sample preparation step in integration with μNMR. A filter membrane cartridge was incorporated to directly isolate and enrich exosomes from blood before labeling with target-specific magnetic nanoparticles for μNMR sensing.62 Im et al. introduced a nanoplasmonic exosome (nPLEX) assay platform that consists of arrays of lattice nanoholes patterned in a gold film within parallel microfluidic channels for label-free, surface plasmon resonance (SPR) sensing (Fig. 4b).63 Ascites samples from ovarian cancer patients and 10 ovarian cancer cell lines were studied by probing 71 exosomal protein markers. Levels of EpCAM and CD24 markers showed significant responses for improving ovarian cancer diagnosis with an accuracy of 97%. The nPLEX platform provided high-throughput screening capability with improved sensitivity, compared to μNMR platform. However, the system complexity of both nPLEX and μNMR requires nanofabrication and skilled operations, which limits the application in routine clinical tests. In 2015, Shao et al. expanded the work by investigating exosomal mRNA in glioblastoma patient blood during treatment, since serial rebiopsy of primary tumors is difficult.64 Real-time PCR analysis of RNA contents released from immunomagnetically captured exosomes was integrated into a microfluidic chip (iMER) for studying key exosomal mRNA markers that could probe epigenetic status of a primary tumor and potentially predict TMZ drug resistance. Results from the above several novel platforms supported the use of exosomes as tumor cellular surrogates. In parallel with CTC research, exosomes were shown to be abundant and stable in circulation, providing significant practical value as surrogates of tumors for liquid biopsy (Fig. 2).
Figure 4.
Microfluidic exosome molecular profiling for monitoring of disease and treatment. (a) μNMR chip with two-step labeling of target proteins on MVs using magnetic nanoparticles. The μNMR microfluidic system is designed to monitor TMZ drug treatment responses for patients with glioblastoma, as MV numbers decrease in proportion to drug concentration. (b) The nPLEX chip evaluated ascites-derived exosomes from ovarian cancer and noncancer patients. Cancer exosomes were captured on EpCAM and CD24-specific sensor sites, which led to intensity changes in the transmitted light (left scheme of principle). The nPLEX chip was integrated with a multichannel microfluidic cell for independent and parallel analyses. Transmission intensities of 12 × 3 nanohole arrays can be measured simultaneously using the imaging setup. (c) The microfluidic iMER chip was developed to integrate (1) capture of cancer exosomes in serum with magnetic microbeads containing affinity ligands, (2) immunoenrichment of exosomal population and lysing for flowing through a glass bead filter and RNA extraction, and (3) elution and reverse transcription for real-time amplification and quantitation. SEM image scale bars are 500 nm, 100 nm (inset). The device has been used for analyzing mRNA levels of drug resistance markers DNA ethyltransferase (MGMT) and alkylpurine-DNA-N-glycosylase (APNG) in enriched tumor exosomes obtained from blood. (Figures are adapted with permission from Nature Publishing Group.)
Discussion
Tumor-derived circulating exosomes have attracted increasing interest for noninvasive cancer diagnosis and monitoring of treatment response, as a promising alternative to liquid biopsy. In addition to microfluidic technology, several other emerging approaches have been implemented to study exosomes and their molecular compositions, such as single exosome detection,65,66 SPR and imaging,67–70 magnetoelectrochemical sensor,71 ExoScreen 96-well plate,72 and EV array.73,74 We summarized detection sensitivity and sensing capability in Table 2, which showed superior performance compared to conventional Western blotting analysis and chemiluminescence ELISA. Such improvements in terms of analytical sensitivity and specificity will greatly address current hurdles in liquid biopsy of cancers. We foresee that a growing number of investigators across disciplines will devote more efforts to advancing exosome research in the next 5 years and beyond.
Table 2.
Detection Performance of Exosome Sensing Technologies.
| Approaches | Detection Sensitivity | Detection Multiplexity | Markers Detected | Reference |
|---|---|---|---|---|
| ExoChip | 0.5 pM | Doi dye staining, no multiplexity | CD63 capture exosomes and extract total RNA | 45 |
| ExoSearch chip | 750 particles/μL | Simultaneous detection of 3 markers | CA125, EpCAM, and CD24 | 46 |
| nPLEX chip | 1000 particles/μL | Parallel detection of 12 potential exosomal markers | EpCAM, CD24, CA125, CA19-9, HER2, MUC18, EGFR, CLDN3, CD45, CD41, D2-40 | 59 |
| GO/PDA nano-IMEX | 50 particles/μL (80 aM) | Single-plex sandwich ELISA | D9, CD63, CD81, EpCAM | 60 |
| iMEX chip | 104 particles/μL | 8 channels for simultaneous detection of 4 markers | EpCAM, CD24, CA125, CD63, HER2, MUC18, EGFR | 67 |
| μNMR chip | 2 × 106 particles/μL | Magnetic nanoparticle labeling | EGFR, PDGFR-α, PDPN, EphA2, EGFRvIII6, IDH1 R132H, HSP90, CD41, MHCII | 57 |
| ExoScreen plate | ELISA grade | 96-well plate | CD9, CD63, CD147, CEA, CA19-9 | 68 |
| Nanoshearing microfluidic approach | 2760 particles/μL | Simultaneous detection of 3 markers | HER2, PSA, CD9 | 56 |
| EV array | 5000 particles/μL | Simultaneous detection of 21 markers | CD9, CD63, CD81, TNF RI, TNF RII, HSAP90, HLA-ABC, GRP78, Mucin16, PLAP, SPA, P53, EGFR, HER2, CD276, Osteopontin, SFTPD, Coilin, NY-ESO-1, EpCAM, PAX-8 | 69 |
| SPR approach | 4.87 × 104 particles/μL, or low picomolar | No multiplexity | CD63, CD9, CD24, CD44, EpCAM, HER2 | 63–66 |
| FLOWER/laser dark-field imaging | Single exosome | No multiplexity | N/A | 61, 62 |
Exosome secretion is a dynamic process, producing diverse populations with 5-fold differences in size and 104-fold differences in concentration;27 as a result, precise measurement and analysis of exosomes is challenging. To date, standardized isolation protocols are still lacking, and the field of exosome research lags behind CTC research because the definition and characterization of exosome types are not yet firmly established. Specific exosomal markers for quantitative evaluation of exosomes with cell origins are urgently needed. In order to increase understanding of exosomes and quantitatively decipher exosomal components, more novel technologies are needed for comprehensive characterization of surface and intravesicular compositions. Interconnections between exosomal RNA, surface protein topography, and posttranslational modification could aid identification of exosome associated with cancer phenotypes. It is worth mentioning that increased MV counts with prolonged storage of blood have been observed, but the protein profile per vesicle in blood displayed negligible changes during blood aging based on Rho et al.’s study.62 These findings point out that sample preparation protocols are critical and need to be standardized. Using highly precise fluid control and automation, microfluidic technology has the capability to address the above issues in sample preparation and isolation, with more power for molecular characterization and sensitive detection. However, in order to achieve clinical utilities in liquid biopsy for cancer, tremendous efforts are still needed to improve the adaptability of the microfluidic technologies to clinical settings and promote the commercialization of the systems. Some of the microfluidic platforms described above require off-chip sample preparation steps, which introduce extensive manual interventions. Microfluidic technologies are still largely developed for operation under the research settings and require expensive setup and well-trained professional to run tests. Simple colorimetric or mobile device–based readout from blood drops is garnering great interest. Other essential research efforts include improving exosome processing throughput and enhancing assay reliability. Overall, in addition to providing new abilities to better elucidate the biology and clinical relevance of exosomes, robustness and ease of use will be the key considerations in microfluidic research to drive the benchtop-to-bedside transition of both technology innovation and scientific advances in exosome biology. Cancer is a complicated and dynamic disease. For accomplishing personalized cancer medicine, the development of a reliable, novel liquid biopsy platform will have tremendous benefit. We anticipate that microfluidic technology will play a game-changing role in exosome analysis and liquid biopsy of cancer.
Acknowledgments
We would like to acknowledge our collaborator, Dr. Andrew Godwin at the University of Kansas Cancer Center, and fund donor Harvey McCarter for supporting our research.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the K-INBRE Developmental Research Project Award from NIH/NIGMS (P20GM103418) and the Innovative Research Award from the Terry C. Johnson Cancer Research Center to M.H., and the J. R. and Inez Jay Award from the University of Kansas, the COBRE Research Project Award under P20GM103638 (NIGMS), and NIH/NCI grant R21CA186846 to Y.Z.
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Pantel K, Alix-Panabieres C. Real-Time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Res. 2013;73(21):6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 2.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 3.Sundar S, Neal RD, Kehoe S. Diagnosis of Ovarian Cancer. BMJ. 2015;351:h4443. doi: 10.1136/bmj.h4443. [DOI] [PubMed] [Google Scholar]
- 4.Hammerschmidt S, Wirtz H. Lung Cancer: Current Diagnosis and Treatment. Dtsch Arztebl Int. 2009;106(49):809–818. doi: 10.3238/arztebl.2009.0809. quiz 819–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menzel C, Kazinski M. Liquid Biopsy: Its Impact on Cancer Diagnostics. MLO Med Lab Obs. 2015;47(7):27–28. [PubMed] [Google Scholar]
- 6.Brock G, Castellanos-Rizaldos E, Hu L, et al. Liquid Biopsy for Cancer Screening, Patient Stratification and Monitoring. Transl Cancer Res. 2015;4(3):280–290. [Google Scholar]
- 7.Sawyers CL. The Cancer Biomarker Problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins MM, Hogarth S. Biomarker Patents for Diagnostics: Problem or Solution? Nat Biotechnol. 2012;30(6):498–500. doi: 10.1038/nbt.2257. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Alix-Panabieres C. Liquid Biopsy: Potential and Challenges. Mol Oncol. 2016;10(3):371–373. doi: 10.1016/j.molonc.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 11.Kowal J, Tkach M, Thery C. Biogenesis and Secretion of Exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Chaput N, Thery C. Exosomes: Immune Properties and Potential Clinical Implementations. Semin Immunopathol. 2011;33(5):419–440. doi: 10.1007/s00281-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 13.Azmi AS, Bao B, Sarkar FH. Exosomes in Cancer Development, Metastasis, and Drug Resistance: A Comprehensive Review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang MK, Wong AS. Exosomes: Emerging Biomarkers and Targets for Ovarian Cancer. Cancer Lett. 2015;367(1):26–33. doi: 10.1016/j.canlet.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Bobrie A, Thery C. Exosomes and Communication between Tumours and the Immune System: Are All Exosomes Equal? Biochem Soc Trans. 2013;41(1):263–267. doi: 10.1042/BST20120245. [DOI] [PubMed] [Google Scholar]
- 16.Simpson RJ, Lim JW, Moritz RL, et al. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Rev Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 17.Mignot G, Roux S, Thery C, et al. Prospects for Exosomes in Immunotherapy of Cancer. J Cell Mol Med. 2006;10(2):376–388. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo M, Raposo G, Thery C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 19.Mora EM, Alvarez-Cubela S, Oltra E. Biobanking of Exosomes in the Era of Precision Medicine: Are We There Yet? Int J Mol Sci. 2015;17(1):13. doi: 10.3390/ijms17010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao H, Chung J, Issadore D. Diagnostic Technologies for Circulating Tumor Cells and Exosomes. Biosci Rep. 2015;36(1):e00292. doi: 10.1042/BSR20150180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko J, Carpenter E, Issadore D. Detection and Isolation of Circulating Exosomes and Microvesicles for Cancer Monitoring and Diagnostics Using Micro-/Nano-Based Devices. Analyst. 2016;141(2):450–460. doi: 10.1039/c5an01610j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinton LT, Sloane HS, Kester M, et al. Formation and Role of Exosomes in Cancer. Cell Mol Life Sci. 2015;72(4):659–671. doi: 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livshts MA, Khomyakova E, Evtushenko EG, et al. Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci Rep. 2015;5:17319. doi: 10.1038/srep17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baranyai T, Herczeg K, Onodi Z, et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One. 2015;10(12):e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita T, Takahashi Y, Nishikawa M, et al. Effect of Exosome Isolation Methods on Physicochemical Properties of Exosomes and Clearance of Exosomes from the Blood Circulation. Eur J Pharm Biopharm. 2016;98:1–8. doi: 10.1016/j.ejpb.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome Isolation for Proteomic Analyses and RNA Profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 27.Raposo G, Stoorvogel W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akers JC, Gonda D, Kim R, et al. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J Neurooncol. 2013;113(1):1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greening DW, Xu R, Ji H, et al. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. Methods Mol Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Rai AJ, Joel DeCastro G, et al. An Optimized Procedure for Exosome Isolation and Analysis Using Serum Samples: Application to Cancer Biomarker Discovery. Methods. 2015;87:26–30. doi: 10.1016/j.ymeth.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Bobrie A, Colombo M, Krumeich S, et al. Diverse Subpopulations of Vesicles Secreted by Different Intracellular Mechanisms Are Present in Exosome Preparations Obtained by Differential Ultracentrifugation. J Extracell Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gyorgy B, Modos K, Pallinger E, et al. Detection and Isolation of Cell-Derived Microparticles Are Compromised by Protein Complexes Resulting from Shared Biophysical Parameters. Blood. 2011;117(4):e39–e48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 33.Müller G. Novel Tools for the Study of Cell Type-Specific Exosomes and Microvesicles. Bioanal Biomed. 2012;4(4):4. [Google Scholar]
- 34.Sokolova V, Ludwig AK, Hornung S, et al. Characterisation of Exosomes Derived from Human Cells by Nanoparticle Tracking Analysis and Scanning Electron Microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 35.van der Pol E, Coumans FA, Grootemaat AE, et al. Particle Size Distribution of Exosomes and Microvesicles Determined by Transmission Electron Microscopy, Flow Cytometry, Nanoparticle Tracking Analysis, and Resistive Pulse Sensing. J Thromb Haemost. 2014;12(7):1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 36.He M, Novak J, Julian BA, et al. Membrane-Assisted Online Renaturation for Automated Microfluidic Lectin Blotting. J Am Chem Soc. 2011;133(49):19610–19613. doi: 10.1021/ja207963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He M, Herr AE. Automated Microfluidic Protein Immunoblotting. Nat Protoc. 2010;5(11):1844–1856. doi: 10.1038/nprot.2010.142. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y, Novak R, Shuga J, et al. High-Performance Single Cell Genetic Analysis Using Microfluidic Emulsion Generator Arrays. Anal Chem. 2010;82(8):3183–3190. doi: 10.1021/ac902683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sackmann EK, Fulton AL, Beebe DJ. The Present and Future Role of Microfluidics in Biomedical Research. Nature. 2014;507(7491):181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 40.Duncombe TA, Tentori AM, Herr AE. Microfluidics: Reframing Biological Enquiry. Nat Rev Mol Cell Biol. 2015;16(9):554–567. doi: 10.1038/nrm4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Y, Wang T. Quantitative Microfluidic Biomolecular Analysis for Systems Biology and Medicine. Anal Bioanal Chem. 2013;405(17):5743–5758. doi: 10.1007/s00216-013-6930-1. [DOI] [PubMed] [Google Scholar]
- 42.Liga A, Vliegenthart AD, Oosthuyzen W, et al. Exosome Isolation: A Microfluidic Road-Map. Lab Chip. 2015;15(11):2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 43.Jo W, Jeong D, Kim J, et al. Microfluidic Fabrication of Cell-Derived Nanovesicles as Endogenous RNA Carriers. Lab Chip. 2014;14(7):1261–1269. doi: 10.1039/c3lc50993a. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Skog J, Hsu CH, et al. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip. 2010;10(4):505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic Device (ExoChip) for On-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab Chip. 2014;14(11):1891–1900. doi: 10.1039/c4lc00136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Z, Yang Y, Zeng Y, et al. A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip. 2016;16(3):489–496. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He M, Crow J, Roth M, et al. Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab Chip. 2014;14(19):3773–3780. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudani JS, Gossett DR, Tse HT, et al. Rapid Inertial Solution Exchange for Enrichment and Flow Cytometric Detection of Microvesicles. Biomicrofluidics. 2015;9(1):014112. doi: 10.1063/1.4907807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Wu HJ, Fine D, et al. Ciliated Micropillars for the Microfluidic-Based Isolation of Nanoscale Lipid Vesicles. Lab Chip. 2013;13(15):2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies RT, Kim J, Jang SC, et al. Microfluidic Filtration System to Isolate Extracellular Vesicles from Blood. Lab Chip. 2012;12(24):5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 51.Lee K, Shao H, Weissleder R, et al. Acoustic Purification of Extracellular Microvesicles. ACS Nano. 2015;9(3):2321–2327. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santana SM, Antonyak MA, Cerione RA, et al. Microfluidic Isolation of Cancer-Cell-Derived Microvesicles from Heterogeneous Extracellular Shed Vesicle Populations. Biomed Microdevices. 2014;16(6):869–877. doi: 10.1007/s10544-014-9891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tauro BJ, Greening DW, Mathias RA, et al. Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line LIM1863-Derived Exosomes. Methods. 2012;56(2):293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Properzi F, Logozzi M, Fais S. Exosomes: The Future of Biomarkers in Medicine. Biomark Med. 2013;7(5):769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 55.Akagi T, Kato K, Kobayashi M, et al. On-Chip Immunoelectrophoresis of Extracellular Vesicles Released from Human Breast Cancer Cells. PLoS One. 2015;10(4):e0123603. doi: 10.1371/journal.pone.0123603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaidyanathan R, Naghibosadat M, Rauf S, et al. Detecting Exosomes Specifically: A Multiplexed Device Based on Alternating Current Electrohydrodynamic Induced Nanoshearing. Anal Chem. 2014;86(22):11125–11132. doi: 10.1021/ac502082b. [DOI] [PubMed] [Google Scholar]
- 57.Taller D, Richards K, Slouka Z, et al. On-Chip Surface Acoustic Wave Lysis and Ion-Exchange Nanomembrane Detection of Exosomal RNA for Pancreatic Cancer Study and Diagnosis. Lab Chip. 2015;15(7):1656–1666. doi: 10.1039/c5lc00036j. [DOI] [PubMed] [Google Scholar]
- 58.Wei F, Yang J, Wong DT. Detection of Exosomal Biomarker by Electric Field-Induced Release and Measurement (EFIRM) Biosens Bioelectron. 2013;44:115–121. doi: 10.1016/j.bios.2012.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu M, Wei F, Yang J, et al. Detection of Exosomal Biomarker by Electric Field-Induced Release and Measurement (EFIRM) J Vis Exp. 2015;(95):52439. doi: 10.3791/52439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang P, He M, Zeng Y. Ultrasensitive Microfluidic Analysis of Circulating Exosomes Using a Nanostructured Graphene Oxide/Polydopamine Coating. Lab Chip. 2016 doi: 10.1039/C6LC00279J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao H, Chung J, Balaj L, et al. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat Med. 2012;18(12):1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rho J, Chung J, Im H, et al. Magnetic Nanosensor for Detection and Profiling of Erythrocyte-Derived Microvesicles. ACS Nano. 2013;7(12):11227–11233. doi: 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Im H, Shao H, Park YI, et al. Label-Free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat Biotechnol. 2014;32(5):490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao H, Chung J, Lee K, et al. Chip-Based Analysis of Exosomal mRNA Mediating Drug Resistance in Glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su J. Label-Free Single Exosome Detection Using Frequency-Locked Microtoroid Optical Resonators. ACS Photonics. 2015;2(9):1241–1245. [Google Scholar]
- 66.Akagi T, Hanamura N, Ichiki T. Measurement of Individual Nanobioparticles on Microfluidic Chips by Laser Dark-Field Imaging. J Photopolym Sci Technol. 2015;28(5):727–730. [Google Scholar]
- 67.Zhu L, Wang K, Cui J, et al. Label-Free Quantitative Detection of Tumor-Derived Exosomes through Surface Plasmon Resonance Imaging. Anal Chem. 2014;86(17):8857–8864. doi: 10.1021/ac5023056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grasso L, Wyss R, Weidenauer L, et al. Molecular Screening of Cancer-Derived Exosomes by Surface Plasmon Resonance Spectroscopy. Anal Bioanal Chem. 2015;407(18):5425–5432. doi: 10.1007/s00216-015-8711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rupert DLM, Lasser C, Eldh M, et al. Determination of Exosome Concentration in Solution Using Surface Plasmon Resonance Spectroscopy. Anal Chem. 2014;86(12):5929–5936. doi: 10.1021/ac500931f. [DOI] [PubMed] [Google Scholar]
- 70.Di Noto G, Bugatti A, Zendrini A, et al. Merging Colloidal Nanoplasmonics and Surface Plasmon Resonance Spectroscopy for Enhanced Profiling of Multiple Myeloma-Derived Exosomes. Biosens Bioelectron. 2016;77:518–524. doi: 10.1016/j.bios.2015.09.061. [DOI] [PubMed] [Google Scholar]
- 71.Jeong S, Park J, Pathania D, et al. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano. 2016;10(2):1802–1809. doi: 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra-Sensitive Liquid Biopsy of Circulating Extracellular Vesicles Using ExoScreen. Nat Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jorgensen M, Baek R, Pedersen S, et al. Extracellular Vesicle (EV) Array: Microarray Capturing of Exosomes and Other Extracellular Vesicles for Multiplexed Phenotyping. J Extracell Vesicles. 2013;2:20920. doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jorgensen MM, Baek R, Varming K. Potentials and Capabilities of the Extracellular Vesicle (EV) Array. J Extracell Vesicles. 2015;4:26048. doi: 10.3402/jev.v4.26048. [DOI] [PMC free article] [PubMed] [Google Scholar]