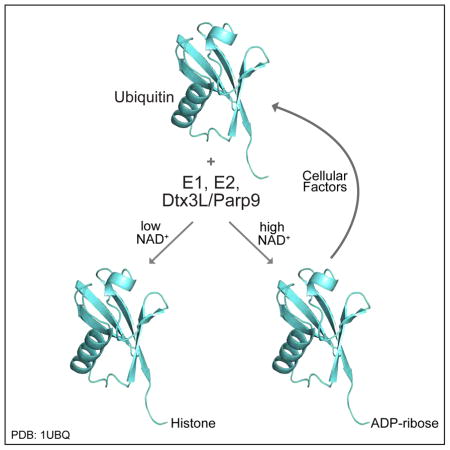
SUMMARY
ADP-ribosylation of proteins is emerging as an important regulatory mechanism. Depending on the family member, ADP-ribosyltransferases conjugate either a single ADP-ribose to a target, or generate ADP-ribose chains. Here we characterize Parp9, a mono-ADP-ribosyltransferase reported to be enzymatically inactive. Parp9 undergoes heterodimerization with Dtx3L, a histone E3 ligase involved in DNA damage repair. We show that the Dtx3L/Parp9 heterodimer mediates NAD+- dependent mono-ADP-ribosylation of ubiquitin, exclusively in the context of ubiquitin processing by E1 and E2 enzymes. Dtx3L/Parp9 ADP-ribosylates the carboxyl group of Ub Gly76. Because Gly76 is normally used for Ub conjugation to substrates, ADP-ribosylation of the Ub carboxyl terminus precludes ubiquitylation. Parp9 ADP-ribosylation activity therefore restrains the E3 function of Dtx3L. Mutation of the NAD+ binding site in Parp9 increases the DNA repair activity of the heterodimer. Moreover, poly-ADP-ribose binding to the Parp9 macrodomains increases E3 activity. Dtx3L heterodimerization with Parp9 enables NAD+ and poly- ADP-ribose regulation of E3 activity.
Graphical Abstract
INTRODUCTION
The ADP-ribosyltransferase (ART) family of enzymes is defined on the basis of an evolutionarily conserved catalytic domain that cleaves NAD+ and produces ADP-ribose (ADPr) and nicotinamide. ADPr generated from this reaction can be covalently attached to protein, and depending on the ART, extended to generate polymers of ADPr (PAR). The founding member of the ART family, Parp1, along with Parp2, Parp3, and tankyrases1 and 2, generate PAR which is used to regulate diverse nuclear pathways including DNA repair, transcription, and maintenance of telomere structure (Kraus, 2015). As the most extensively studied of these enzymes, Parp1 has become an important drug target in malignancies such as prostate cancer (Feng et al., 2015). Most of the 17 ART family members do not produce poly-ADPr, rather, they catalyze addition of a single ADPr to a protein substrate (mono-ADP-ribosylation). Mono-ADP-ribosylation by mammalian ARTs is analogous to reactions catalyzed by certain bacterial toxins (Hottiger, 2015b; Hottiger et al., 2010). But in contrast to the wealth of information on bacterial toxins that have been characterized in detail both biochemically and structurally (Deng and Barbieri, 2008), our understanding of the reactions that generate mono-ADPr and effects on protein function is very limited. Given the number of mono-ARTs expressed in mammals, one can anticipate that mono-ARTs contribute to a variety of cellular pathways (Hottiger, 2015a; Scarpa et al., 2013). To date, however, only a few direct substrates of mono-ADPr addition have been identified.
The ART member known as BAL1/Parp9/ARTD9 (referred to here as Parp9) contains a putative catalytic domain that is highly similar to several ART family members. For example, the Parp9 catalytic domain is 30% identical to that of Parp10/ARTD10, one of the few ARTs for which a substrate has been identified (Feijs et al., 2013; Verheugd et al., 2013). Parp9, however, is thought to be enzymatically inactive because it does not undergo auto-ADP-ribosylation, a common property of these enzymes (Vyas et al., 2014). Parp9 was discovered by Shipp and colleagues as a risk-associated gene in diffuse large B cell lymphoma and subsequently shown to physically interact with the ubiquitin (Ub) E3 ligase Dtx3L (Aguiar et al., 2000; Takeyama et al., 2003). Parp9 and Dtx3L are co-expressed from a bidirectional promoter and assemble into a heterodimer (Takeyama et al., 2003). Dtx3L contains a RING domain and mediates mono-ubiquitylation (mono-Ub) of core histones, which in the context of a DNA damage response includes histone H4 (Yan et al., 2009). The exact function of Parp9 in this setting is not entirely clear, but its contribution likely involves tandemly-arranged modules in Parp9 termed macrodomains that bind ADPr and PAR. Micro-irradiation experiments were used to show the Parp9 macrodomains are sufficient to rapidly localize a GFP reporter to DNA damage sites in cells, suggesting that Parp9 acts as a targeting factor that directs the heterodimer to PAR-containing sites generated by Parp1. Given this occurs within minutes of DNA damage, Dtx3L/Parp9 was proposed to contribute to the DNA damage response through Histone H4 mono-ubiquitylation (Yan et al., 2013).
Dtx3L and Parp9 are relatively abundant proteins in prostate cancer cells where they can promote survival and chemo-resistance (Bachmann et al., 2014). We posited that the complex of Dtx3L and Parp9 might function in a manner that couples the two enzyme activities of the proteins. Although Parp9 is thought to lack catalytic activity, our biochemical analysis indicates that the recombinant Dtx3L/Parp9 heterodimer can cleave NAD+ and generate nicotinamide and ADPr. We found that the ADPr produced from this reaction is transferred to Ub, thereby generating Ub-ADPr. In addition to requiring NAD+, ADP-ribosylation of Ub is dependent on E1 and E2 enzymes, ATP, and ambient temperature, all indicating the reaction requires Ub processing. Using an antibody that recognizes the free C-terminus of Ub, reagents to probe the chemical sensitivity of the ADP-ribose-Ub linkage, and mass spectrometry, we show that the site of ADP-ribosylation is the carboxyl terminus of Ub Gly76. Because the carboxyl group of Gly76 is used to create the linkage between Ub and Lys residues in target proteins, and with Lys residues in Ub for chain formation, ADP-ribosylation of Ub precludes conjugation. ADP-ribosylation of Ub and Ub conjugation are mutually exclusive reactions, and in vitro, the outcome of the reaction is determined by NAD+ concentration. Dtx3L/Parp9 contributes to DNA repair by non-homologous end joining (NHEJ) and its activity is restrained by Parp9 catalytic function. We propose that NAD+ levels regulate the E3 activity of Dtx3L/Parp9 in the context of DNA repair.
RESULTS
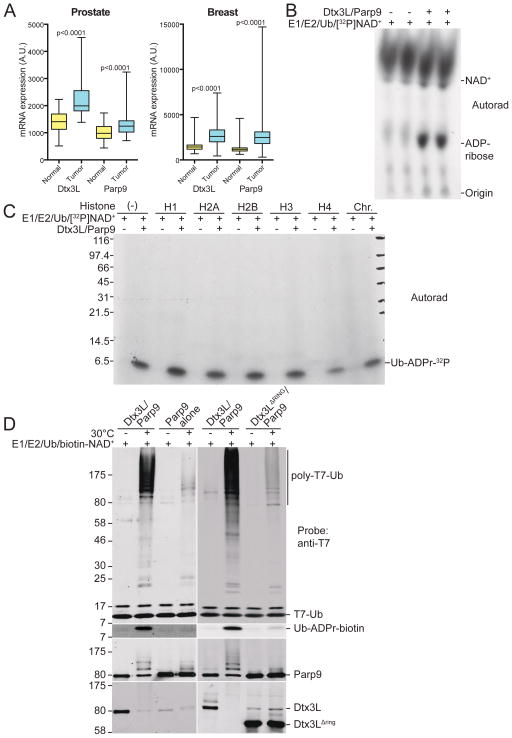
Data from The Cancer Genome Atlas (TCGA) revealed the expression levels of the E3 ligase Dtx3L and its binding partner, Parp9, are significantly higher in cancers from prostate (Gleason grade 7–10) and breast (Figure 1A), as well as other solid tumors (Figure S1). The genes encoding Dtx3L and Parp9 proteins share a bidirectional promoter and can be induced by interferon (IFN) (Bachmann et al., 2014; Juszczynski et al., 2006). Accordingly, IFN-α treatment of VCaP cells, a highly aggressive metastatic prostate cancer line, induced the expression of Dtx3L and Parp9, and the proteins co-eluted from a Superdex200 column with a molecular size in excess of 400 kDa (Figure S2A). Given the absence of monomeric Dtx3L or Parp9 in the gel filtration profile, and the fact that the two proteins co-immunoprecipitate (Figure S2B), Dtx3L and Parp9 form a stable complex in prostate cancer cells. To analyze the properties of the Dtx3L/Parp9 complex, we co-expressed the proteins in bacteria and purified the heterodimer via a His-tag on Parp9. Dtx3L contains a RING domain and can function as an E3 ligase for histones (Takeyama et al., 2003), so we first tested the complex for histone mono-ubiquitylation activity. Reactions containing E1 (Ube1), E2 (UbcH8), biotin-tagged Ub, ATP, and E3 (Dtx3L/Parp9 complex) generated Ub-modified forms of recombinant histones including H4 (Figure S3A), which is consistent with a previous study that showed Dtx3L has E3 activity (Yan et al., 2009). Dtx3L/Parp9-dependent mono-ubiquitination of Histone H2B, both as a recombinant protein and within a core histone preparation (Figure S3B), was detected using an antibody specific for H2B-Lys120-Ub, a modification linked to nucleosome structure and stability (Fuchs and Oren, 2014; Minsky et al., 2008). These data show that recombinant Dtx3L/Parp9 heterodimer is biochemically active as a histone E3 ligase.
Figure 1. Expression and activity of the Dtx3L/Parp9 heterodimer.
(A) Box and whisker plots showing expression of Dtx3L and Parp9 in cancer (blue) compared to adjacent normal tissue (yellow) for prostate and breast cancer, and nine additional cancers (Figure S1). RNA-Seq by Expectation-Maximization (RSEM) values from TCGA were used to plot normalized expression, whiskers represent the minimum and maximum data values, and significance was calculated using a paired t-test.
(B) Recombinant Dtx3L/Parp9 cleaves NAD+. Ubiquitylation reactions containing 32P-NAD+ were analyzed by TLC and autoradiography, with unlabeled ADP-ribose as a migration standard.
(C) Dtx3L/Parp9 ADP-ribosylates Ub. Ubiquitylation reactions with purified Dtx3L/Parp9, histones, and chromatin in the presence of 32P-NAD+. The reactions were subjected to SDS-PAGE, Coomassie Blue (CB) staining (Figure S3D), and autoradiography.
(D) Ubiquitylation reactions with different combinations of human Dtx3L and Parp9 purified from insect cells. Biotin-labeled NAD+ was added to monitor ADP-ribosylation of Ub (FL-Neutra detection).
Dtx3L/Parp9 has Dual Activities
Parp9 function is not well understood, though its localization to DNA damage sites in HeLa cells clearly suggests a role in DNA repair (Yan et al., 2013). Parp9 contains a ~220 amino acid catalytic domain with homology to other ARTs, which includes 35% identity to its closest family member Parp14. In a family-wide analysis of ARTs, Parp14 displayed mono-ADP-ribosyl transferase activity, but Parp9 did not undergo auto-modification, which led to the conclusion it lacks enzyme activity (Aguiar et al., 2005). Given that Parp9 forms a heterodimer with Dtx3L, we considered whether its enzyme activity might require dimerization with Dtx3L or that ADP-ribosylation activity might be apparent in the context of E3 function. We also considered whether Parp9 might mono-ADP-ribosylate histones, a function demonstrated for Parp10 (Kleine et al., 2008). Because ARTs cleave NAD+ to generate ADP-ribose (ADPr), we added 32P-NAD+ to Ub reactions and analyzed the products by thin layer chromatography (TLC). Purified Dtx3L/Parp9 cleaved 32P-NAD+ and generated a product with the same mobility as unlabeled ADPr (Figure 1B). Using an assay that measures nicotinamide production through a coupled reaction (Smith et al., 2009), we determined that the Km of Dtx3L/Parp9 for NAD+ (197±64 μM; Figure S3C) is comparable to other ART family members (Kleine et al., 2008; Mendoza-Alvarez and Alvarez-Gonzalez, 1993). Also like other mono-ARTs (Wahlberg et al., 2012), Dtx3L/Parp9 cleavage of NAD+ was insensitive to the Parp1 inhibitor olaparib (data not shown). From these data, we conclude that recombinant Dtx3L/Parp9 heterodimer has an associated NADase activity that generates nicotinamide and ADPr.
Dtx3L/Parp9 Modifies Ub
To test whether Dtx3L/Parp9 has catalytic activity towards histones, and possibly other components of the Ub reaction, we performed ubiquitylation assays using 32P-NAD+ and analyzed the products by SDS-PAGE and autoradiography. Unexpectedly, we found that Dtx3L/Parp9 mediated ADP-ribosylation of Ub (Ub-ADPr), both in the absence and presence of histones and chromatin (Figure 1C). There was no evidence of Parp9 automodification, Ub-ADPr conjugation to histones, or ADP-ribosylation of H1 or core histones, all of which were visible by Coomassie blue staining (Figure S3D). Thus, the Dtx3L/Parp9 heterodimer can use NAD+ to produce ADPr and nicotinamide, and in the presence of the Ub machinery, selectively ADP-ribosylate Ub.
We characterized the ADP-ribosylation of Ub using biotin-NAD+ as the source of ADPr and FL-Neutravidin detection of the reaction product, Ub-ADPr. We also prepared and tested recombinant Dtx3L/Parp9 heterodimer and Parp9 monomer from insect cells. The Dtx3L/Parp9 heterodimer produced in insect cells had robust E3 activity and it mediated ADP-ribosylation of Ub (Figure 1D, lane 2). By contrast, Parp9 alone, and Dtx3L/Parp9 heterodimer lacking the RING domain, were both defective for Ub ADP-ribosylation (Figure 1D, lanes 4, 8). Thus, Parp9 is necessary but not sufficient for ADP-ribosylation of Ub, and the RING domain of Dtx3L is required for the reaction.
Ub ADP-ribosylation Requires E1/E2/E3 Processing
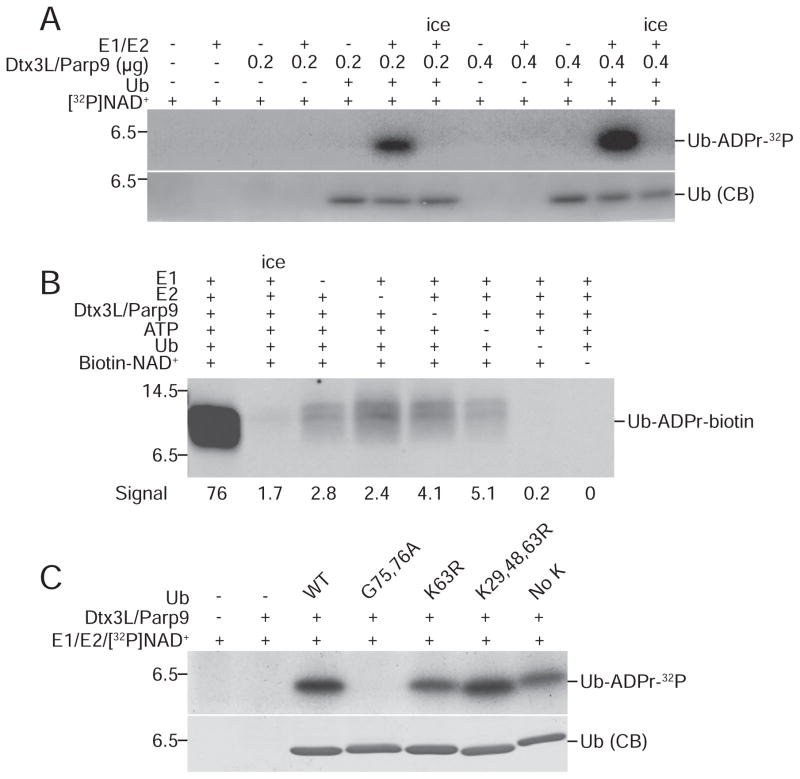
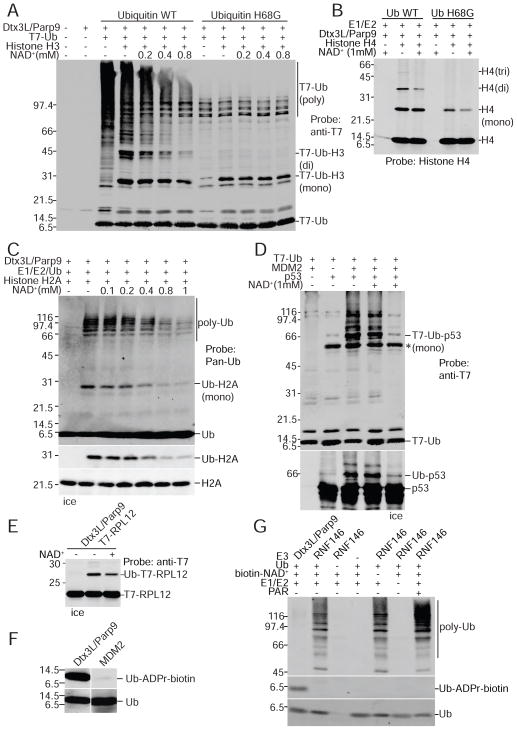
RING domains in E3 enzymes play essential roles in ubiquitylation through E2~Ub thioester binding and Ub transfer to substrates (Deshaies and Joazeiro, 2009). Because Ub-ADPr formation was dependent on the RING domain of Dtx3L, we examined the contribution of each component in the ubiquitylation reaction. Using 32P-NAD+ to monitor the reaction, we found that upon E1 and E2 enzyme omission there was a complete loss of Ub ADP-ribosylation, while increasing the amount of Dtx3L/Parp9 in the reaction caused a proportionate increase in Ub-ADPr formation (Figure 2A). With biotin-NAD+ as the co-factor and FL-Neutravidin detection, dropout of individual components (lanes 3–6) reduced Ub-ADPr-biotin generation by 15 to 40-fold (Figure 2B). These data indicate that Dtx3L/Parp9-dependent ADP-ribosylation of Ub requires canonical processing by the Ub E1 and E2 enzymes. This conclusion was further corroborated using mutant forms of Ub. The processing deficient Ub mutant G75, 76A was not ADP-ribosylated by Dtx3L/Parp9, while various Lys mutants were modified to about the same extent as WT Ub (Figure 2C).
Figure 2. ADP-ribosylation of Ub by Dtx3L/Parp9 requires E1 and E2 processing.
(A) ADP ribosylation reactions were performed using 32P-NAD+ and analyzed by SDS-PAGE and autoradiography with the protein combinations indicated.
(B) ADP-ribosylation reactions were performed using biotin-NAD+ with drop-out of individual components as indicated. ADP-ribosylation of Ub was measured by probing with FL-Neutra.
(C) ADP-ribosylation reactions were performed using 32P-NAD+ and Ub mutants bearing amino acid substitutions that affect processing (G75, 76A) and conjugation (K63R; K29, 48, 63R; No K).
Parp9 Catalytic Domain Function is required for ADP-ribosylation
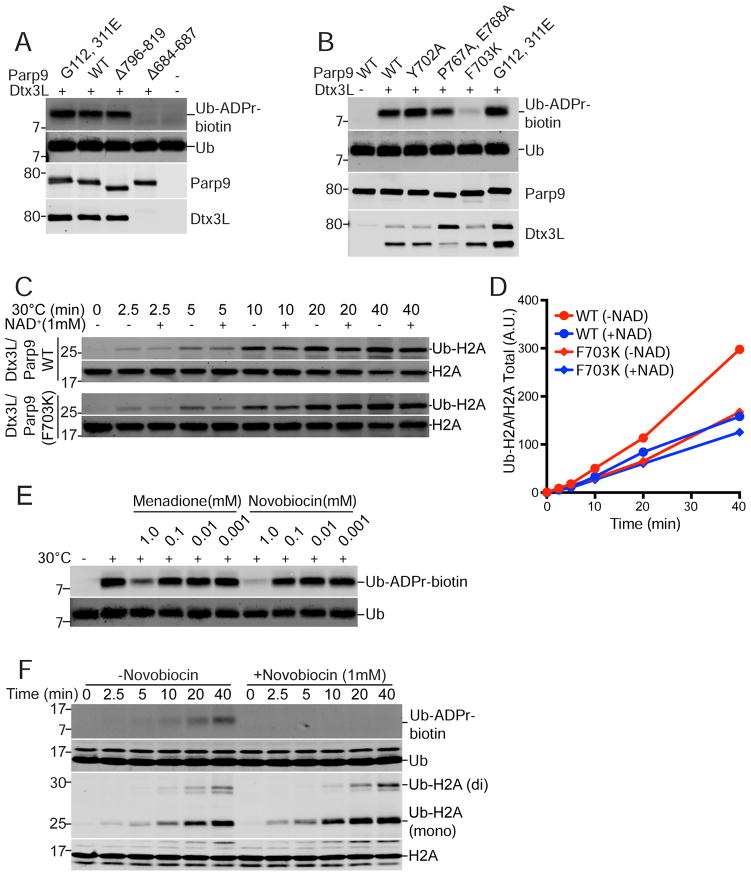
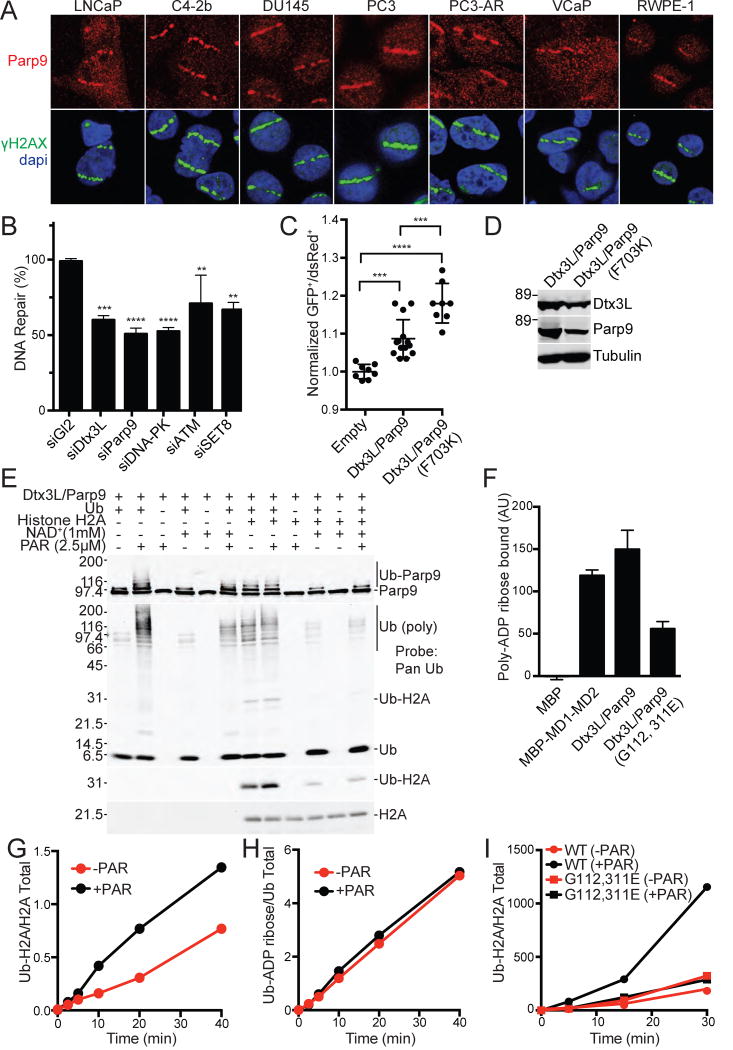
We used mutational approaches to determine which domain of Parp9 is responsible for Ub ADP-ribosylation. Since we found that heterodimerization with Dtx3L is required for Ub ADP-ribosylation (Figure 1D), we also examined if changes introduced into Parp9 affected binding to Dtx3L. Parp9 contains tandem copies of the macrodomain, a module that in some proteins displays enzymatic function (Rosenthal et al., 2013). Introduction of point mutations into the two Parp9 macrodomains (G112, 311E; denoted MUT1) reduced PAR binding by ~50% but did not affect Ub ADP-ribosylation or heterodimerization with Dtx3L in preparations from mammalian cell and E. coli (Figures 3A, B, C, S4A, B). Next, we introduced amino acid changes in the Parp9 catalytic domain modeled on loss-of-function mutations characterized in Parp1 (Rolli et al., 1997) (Figures S4C, S5A,B,C). Removing 24 residues from the C-terminal region of Parp9 had no appreciable effect on ADP-ribosylation of Ub or heterodimerization with Dtx3L, but deleting 50 amino acids from the C-terminus abolished both activities (Figures 3A, S5C). Small internal deletions in the Parp9 catalytic domain also resulted in loss of both activities, presumably due to an altered catalytic domain structure incompatible with heterodimerization (Figure S5C). The loss-of-function resulting from a six amino acid deletion in Parp9 (Δ698-703; Figure S4C) attracted our attention because based on modeling this segment is part of a beta-strand within the NAD+ binding site (Figure S4D). We generated individual point mutations within the predicted beta-strand, expressed the mutants in E coli, and tested them in our assays. The Parp9 F703K substitution (MUT 13) had a strong reduction in Ub ADP-ribosylation while maintaining heterodimeration with Dtx3L (Figure 3B; structure depicted in Figure S4D). Altering the adjacent amino acid (Y702A), which is conserved in Parps, did not affect Dtx3L/Parp9 activity (Figures 3B, S5C). Importantly, the Parp9 F703K substitution did not inhibit the E3 activity of the heterodimer towards Histone H2A (Figure 3C, D), consistent with the mutation selectively affecting ADP-ribosylation mediated by the Parp9 catalytic domain.
Figure 3. Ub ADP-ribosylation mediated by the Parp9 catalytic domain.
(A) Parp9 mutations engineered in the macrodomains (G112, 311E) and catalytic domain (Δ796-819; Δ684-687) expressed in E. coli and tested for Ub ADP-ribosylation (ADPr-biotin) and heterodimerization with Dtx3L. The locations of the mutations are depicted within a structural model of Parp9 (Figure S4).
(B) Parp9 mutations engineered in the catalytic domain (Y702A; P767A, E768A; F703K) expressed in mammalian cells and tested for Ub ADP-ribosylation (ADPr-biotin) and heterodimerization with Dtx3L.
(C, D) E3 activity of WT Parp9 and F703K Parp9 using recombinant heterodimers with Dtx3L.
(E) Effect of non-selective Parp inhibitors Menadione and Novobiocin on Ub ADP-ribosylation.
(F) Time course of Ub-ADP-ribosylation and H2A ubiquitylation in the presence of Novobiocin.
Additional evidence that Parp9 catalytic function is required for ADP-ribosylation of Ub was obtained using selective and non-selective Parp inhibitors (Banasik et al., 1992; Wahlberg et al., 2012). Two Vitamin K analogues (Menadione, Phylloquinone) reduced Ub ADP-ribosylation, as did the antibiotic Novobiocin (Figures 3E, S5D). In a time course experiment, Novobiocin inhibited Dtx3L/Parp9-mediated ADP-ribosylation of Ub without reducing mono-ubiquitylation of Histone H2A (Figure 3F). The mutagenesis and inhibitor data show that ADP-ribosylation of Ub requires the catalytic function of Parp9, and that the E3 and ADP-ribosylation activities of the Dtx3L/Parp9 heterodimer are separable.
Ub is mono-ADP-ribosylated
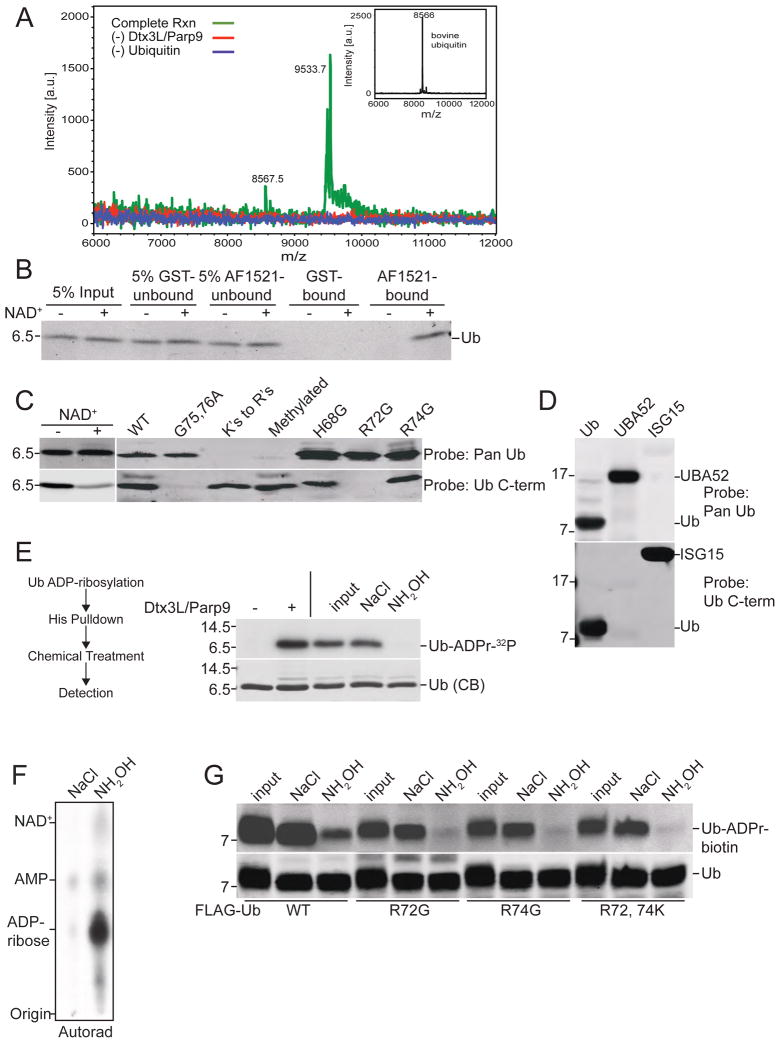
We next used mass spectrometry (MS) to characterize the Ub-ADPr product generated by Dtx3L/Parp9. Because under low NAD+ concentrations a small fraction of the total Ub appeared to be modified by ADPr, we performed Ub reactions with biotin-NAD+ and enriched for the Ub-ADPr-biotin product on streptavidin beads. The spectra generated from a complete reaction, and from reactions performed in the absence of Dtx3L/Parp9 and Ub revealed clear mass differences. The Ub product from the complete reaction had a mass (9533.7) that was larger (+962.8) than Ub alone (8570.1) and consistent with addition of a single molecule of biotin-ADPr (predicted total mass: 960.8) (Figure 4A, green tracing). To corroborate our finding that Ub was modified by ADPr, we took advantage of the ADP-ribose binding macrodomain from Archaeoglobus fulgidus (Af1521) (Karras et al., 2005). Recombinant Af1521 shows 1000-fold selectivity for ADP-ribose over NAD+, and can bind ADP-ribose conjugated to proteins (Dani et al., 2009). For our application, the Af1521 macrodomain was immobilized on beads as a GST fusion protein, and used in pull-down experiments with Ub from ADP-ribosylation reactions. Ub binding to the Af1521 macrodomain was only observed if the reaction contained NAD+ (Figure 4B). Given the selectivity of Af1521 for ADPr (Karras et al., 2005), the +962.8 mass difference compared to Ub observed by MS, and the fact that in both assays Dtx3L/Parp9 addition was required to generate ADPr, we conclude the E3 heterodimer mediates covalent attachment of a single ADPr to Ub, and that this occurs specifically in the context of Ub processing by E1 and E2.
Figure 4. Dtx3L/Parp9 ADP-ribosylates the C-terminus of Ub.
(A) Spectra from MALDI-TOF analysis of Ub. ADP-ribosylation reactions were performed with biotin-NAD+ with drop-out of Dtx3L/Parp9 and purified bovine Ub (inset shows unmodified Ub). ADPr-biotin labeled reaction product was isolated on streptavidin beads and analyzed by MS.
(B) Recovery of ADP-ribosylated Ub on recombinant Af1521 macrodomain. Ub ADP-ribosylation reactions performed in the absence and presence of unlabeled NAD+ were dialyzed and then combined with GST- and GST-Af1521 beads. The input, unbound, and bound fractions were analyzed by immunoblotting with a pan-Ub Ab.
(C) ADP-ribosylation of Ub reduces C-term antibody binding. ADP-ribosylation reactions were performed in the absence and presence of unlabeled NAD+ and Ub immunoblotting performed with Pan-Ub and Ub C-term Abs (left panel). Epitope mapping of Pan-Ub and Ub C-term Abs using WT and mutant Ub proteins (right panel).
(D) Mapping the C-term Ab epitope with Ub proteins. Recombinant Ub, Uba52, or ISG15 (mature form) was separated via SDS-PAGE and probed with Pan-Ub antibody or Ub C-term antibody. Alignments of the C-terminal domains of the Ub proteins are provided (Figure S6C).
(E) Chemical sensitivity of ADP-ribose conjugated to Ub. ADP-ribosylation reactions containing His-Ub and 32P-NAD+ were recovered on cobalt beads, treated as indicated, and analyzed by SDS-PAGE and autoradiography.
(F) Release of 32P-ADPr from Ub by NH2OH treatment analyzed by TLC.
(G) WT and Ub mutants were ADP-ribosylated using biotin-NAD+ and ADPr release determined by probing with FL-Neutra. ADPr release from Ub is CHES-resistant (Figure S6E).
The Ub C-terminus is the Site of ADP-ribosylation
Modification by ADPr occurs on a variety of amino acids, including Lys, Arg, Glu, Asp, and Cys (Hottiger et al., 2010). Amino acid assignment of ADP-ribosylation sites by MS is challenging due to chemical lability of the protein-ribose linkage and inadvertent fragmentation of ADPr (Hengel and Goodlett, 2012). Fortuitously, existing Ub antibodies (Abs) were useful for deducing the site of ADPr modification. A pan-Ub Ab, whose epitope includes one or more Lys residues but maps outside of the C-terminus, showed similar levels of binding to unmodified and ADPr-modified Ub (Figure 4C, lane 1, 2 top panel). By contrast, a monoclonal Ab to the C-terminal region of Ub (Ub C-term) showed reduced binding to ADPr modified Ub (Figure 4C, lane 1,2 bottom panel). Although the exact epitope for the Ub C-term Ab has not been described, we determined that binding is strictly dependent on Arg72, Gly75, and Gly 76 in Ub (Figure 4C), moreover it binds the C-terminal sequence in the Ub-related protein ISG15 (Arg-Leu-Arg-Gly-Gly; Figure 4D). Ub C-term Ab binding is blocked in unprocessed Ub (UBA52; Figure 4D), and when Ub and ISG15 are conjugated to their respective substrates through the C-terminal Gly (Figure S6A, B). The epitope mapping data (summarized in Figure S6C) shows unequivocally that Ub C-term Ab recognition requires a free C-terminus in Ub. The NAD+- dependent reduction in Ub C-term Ab reactivity (Figure 4C, lane 2) is consistent with ADP-ribosylation of the Ub C-terminus on Gly76. Mutation of Gly76 to Ala eliminates ADP-ribosylation of Ub (Figure S6D).
Corroborating evidence that the C-terminus is the site of ADPr addition to Ub was obtained by analyzing the chemical sensitivity of the modification. Based on chemical reactivity three amino acids within the C-term Ab epitope could, theoretically, be sites of ADPr addition; the guanidino groups on the side chains of Arg72 and Arg74, and the carboxyl group on Gly76. Neutral hydroxylamine (NH2OH) rapidly releases ADPr from carboxyl groups, whereas 2- (cyclohexylamino)ethanesulphonic acid (CHES) causes a slow release (Cervantes-Laurean et al., 1997). ADPr-modified Arg is released slowly by NH2OH and resistant to CHES (Cervantes-Laurean et al., 1997). We prepared ADPr-modified Ub using 32P-NAD+, added neutral NH2OH, and used SDS-PAGE and autoradiography to show that ADPr was efficiently released by the treatment (Figure 4E, F, G). Incubation with CHES resulted in a slow release of ADPr detectable by 2 hrs. (Figure S6E). To further eliminate Arg as the potential site of ADP-ribosylation, we generated recombinant forms of Ub with substitutions in Arg residues near the C-terminus of Ub. Analysis of Ub Arg72Lys, Ub Arg74Lys, and Ub Arg72, 74Lys showed that all three mutant Ub proteins underwent ADP-ribosylation, and, like WT Ub, ADPr was released from the mutant Ub proteins with neutral NH2OH (Figure 4G). The epitope mapping and epitope masking data, the NH2OH-sensitivity and relative CHES-resistance, and the Ub point mutant results are all consistent with ADPr addition to the carboxyl group of Gly76 on the C-terminus of Ub.
ADP-ribosylation of Ub is Reversible
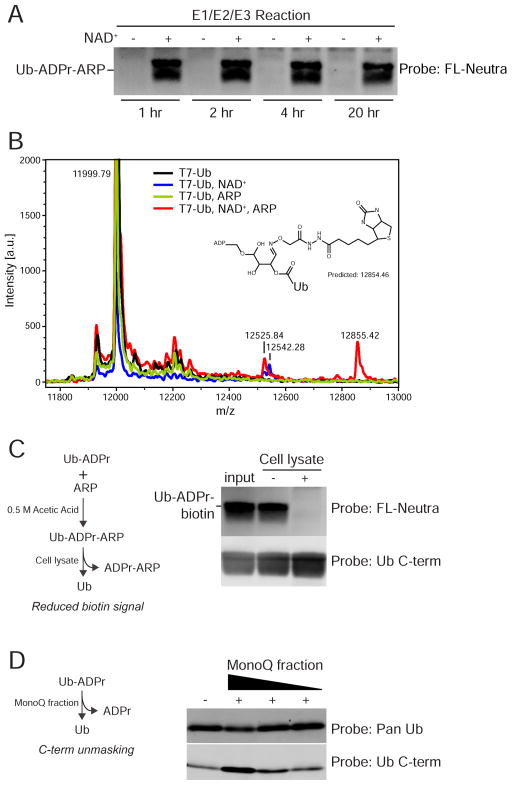
Neutral NH2OH releases ADPr from an acidic R-group by attacking the carbonyl carbon (Zhang et al., 2013). We explored whether a similar reaction could be performed with the probe N-(Aminooxyacetyl)-N'-biotinylhydrazine (ARP) that contains a terminal O-NH2. E1/E2/E3 reactions were performed in the absence and presence of NAD+, and subsequently acidified and incubated with ARP. By probing for the biotin moiety, we determined that ARP reactivity with Ub is specific for ADP-ribosylated Ub since the Ub-ARP product was not generated if the E1/E2/E3 reaction was performed without NAD+ (Figure 5A). We analyzed the products by MS and determined that ARP modification of Ub-ADPr generates an adduct (12855) that retains the ADPr, and consistent with the blotting, is dependent on NAD+ in the reaction (Figure 5B, red tracing). A simple explanation for adduct formation is that the ribose ring in ADPr undergoes a rearrangement that creates an aldehyde (Morgan and Cohen, 2015), which is reactive with ARP (Figure S6F). We used Ub- ADPr-ARP as the substrate and biotin detection to determine whether cell extracts contain an activity that removes ADPr from Ub. Addition of PC-3 cell lysate abolished the biotin signal, consistent with removal of ADPr-ARP by a cellular factor (Figure 5C). Addition of MonoQ fractionated cytosol to ADP-ribosylated Ub increased immunoreactivity with the Ub C-term Ab (Figure 5D). This indicates that release of ADPr by a cellular factor restores the carboxyl group on Gly76. These data show that cells have the capacity to reverse ADP-ribosylation of Ub by Dtx3L/Parp9.
Figure 5. ADP-ribosylated Ub is reactive with N-(aminooxyacetyl)-N'-(D-Biotinoyl) hydrazine (ARP).
(A) ARP modification of Ub requires initial ADP-ribosylation. The E1/E2/E3 reactions were performed in the absence and presence of NAD+. In a second step, ARP reactions were performed at room temperature for the times indicated, followed by FL-Neutra detection of the biotin moiety in ARP.
(B) Spectra of reactions containing combinations of Ub, NAD+, and ARP. The 12855.42 Da mass is unique to T7-Ub-ADPr labeled with ARP. A putative structure consistent with the mass is shown, along with a potential mechanism (Figure S6F).
(C) ADP-ribosylation of Ub is reversible. T7-Ub-ADPr-ARP was generated, incubated with cell buffer or cell lysate, and subsequently assayed for the loss of ADPr-ARP by streptavidin detection of biotin.
(D) Removal of ADPr from Ub restores C-term Ab reactivity. Ub was ADP-ribosylated, incubated with a MonoQ fraction, subsequently immunoblotted with Pan and Ub C-term antibodies.
NAD+ Inhibits Ub Conjugation by Dtx3L/Parp9
The C-terminus of Ub is required for processing by E1 and E2, and following its activation, the carboxyl group of Gly76 is used to generate a covalent bond using the epsilon amino group of Lys residues both in acceptor proteins and in Ub itself (Schulman and Harper, 2009). ADPr attachment to Ub is therefore predicted to inhibit Ub conjugation to protein substrates, though we considered the possibility that Ub-ADPr might be an unusual intermediate in the Ub conjugation pathway (Berndsen and Wolberger, 2014; Komander and Rape, 2012). We explored these possibilities by performing ubiquitylation assays with NAD+ concentrations comparable to those reported in human tissue (Zhu et al., 2015). We then analyzed the Ub conjugation products by immunoblotting. We found that including NAD+ in the reaction strongly reduced poly-Ub formation (Figure 6A). Increasing the concentration of NAD+ also reduced the level of Histone H3 modified with two copies of Ub, and to some extent, the level of Histone H3 modified by a single Ub (Figure 6A). Using a Ub mutant that is active for mono-ubiquitylation but defective for chain formation (Ub H68G), NAD+ had no obvious effect on mono-Ub of Histone H3 (Figure 6A). Thus, with Histone H3, NAD+ does not appear to interfere with ATP-dependent activation of Ub and its transfer to substrate. NAD+ reduced the levels of di-modified Histone H4 with WT Ub, and mono-modified with Ub H68 (Figure 6B). NAD+ reduced Ub conjugation to Histone H2A (Figure 6C), but had only a slight effect on ribosomal protein RPL12 (Figure 6E). By contrast, NAD+ did not affect MDM2-mediated Ub conjugation to p53 (Figure 6D), underscoring the fact that NAD+ is not a general inhibitor of E1/E2/E3 function. Importantly, Ub did not undergo ADP-ribosylation in the presence of MDM2 or RNF146 under conditions where these E3s are biochemically active (Figures 6F, G, S6H). This suggests that Ub reactivity towards NAD+ as a consequence of processing by E1/E2. ADP-ribosylation by Dtx3L/Parp9 is selective for Ub since the Ub-related protein NEDD8 underwent conjugation but not ADP-ribosylation (Figure S6G). Our data shows that ADPr transferred to the Ub C-terminus by Dtx3L/Parp9 action inhibits Ub conjugation, and that the reaction is responsive to NAD+ concentration. By extension, the concentration of free NAD+ is predicted to influence Ub conjugation to substrates of Dtx3L/Parp9.
Figure 6. NAD+ inhibits Ub conjugation mediated by the E3 Dtx3L/Parp9 heterodimer.
(A) Dtx3L/Parp9 ubiquitylation reactions performed with T7 epitope-tagged Ub (WT and H68G), the protein combinations indicated, and unlabeled NAD+. The reaction products were analyzed by anti-T7 immunoblotting.
(B) Reactions (unlabeled NAD+) with Histone H4 analyzed by immunoblotting with an H4-specific antibody.
(C) Reactions (unlabeled NAD+) with Histone H2A analyzed by immunoblotting (Pan-Ub, Ub-H2A, H2A).
(D) E3 MDM2-dependent p53 ubiquitylation is insensitive to NAD+. Reactions were performed with the proteins indicated (all recombinant) in the absence and presence of 1 mM NAD+, and the products analyzed by immunoblotting (T7-epitope, p53).
(E) NAD+ effect on ubiquitylation of the substrate RPL12. Ubiquitylation assays supplemented with epitope-tagged ribosomal protein L12 (RPL12) were analyzed by SDS-PAGE and immunoblotting.
(F) Ub does not undergo ADP-ribosylation in a ubiquitylation reaction containing the E3 MDM2. Reactions containing Ub, E1, E2, and E3 (Dtx3L/Parp9 or MDM2) were performed in the presence biotin-NAD+. Ub products were analyzed by probing with Pan-Ub Ab and FL-Neutra.
(G) Ub is not ADP-ribosylated by the E3 RNF146. ADP-ribosylation assays were performed using biotin-NAD+ and ADP-ribosylation of ubiquitin was detected with FL-Neutra. Ubiquitylation was examined using the pan-Ub Ab.
DNA Repair Function of Dtx3L/Parp9
Dtx3L/Parp9 has been shown to be dependent on PAR formation by Parp1 for its recruitment to DNA damage sites, with depletion of these components reducing an early phase of 53BP1 recruitment (Yan et al., 2013). We tested whether endogenous Dtx3L/Parp9 is recruited to laser-induced damage sites in a panel of prostate cancer cell lines by co-staining for Parp9 and γH2AX. We found that Parp9 was recruited to DNA damage sites in prostate cancer cells that have a range of tumorgenic properties including metastatic capability and androgen receptor status (Figure 7A). Recruitment was also detected in a normal prostate cell line RWPE-1 (HPV immortalized). We tested whether Dtx3L/Parp9 is important for DNA repair via NHEJ using an integrated reporter that undergoes Cas9-dependent cleavage, and upon repair, loss of mCherry+ cells. Depletion of Dtx3L and Parp9 reduced NHEJ-mediated DNA repair to the same extent as core components including DNA PK (Figure 7B). We used a plasmid repair assay to address the role of the Parp9 catalytic domain in DNA repair. Because Parp9 restrains the E3 output of the heterodimer by catalyzing ADP-ribosylation of Ub, we predicted that mutation of Parp9 might enhance the DNA repair function of the complex. We tested this by comparing the level of repair obtained with WT Parp9 and Parp9 F703K, each co-transfected with WT Dtx3L since heterodimerization is critical for functionality. The data from three experiments shows that WT Dtx3L/Parp9 promotes DNA repair via NHEJ, and that the activity is further increased using Parp9 that has reduced ADP-ribosylation activity (Figure 7C, D). These data show that Dtx3L/Parp9 participates in NHEJ and that its activity is at least partially restrained by Parp9 catalytic activity. PAR has been shown to stimulate Ub conjugation by the E3 ligase RNF146 (DaRosa et al., 2015; Wang et al., 2012). This involves PAR contact with the Trp-Trp-Glu (WWE) domain in RNF146, which appears to trigger an allosteric activation mechanism. The WWE domain is also found in the mARTs Parp11 and Parp14(He et al., 2012). Though Parp9 does not contain a WWE domain in RNF146, it does contain tandem macrodomains (Yan et al., 2013), each of which can bind PAR (Figures 7F, S7A–D). We tested whether addition of purified PAR affects the E3 activity of the Dtx3L/Parp9 heterodimer. PAR addition enhanced the formation of poly-Ub forms of Parp9 and other components of the reaction (Figure 7E, lanes 1, 2). PAR modestly enhanced mono Ub of Histone H2A at Lys120 (Figure 7E, lane 8), an effect that was more evident during a time course of Histone H2A ubiquitylation (Figure 7G; Figure S7E). In these reactions, PAR did not affect ADPr addition to Ub (Figure 7H; Figure S7E). The PAR enhancement was lost upon mutation of the Parp9 macrodomains (Figure 7I; Figure S7F). These data suggest that both free NAD+ and its product PAR contribute inputs that influence the E3 activity of Dtx3L/Parp9, and that interactions within the heterodimer are critical for activity of the complex both in vitro and for DNA repair in cells.
Figure 7. DNA repair and PAR binding activities of Dtx3L/Parp9.
(A) Immunofluorescent localization of endogenous Dtx3L/Parp9 heterodimer to microirradiated DNA stripes.
(B) Depletion of Dtx3L/Parp9 by siRNA reduces DNA repair by NHEJ. Depletion of DNA PK, ATM, and SET8 were performed in parallel as positive controls. DNA repair was measured in 293T cells containing an integrated reporter that undergoes Cas9/sgRNA mediated cleavage and subsequent repair by NHEJ. Bar graphs are represented as the mean and SD. **P≤0.01, ***P≤0.001, P≤0.0001.
(C) The DNA repair function of Dtx3L/Parp9 is increased in a loss-of-function Parp9 catalytic domain mutant (F703K). DNA repair by NHEJ was assayed using a plasmid-based reporter. Data points are pooled from three experiments, and the mean and SD for each condition are indicated.
(D) Expression levels of Dtx3L/Parp9 proteins in the plasmid-based reporter assay (in Figure 7C).
(E) PAR stimulates the E3 activity of the Dtx3L/Parp9 heterodimer.
(F) PAR binding to Dtx3L/Parp9 heterodimer is reduced by single point mutations in each of the macrodomains. Proteins were spotted on nitrocellulose, incubated with biotin-tagged PAR, and binding detected with FL-Neutra (Figure S7).
(G, H) PAR stimulates ubiquitylation of Histone H2A without ADP-ribosylation of Ub (blots in Figure S7E).
(I) PAR enhancement of histone ubiquitylation is lost upon mutation of the Parp9 macrodomains (blots in Figure S7F).
DISCUSSION
ADP-ribosylation by the ART family contributes to the regulation of a diverse set of biological pathways (Hottiger, 2015a). Parp1 regulates nuclear activities including the DNA damage response and transcription by generating polymers of ADPr that affect chromatin structure and co-factor recruitment (Ryu et al., 2015; Tallis et al., 2014). Tankyrase promotes telomere elongation through a pathway involving ADP-ribosylation of TRF1 (Smith and de Lange, 2000). With the notable exceptions of studies done on bacterial toxins, however, the pathways and mechanisms utilized by ART enzymes that mediate mono-ADPr attachment have been mostly elusive. Of the 17 human enzymes that contain a Parp catalytic domain, 11 have mono-ADP ribosyltransferase activity based on the criteria of automodification (Hottiger, 2015b). For these 11 enzymes, only a few substrates have been identified (Barkauskaite et al., 2015).
While Parp9 is a member of the ART family by virtue of a catalytic domain that has significant protein sequence identity, its lack of auto-modification suggested it lacks catalytic activity (Aguiar et al., 2005). We reasoned that Parp9 activity might depend on Dtx3L since the proteins are coordinately expressed and undergo hetero-dimerization. Moreover, because Dtx3L is an E3 Ub ligase (Aguiar et al., 2005), we hypothesized that Parp9 function as a potential ART might occur in the context of the Ub pathway. Using purified Dtx3L/Parp9 heterodimer, we made six observations that helped establish that Parp9 has catalytic function and acts as a mono-ART towards a defined substrate, Ub, and that Parp9 modifies Ub in the context of its processing pathway. (i) The Dtx3L/Parp9 heterodimer cleaves NAD+ and generates nicotinamide and ADPr; (ii) In a ubiquitylation reaction, Dtx3L/Parp9 transfers ADPr to Ub; (iii) Parp9 mutation and non-selective Parp inhibitors prevent Ub ADP-ribosylation; (iv) Ub-ADPr production requires E1 and E2 enzymes; (v) ADPr transfer to Ub requires ambient temperature, ATP, and a C-terminal Gly in Ub; (vi) The mass addition measured by MALDI-TOF reflects addition of a single ADPr to Ub, consistent with mono-ADP-ribosylation by Parp9.
In ubiquitylation assays using histones as substrates and radiolabeled NAD+ to monitor ADP-ribosylation, the only protein labeled by 32P-ADPr was Ub. This key result suggests that under these assay conditions, the Ub-ADPr produced by E1/E2/E3 action was not added to histones. Moreover, our data is not readily explained by a model whereby Dtx3L/Parp acts as an NADase that generates a chemically-reactive ADPr that labels proteins indiscriminately. In these same reactions containing E1/E2/E3 enzymes, the histones underwent Ub conjugation. This result implied that ADPr modification of Ub, and Ub conjugation to protein, are probably mutually exclusive outcomes of the pathway. This led us to examine whether NAD+ concentration affects histone ubiquitylation in this assay. Addition of 0.2–0.8 mM NAD+ strongly reduced the poly-Ub and di-Ub of reaction components, and to some extent, mono-Ub of Histones H3 and H2A. Thus, in a purified system, the NAD+ concentration significantly influences the efficiency of Ub conjugation. In reactions using the same E1 and E2, NAD+ addition (1 mM) did not affect the E3 activity of MDM2, nor was ADPr conjugated to Ub in the presence of MDM2 or another E3 RNF146. Thus, the NAD+ effects on Ub reported here are mediated through Dtx3L/Parp9 and are not general effects on Ub activation and processing by E1/E2/E3. This pathway appears to be selective for Ub since the Ub-related protein Nedd8 does not undergo ADP-ribosylation under conditions where conjugation is stimulated by the presence of Dtx3L/Parp9. Using a battery of approaches including an antibody specific for Ub that has a free C-terminus, we showed that a single ADPr is added to the carboxyl group of Gly76, which renders Ub inactive for conjugation through this site.
With regard to Parp9-mediated ADP-ribosylation, defining how the E2-Ub thioester is transferred to ADP-ribose generated from NAD+ will be critical for understanding the mechanism. Presumably this reflects the spatial coordination between the C-terminal catalytic domain of Parp9 and the active site of the E2. On this note, core histones undergo E1/E2/Dtx3L/Parp9-dependent ubiquitylation, but there is no indication that histones, or proteins other than Ub, undergo ADP-ribosylation in our assays. Again, this emphasizes the selectivity of ADP-ribosylation, and the fact that the complex decides between Ub transfer to a substrate and Ub transfer to ADP-ribose based on NAD+ concentration.
One striking feature of ARTs is the variable domain architecture outside of the Parp homology domain that contributes to the structural and functional diversity of family members (Barkauskaite et al., 2015). Three members of the ART family including Parp9 contain macrodomains, modules that can bind both free and polymerized ADPr. We found that PAR addition to Ub assays stimulated the E3 activity of Dtx3L/Parp9 towards Histone H2A, and that the individual macrodomains of Parp9 can bind to PAR. Our finding is reminiscent of recent work on RNF146, an E3 that undergoes allosteric activation by PAR. In this case, PAR binding occurs through a WWE domain in RNF146, and E3 activation promotes mono-Ub conjugation to axin1 (DaRosa et al., 2015; Wang et al., 2012). The WWE domain is also found in five ARTs (Barkauskaite et al., 2015). Thus, multiple ART family members are endowed with readers of ADPr and PAR that could modulate targeting, catalytic activity, or both.
A conceptual model suggested by our data is that the E3 activity of Dtx3L/Parp9 is controlled by the concentration of NAD+, which at high levels would promote Ub modification by ADPr and reduce Dtx3L/Parp9-dependent Ub conjugation to substrates. Our data using Histone H3 as a substrate raises the additional possibility that under normal conditions, the effect of NAD+ is not necessarily to reduce mono- Ub conjugation, but rather to suppress the formation of poly-Ub by restricting the reaction to a single round of Ub transfer. Given the chemical lability of the ADPr-protein bond and the presence of an activity that can remove ADP-ribose from Ub, it is likely that Ub-ADPr is recycled to generate unmodified Ub, even in the face of a high concentration of NAD+. Finally, NAD+ levels are reduced during aging (Imai and Guarente, 2014), and NAD+ is depleted during the DNA damage response because Parp1 generates large amounts of PAR (Schreiber et al., 2006). In settings where NAD+ levels are reduced, Dtx3L/Parp9 could mediate Ub conjugation to target proteins such as histones. The combination of reduced levels of free NAD+ and Dtx3L/Parp9 binding to PAR, which also enhances E3 activity, might all be important for directing its E3 activity to Histone H4 or other substrates at break sites during the DNA damage response (Yan et al., 2009). Our data indicates that the DNA repair function of Dtx3L/Parp9 is important for efficient NHEJ, and that the contribution is restrained by Parp9 catalytic activity.
It is now apparent that certain E3’s can cooperate with Ub through mechanisms that are more complex than simply acting as scaffolds that facilitate interactions between substrates and Ub-E2 complexes (Berndsen and Wolberger, 2014; Deshaies and Joazeiro, 2009). Moreover, these emerging forms of E3-Ub crosstalk make contributions in diverse biological contexts via mechanisms that were not anticipated (Herhaus and Dikic, 2015). Ub phosphorylation on Ser65 promotes allosteric activation of Parkin (Wauer et al., 2015), an E3 that helps control in mitophagy in response to mitochondrial damage. Members of the SidE effector family in L. pneumophila mediate ADP-ribosylation of host cell Ub on Arg42 as part of a unique activation mechanism en route to E1, E2, and ATP-independent Ub conjugation to Rab GTPases (Qiu et al., 2016). These studies together with our finding that Ub can be ADP-ribosylated on Gly76 by Dtx3L/Parp9 illustrates how post-translational modification of Ub itself can contribute to protein regulation (Herhaus and Dikic, 2015).
STAR Method
CONTACT FOR REAGENT AND RESOURCE SHARING
Information and requests should be made to the corresponding author Bryce M. Paschal (paschal@virginia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Human 293T cells were cultured in DMEM supplemented with 5% FBS, 1% non-essential amino acids, and 1% sodium pyruvate. RWPE-1 prostate cells were cultured in Keratinocyte SFM and supplemented with the provided EGF and BPE factors. VCaP prostate cells were maintained in DMEM and 10% FBS, and all other cells were grown in RPMI supplemented with 5% FBS. Cells were incubated at 5% CO2 and 37°C.
METHOD DETAILS
Recombinant proteins
Standard methods were used for protein expression in E. coli and purification on TALON (Clontech) and Glutathione (Sigma) beads. The pET-DUET1 plasmid (Novagen) was used for co-expression of human His6-Parp9 and untagged human Dtx3L in the strain BL21(DE3)pLysS at 18°C overnight. Expression of His-Parp9 and Dtx3L in Sf9 insect cells was performed using the pBacPAK8 plasmid (Clontech). Plasmids were co-transfected with linearized BacPAK6 DNA to prepare the initial baculovirus, which was amplified using standard protocols. The pET28a plasmid was used to make His-T7-Ub and its mutant derivatives (H68G, R72G, R74G). PCR mutagenesis was performed using Pfu Ultra II (Agilent). The pGEX plasmid 4T-2 was used to express AF1521 protein. MDM2, p53, RNF146, ISG15, and Uba52 were expressed and purified by standard methods. Recombinant histones were purified by column chromatography following published procedures (Luger et al., 1999). Proteins purchased from commercial sources included WT-Ub (Sigma U6253 and U5507, AbD serotec 94001502, or R & D system U-530), K29,48,63R-Ub (Boston Biochem UM-3KTR), G75,76A-Ub (Boston Biochem UM-HAA), E1 Ube1 (R&D System E304), E2 UbcH8 (R&D System E2-616), and biotin-tagged WT-Ub (R&D U-570–100).
NAD+ cleavage assays
NAD+ cleavage was assessed by thin-layer chromatography of reactions that contained 32P-NAD+ (0.1 μCi/assay; 100 μM unlabeled NAD+) using cellulose PEI plates in LiCl2 and formic acid (0.15 M each). Nicotinamide generated by Parp9 cleavage of NAD+ was measured in 96-well assays. Reactions contained 100 μl potassium phosphate (pH 7.5), 0.2 mM NAD(P)H, 1 mM DTT, 3.3 mM α-ketoglutarate, 2 mM PncA (nicotinamidase), 0.1 unit/ml of glutamate dehydrogenase from bone liver (Sigma), 20–500 μM NAD+, and 10 μg/ml of recombinant Dtx3L/Parp9. Assay components except NAD+ were pre-incubated at 30°C for 5 min or until absorbance at 340 nM stabilized, and the reaction was initiated by addition of NAD+. The absorbance at 340 nM was measured in 15 sec intervals. The background rates of reactions lacking Dtx3L/Parp9 resulting from the spontaneous formation of nicotinamide or ammonia were subtracted from the initial velocities of the reactions.
ADP-ribosylation and ubiquitylation assays
Non-radioactive ADP-ribosylation assays were typically performed in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, containing 1 mM ATP, 1 mM DTT, 0.1 mg/ml ubiquitin, 0.01 mg/ml each of E1 and E2, 0.004–0.025 mg/ml Dtx3L/His6-Parp9, 0.1 mM unlabeled NAD+, and 0.0125 mM biotin-NAD+. To test for Ub ADP-ribosylation by Dtx3L/Parp9 complexes from mammalian cells, untagged Dtx3L and HA-tagged Parp9 were co-transfected and isolated by immunoprecipiation. Assays were assembled on ice, incubated at 30°C for 30 min, and subjected to SDS-PAGE. After transfer to nitrocellulose, blots were probed with primary and fluorescently-labeled secondary antibodies, fluorescently-labeled Neutravidin, and scanned and quantified on the Odyssey Infrared Imager (LICOR). Radioactive ADP-ribosylation assays were similar except 32P-NAD+ was added (0.1 μCi/assay; 25 μM unlabeled NAD+), and autoradiography was used to detect ADP-ribosylation. Ubiquitylation assays were essentially the same as ADP-ribosylation assays except E3 substrates (recombinant histones) were added to a final concentration of 0.1–0.25 mg/ml. Unless stated otherwise, ubiquitylation and ADP-ribosylation assays contained E1, E2, E3, Ub, substrate, ATP, and NAD+. Chemical release experiments of ADPr from Ub were performed with NH2OH (0.4 M, pH 7.5) and CHES (0.1 M, pH 9.0).
Ub modification with N-(Aminooxyacetyl)-N'-biotinylhydrazine (ARP)
ARP dissolved in DMSO was used at a final concentration of 1 mM. In brief, ADP-ribosylated Ub was incubated ARP in the presence of 0.5 M acetic acid for the time points indicated. The sample was then neutralized with NaOH, incubated with cobalt beads, and eluted with imidazole. The T7-Ub-ADPr-ARP was used as a substrate for cleavage by cellular factors.
Mass Spectrometry
ADP-ribosylation reactions were carried out in the presence of biotin-NAD+, as a complete reaction and with dropout of Dtx3L/Parp9 or Ub. The reaction products were dialyzed (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA and 5 mM β-mercaptoethanol), and bound to Streptavidin-Sepharose (GE Healthcare). After extensive washing, the beads were resuspended in 10 μL 0.1% TFA for a protein concentration of ~1 pmol/μL. Each sample was desalted using C4 ZipTips (EMD Millipore) and eluted with sinapinic acid (Sigma-Aldrich) as a sample matrix. Samples were spotted onto a stainless steel target plate and air-dried at room temperature. Mass spectrometric analysis was performed using a MALDI-TOF MicroFlex (Bruker Daltonics). Positive ions were generated by laser desorption at 337nm (N2 laser) using linear mode with an acceleration voltage of 20kV. Calibration of the instrument was performed using external protein calibration standard I, mass range 5,700–16,900 Da (Bruker Daltonics, part no. 206355). Approximately 200–300 laser shots were accumulated per sample and spectra were processed using FlexAnalysis version 3.4 (Bruker Daltonics).
Macrodomain binding assays
ADP-ribosylated Ub was dialyzed into binding buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT) prior to incubation with recombinant Af1521 (10 μg GST-Af1521/10μl Glutathione-agarose beads). Binding reactions were supplemented with 0.5% Triton X-100, and mixed end-over-end for 4 hrs at 4°C. The beads were washed 5 times with binding buffer, and fractions examined by immunoblotting with Pan-Ub antibody. PAR binding assays were performed by spotting recombinant proteins (0.1 to 2.0 μg) on nitrocellulose, blocking in 5% BSA in PBS overnight, and incubation with biotin-tagged PAR (20 nM) for 4 hrs at room temperature. The nitrocellulose membrane was washed, and biotin-PAR detected with FL-Neutra.
Protein Analysis and Mutagenesis
Coding sequences of the catalytic domains from select Parp family members were aligned using Clustal-Omega. In silico protein modeling was performed by threading sequence from the Parp9 catalytic domain onto the deposited Parp14 protein structure (PDB: 3SE2). Threading and further refinement was achieved using Swiss-PDBViewer, SwissModel, and PyMol. Site-directed mutagenesis of Dtx3L and Parp9 was performed by QuikChange.
DNA damage and Repair Assays
Microirradiation was performed on cells plated onto Nunc glass-bottom dishes and pre-sensitized by overnight treatment with 10 μM BrdU. DNA stripes (~0.25 μm wide) were generated in each field of cells with a 405nm laser at 90% power for 10–15 iterations on a Zeiss LSM 880 confocal microscope. Cells were allowed to recover for 15 minutes before fixation. Cells were stained with antibodies to γH2AX and Parp9. Integrated reporter assays were performed in 293T cells engineered to constitutively express mCherry. Upon transfection of Cas9 and sgRNA targeting the first codon of mCherry, repair via error-prone NHEJ results in loss of expression, which is quantified by flow cytometry. Plasmid-based NHEJ DNA repair was measured by transfection of I-SceI-linearized reporter and DsRed plasmid as transfection control and quantification of GFP+ cells by flow cytometry. The ratio of GFP+/DsRed+ cells was used as a measure of NHEJ efficiency. Flow cytometry results were analyzed Tukey’s multiple comparisons test.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details for the significance tests can be found within the text and was performed using GraphPad Prism6.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pan-Ubiquitin (Pan-Ub) | Cell Signaling Technology | 3933S |
| Ub C-term | Epitomics abcam | 1646-1 Ab33893 |
| T7 epitope tag | Novagen | 69522-3 |
| M2 (flag epitope tag) | Sigma-Aldrich | A2220 |
| Parp9 | This paper ProteinTech |
N/A 17535-1-AP |
| Dtx3L | This paper GeneTex |
N/A GTX116369 |
| γH2AX | EMD Millipore | 05-636 |
| Tubulin | Sigma-Aldrich | T 9028 |
| p53 | Sigma-Aldrich | P6749 |
| Histone H3 | Cell Signaling Technology | 9715 |
| Histone H4 | Millipore | 04-858 |
| Ub-H2A | Cell Signaling Technology | 8240 |
| Ub-H2B | EMD Millipore | 17-650 |
| ISG15 | pbl assay science - Dr. Ernest Borden | N/A |
| Nedd8 | Cell Signaling Technology | 2745S |
| MBP | New England BioLabs | E8032S |
| Neutravidin-DyLight-800 | Pierce | 22853 |
| Glutathione-agarose beads | Sigma-Aldrich | G4510 |
| Streptavidin-Sepharose beads | GE Healthcare | 17-5113-01 |
| Amylose resin | New England BioLabs | E8021S |
| Talon metal affinity resin | Clontech | 635502 |
| Bacterial and Virus Strains | ||
| DH5α (general plasmid preparation) | New England BioLabs | C2987P |
| BL21(DE3)pLysS | EMD Millipore (Novagen) | 69451 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Parp9-Dtx3L | This paper | N/A |
| His-T7-Ub and mutant peptides | This paper | N/A |
| AF1521 | This paper | N/A |
| MDM2 | This paper | N/A |
| p53 | This paper | N/A |
| WT-Ub | Sigma-Aldrich AbD Serotec (BioRad) R&D Systems |
U6253 9400-1502 U-530 |
| Ub mutant (G75,76A) | Boston Biochem | UM-HAA |
| E1 Ube1 | R&D Systems | E304 |
| E2 UbcH8 | R&D Systems | E2-616 |
| WT-Ub (biotin tagged) | R&D Systems | U-570-100 |
| T7-RPL12 | This paper | N/A |
| ISG15 | This paper | N/A |
| E1 UBA3 | Boston Biochem | E-313 |
| E2 UBE2M | Boston Biochem | E2-656 |
| Nedd8 | Boston Biochem | UL-812 |
| Core Histones | This paper | N/A |
| Histone H3 | New England BioLabs | M2503S |
| Histone H4 | New England BioLabs | M2504S |
| RNF146 | This paper | N/A |
| IFNγ | PromoKine | c-60724 |
| IFNα | PromoKine | c-60581 |
| Menadione | Selleckchem | S1949 |
| Novobiocin | EMD Millipore Calbiochem | 49-120-71GM |
| Phylloquinone | MP Biomedicals | 0210328301 |
| PJ34 | Selleckchem | S7300 |
| BMN-673 | MedChem Express | HY-16106 |
| Rucaparib | Selleckchem | S1098 |
| NAD+ | Sigma-Aldrich | N1511 |
| Biotin-NAD+ | R&D Systems | 4670-500-01 |
| 32P-NAD+ | Perkin Elmer | BLU023X250UC |
| NAD(P)H | Sigma-Aldrich | N6505 |
| α-ketoglutarate | Sigma-Aldrich | K1128 |
| PncA | Dr. Jeff Smith (UVa) | N/A |
| Glutamate dehydrogenase | Sigma-Aldrich | G2626 |
| Sinapinic acid | Sigma-Aldrich | 49508 |
| PAR | Trevigen | 4336-100-01 |
| PAR (biotin-tagged) | Trevigen | 4336-100-02 |
| ARP | Cayman Chemical | 10009350 |
| I-SceI | New England BioLabs | R0694S |
| Critical Commercial Assays | ||
| PCR mutagenesis – Pfu Ultra II | Agilent | 600670 |
| QuikChange | Agilent | 200515 |
| Deposited Data | ||
| The Cancer Genome | Atlas http://cancergenome.nih.gov/ | |
| Protein Data Bank | www.rcsb.org | 3SE2 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | CRL-3216 |
| PC3 prostate cells | ATCC | CRL-1435 |
| VCaP prostate cells | ATCC | CRL-2876 |
| LNCaP prostate cells | ATCC | CRL-1740 |
| C4-2b prostate cells | Dr. Michael Weber (UVa) | N/A |
| DU145 prostate cells | ATCC | HTB-81 |
| PC3-AR prostate cells | This paper | N/A |
| RWPE-1 prostate cells | Dr. Dan Gioeli (UVa) | N/A |
| Sf9 insect cells | ATCC | CRL-1711 |
| Oligonucleotides | ||
| siRNA targeting sequence: Parp9:CCUUACCUUGGGUGAACUAAC | This paper | N/A |
| siRNA targeting sequence: Dtx3L: CGCGUAUUAGGAGUCUCAGAU | This paper | N/A |
| siRNA control: Gl2: AACGUACGCGGAAUACUUCGA | This paper | N/A |
| siRNA targeting sequence: DNA-PK: GAUCGCACCUUACUCUGUU | This paper | N/A |
| siRNA targeting sequence: ATM: GCGCCUGAUUCGAGAUCCU | Lin and Dutta, 2007 | N/A |
| siRNA targeting sequence: SET8: GAUUGAAAGUGGGAAGGAA | Benamar et al., 2016 | N/A |
| Recombinant DNA | ||
| pET-DUET1 plasmid: Dtx3L and His6-Parp9 (or Parp9 mutants) | This paper | N/A |
| BacPAK8 plasmid: His6-Parp9 | This paper | N/A |
| BacPAK8 plasmid: Dtx3L and mutant | This paper | N/A |
| pGEX 4T-2 plasmid: GST-AF1521 | This paper | N/A |
| pET30b H2A | Dr. Ben E. Black (U. Penn) | N/A |
| pET12 H2B | Dr. Ben E. Black (U. Penn) | N/A |
| pET12 H3.1 | Dr. Ben E. Black (U. Penn) | N/A |
| pET28a: His-T7-Ub and mutants | This paper | N/A |
| pGEX-4T1: MDM2 | This paper | N/A |
| pET15b: p53 | This paper | N/A |
| pET28a: RNF146 | This paper | N/A |
| pKH3 plasmid (Empty) | Dr. Ian Macara | (now addgene 12555) |
| pKH3 plasmid: Parp9, epitope tagged, and mutants | This paper | N/A |
| pKH3 plasmid: Dtx3L | This paper | N/A |
| pKH3 plasmid: co-expression of Dtx3L and Parp9 OR Dtx3L and Parp9 (F703K) | This paper | N/A |
| pET28a: ISG15 | This paper | N/A |
| pET28a: UBA52 | This paper | N/A |
| Parallel 1 MBP-His plasmid: Parp9 Macrodomains (MBP-MD1-MD2) | This paper | N/A |
| NHEJ reporter plasmid | Mao et al., 2008 | N/A |
| pDSRed2-N1 | Mao et al., 2008 | N/A |
| pET28a: T7-RPL12 | This lab | N/A |
| Software and Algorithms | ||
| FlexAnalysis version 3.4 | Bruker Daltonics | N/A |
| Odyssey Infrared Imager | LICOR | N/A |
| Clustal-Omega | Sievers et al., 2011 | N/A |
| Swiss-PDB Viewer | The SIB Swiss Institute of Bioinformatics | N/A |
| SwissModel | Guex and Peitsch, 1997 | N/A |
| PyMol version 1.8 | The PyMOL Molecular Graphics System, Schrodinger, LLC | N/A |
| Prism6 | Graphpad Software, San Diego, CA | N/A |
| Zen systems software (confocal acquisition) | Carl Zeiss Inc. | N/A |
| FlowJo Collector’s Edition Acquisition Software (flow cytometry acquisition) | Cytek Development | N/A |
| ModFit LT Software (flow cytometry analysis) | Verity Software House, Topsham, ME | N/A |
| Other | ||
| Cellulose PEI plates | Select Scientific | 10078 |
| C4 ZipTips | EMD Millipore | ZTC04S008 |
Supplementary Material
Acknowledgments
We thank Dr. Jeff Smith (Virginia) for help with the NADase assays and Dr. Ernest Borden (Cleveland Clinic) for ISG15 antibody. Our studies were made possible by financial support from the NCI #CA214872 (BMP), NIA #AG047200 (VG), and the Paul Mellon Urologic Cancer Institute at the University of Virginia (BMP).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, BMP, CSY, KJ; Investigation, CSY, KJ, AS, ND, LN, LTO, MC, BK, MP, EJ, NS; Formal analysis, CSY, KJ, BR; Resources, Writing – Original draft, BMP; Writing – Review and Editing, CSY, KJ, VG, TA, BMP; Visualization, KJ, CSY, EJ, LTO; Supervision, VG, TA, BMP; Funding acquisition, VG, TA, BMP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- Bachmann SB, Frommel SC, Camicia R, Winkler HC, Santoro R, Hassa PO. DTX3L and ARTD9 inhibit IRF1 expression and mediate in cooperation with ARTD8 survival and proliferation of metastatic prostate cancer cells. Mol Cancer. 2014;13:125. doi: 10.1186/1476-4598-13-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- Barkauskaite E, Jankevicius G, Ahel I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Benamar M, Guessous F, Du K, Corbett P, Obeid J, Gioeli D, Slingluff CL, Abbas T. Inactivation of the CRL4-CDT2-SET8/p21 ubiquitylation and degradation axis underlies the therapeutic efficacy of pevonedistat in melanoma. EBioMedicine. 2016;10:85–100. doi: 10.1016/j.ebiom.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D, Jacobson EL, Jacobson MK. Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods Enzymol. 1997;280:275–287. doi: 10.1016/s0076-6879(97)80119-x. [DOI] [PubMed] [Google Scholar]
- Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci U S A. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. 2015;517:223–226. doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–288. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, Linzen U, Kremmer E, Luscher B. ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell Commun Signal. 2013;11:5. doi: 10.1186/1478-811X-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol Cell. 2015;58:925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- He F, Tsuda K, Takahashi M, Kuwasako K, Terada T, Shirouzu M, Watanabe S, Kigawa T, Kobayashi N, Guntert P, et al. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Hengel SM, Goodlett DR. A Review of Tandem Mass Spectrometry Characterization of Adenosine Diphosphate-Ribosylated Peptides. Int J Mass Spectrom. 2012;312:114–121. doi: 10.1016/j.ijms.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu Rev Biochem. 2015a;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- Hottiger MO. SnapShot: ADP-Ribosylation Signaling. Mol Cell. 2015b;58:1134– 1134. e1131. doi: 10.1016/j.molcel.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczynski P, Kutok JL, Li C, Mitra J, Aguiar RC, Shipp MA. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol Cell Biol. 2006;26:5348–5359. doi: 10.1128/MCB.02351-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203– 229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kraus WL. PARPs and ADP-Ribosylation: 50 Years ... and Counting. Mol Cell. 2015;58:902–910. doi: 10.1016/j.molcel.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Dutta A. ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J Biol Chem. 2007;282(42):30357–30362. doi: 10.1074/jbc.M705178200. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7(18):2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268:22575–22580. [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Morgan RK, Cohen MS. A Clickable Aminooxy Probe for Monitoring Cellular ADP-Ribosylation. ACS Chem Biol. 2015;10:1778–1784. doi: 10.1021/acschembio.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo ZQ. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli V, O'Farrell M, Menissier-de Murcia J, de Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- Ryu KW, Kim DS, Kraus WL. New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem Rev. 2015;115:2453–2481. doi: 10.1021/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa ES, Fabrizio G, Di Girolamo M. A role of intracellular mono- ADP-ribosylation in cancer biology. FEBS J. 2013;280:3551–3562. doi: 10.1111/febs.12290. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins D. Fast, scalable generation of high–quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal Biochem. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Takeyama K, Aguiar RC, Gu L, He C, Freeman GJ, Kutok JL, Aster JC, Shipp MA. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278:21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]
- Tallis M, Morra R, Barkauskaite E, Ahel I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma. 2014;123:79–90. doi: 10.1007/s00412-013-0442-9. [DOI] [PubMed] [Google Scholar]
- Verheugd P, Forst AH, Milke L, Herzog N, Feijs KL, Kremmer E, Kleine H, Luscher B. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat Commun. 2013;4:1683. doi: 10.1038/ncomms2672. [DOI] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Xu R, Zhu L, Cheng X, Wang Z, Manis J, Shipp MA. BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol Cell Biol. 2013;33:845–857. doi: 10.1128/MCB.00990-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci U S A. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.