Abstract
Hydrogen/deuterium exchange followed by trapping of the labeled species in the aprotic solvent DMSO has been used to elucidate structure in both the burst-phase molten globule-folding intermediate of apomyoglobin and in an equilibrium intermediate that models the kinetic intermediate. Precise estimates can be made of exchange times in an interrupted exchange-out experiment at pH 4 followed by analysis in DMSO solution, giving extensive sequence-specific information about the structure of the equilibrium intermediate. In addition, the use of DMSO as a solvent for NMR measurements after quench-flow pH-pulse labeling experiments gives a greatly increased data set for the elucidation of the kinetic folding pathway. Interestingly, differences are observed in some regions of apomyoglobin between the equilibrium and kinetic intermediates. These differences are quantitative rather than qualitative; that is, the overall patterns of labeling and secondary structure formation remain similar between the two species. However, local differences are observed, which probably reflect the difference in the solution conditions for the equilibrium experiment (pH 4) vs. the kinetic experiment (pH 6) and the change in the status of the stabilizing hydrogen bond between the side chains of His-24 and His-119.
Keywords: apomyoglobin, NMR, molten globule
Elucidation of the detailed mechanism of protein folding remains one of the major challenges in structural biology. Information on the kinetic folding process can be obtained by various biophysical techniques, but the only method that simultaneously provides information on folding at individual sites throughout the protein is the well known quench-flow hydrogen exchange technique coupled with NMR spectroscopy (1, 2). In this method, amides are labeled by hydrogen/deuterium (H/D) exchange at multiple time points during the folding process; exchange is then quenched, the refolding is allowed to go to completion, and the H/D population at individual amides is determined from NMR spectra of the folded protein. A major shortcoming is that kinetic information on the folding process is limited to those residues for which amide proton exchange rate in the native folded form of the protein is slow. For various reasons, including rapid exchange rates in folding intermediates, the number of useful amide proton “probes” for quench-flow experiments may be even further reduced. Information on the extent and rate of folding is therefore unavailable for a substantial portion of the polypeptide chain. In practical terms, the exchange rates of amide protons to be used as probes in the standard quench-flow experiment must be slower than ≈10-4 s-1 (3). A few additional amides can sometimes be observed by the careful use of lowered pH and temperature. However, for most proteins studied by the quench-flow method, only 30-50% of the backbone amides in the molecule can reliably be used as probes in standard quench-flow folding experiments.
Here, we describe a previously unreported method employing the aprotic organic solvent DMSO for NMR measurements on the partially exchanged species that define progress along the folding pathway. This method, which involves measurement of the NMR spectrum of the unfolded protein rather than the refolded, native protein, provides information on a much larger number of amide proton probes, because the detection and quantitation of the extent of H/D exchange of a given amide during the folding process does not depend on the presence of slow exchange in the native form of the protein. DMSO has been used extensively as a solvent for peptides and has advantages for NMR in that the signals of exchangeable protons, which are often attenuated or missing in spectra recorded in aqueous solvents, are readily observable in DMSO. Many proteins are also soluble in DMSO, but they are invariably unfolded. The favorable exchange properties of DMSO were recently used to map the core of β2-microglobulin amyloid fibrils (4, 5).
Apomyoglobin (apoMb) is a paradigm for studies of protein folding. It undergoes a folding transition from low to intermediate pH with formation of an equilibrium intermediate state that has been extensively characterized by CD, NMR, and fluorescence spectroscopy (6-8). Quench-flow amide proton H/D exchange pulse labeling (detected by both NMR and mass spectrometry) and stopped-flow spectroscopy have been used to characterize the kinetic folding of apoMb from states unfolded in urea, guanidine, and acid (9-13). The kinetic experiments also reveal the presence of an intermediate state, the so-called “burst phase intermediate,” which is formed within the dead time of the flow apparatus and that subsequently folds into the native state on a slower time scale. The existence of multiple folding intermediates has been observed under some conditions in equilibrium (14) and kinetic (15, 16) experiments. The kinetic and equilibrium intermediates appear to share a number of characteristics, and it has long been thought that the two states are very similar (9). We present in this work an examination of this hypothesis, by using the DMSO quench-flow technique to give kinetic information on a larger subset of the amino acid residues than was formerly possible.
Materials and Methods
Preparation of Proteins. Recombinant sperm whale apoMb was produced by overexpression in Escherichia coli BL21-DE3 cells. The 15N-labeled and 15N,13C-double-labeled protein were prepared by using M9 minimal medium containing 15N-ammonium sulfate and 13C-glucose and were purified by using methods described in ref. 17. The purified apoMb was dialyzed against H2O before the experiments. The protein concentration of apoMb was calculated by using the extinction coefficient of apoMb (15,900 M-1·cm-1 at 280 nm) at pH 6.1.
Interrupted H/D Exchange. The apoMb solution (≈1.5 mg/ml) in 10 mM sodium acetate buffer in H2O at pH 6 was manually diluted into a 10-fold vol of the same buffer in D2O at 4°C. After incubation for various times between 30 s and 24 h at 4°C, the H/D exchange was quenched by adding 0.1% (vol/vol) trifluoroacetic acid solution in D2O, to give a final pH* (measured pH value in a D2O soluton, uncorrected for the deuterium isotope effect) of 2.5 for the solution. This solution was quickly frozen in liquid N2 and lyophilized. The H/D exchange experiment at pH 4 was conducted in a similar manner, by using 2 mM sodium acetate buffer. Eighteen samples with different exchange times were independently prepared at pH values of 4 and 6, and were analyzed by NMR in DMSO.
pH-Pulse Labeling. The H/D-exchange pH-pulse labeling was conducted during the refolding reaction of apoMb induced by pH-jump at 8°C with a Biologic QFM5 Quench Flow instrument (Grenoble, France). The fully protonated 15N-labeled apoMb (≈2.0 mg/ml in H2O, unbuffered) was unfolded by addition of HCl to bring the pH to 2.2. The refolding reaction was initiated by dilution into 7.5-fold vol of 20 mM sodium acetate buffer in D2O, to a final pH* of 6.0. At each time point during the apoMb refolding to native state, the pH-pulse labeling at pH* 10.3 was used by the addition of 100 mM 3-[cyclohexylamino]-1-propanesulfonic acid buffer in D2O. After 35 ms at pH* 10.3, the pH was reduced to 6.0 with 300 mM 3-(N-morpholino)propanesulfonic acid buffer in D2O. In this work, because we focus on the calculation of the proton occupancy (A0) for the burst phase intermediate free from the effect of pH-pulse labeling, the experiment was conducted at the shortest refolding time (6.4 ms) with different time durations of pH-pulse labeling (12, 20, 35, 65, and 95 ms) (13). This pH-pulse-labeled solution was flushed from the quench-flow device into a trifluoroacetic acid solution (D2O) in a 50-ml tube (final pH* 2.5) and immediately frozen in liquid N2 followed by lyophilization. The lyophilized protein powder was tightly sealed in 50-ml tubes with parafilm and kept in the -80°C freezer until the NMR measurements were made.
NMR Measurements. The NMR spectra for the partly exchanged samples after pH-pulse labeling and H/D exchange were recorded at 30°C on a Bruker DRX600 spectrometer equipped with a cryoprobe. The lyophilized protein was dissolved to a concentration of ≈0.2 mM in 99.6% deuterated DMSO [a mixture of 99.6% (vol/vol) d6-DMSO (99.96% D; Cambridge Isotope Laboratories, Andover, MA) and 0.4% (vol/vol) D2O (99.96% D; Cambridge Isotope Laboratories)] before the NMR measurements. Acquisition of NMR data was started precisely 8 min after the addition of DMSO to the lyophilized protein. The 15N-1H heteronuclear single quantum correlation (HSQC) spectra of pulse labeled and H/D-exchanged protein samples were collected with spectral widths of 9,615 Hz and 2,048 points in the 1H dimension and 1,538 Hz and 256 points in the 15N dimension. For each sample, a series of NMR spectra was recorded every 20 min, giving a total of 15-20 spectra. The intensity of the peak at each position in the HSQC spectra was plotted as a function of time after addition of solvent to the lyophilized protein, and the plot was curve-fitted to an exponential decay. Extrapolation of this curve to zero time allowed calculation of the peak intensity without the small amount of H/D exchange that occurred as a result of the presence of 0.4% D2O in the DMSO. The intensity of the cross peaks in data sets with different refolding and exchange times or pulse durations was calibrated with the signal volumes of the methyl proton signals observed in 1D proton spectra. The proton occupancy for each residue was calculated in the pH-pulse-labeling experiments; the signal intensity at each time point was normalized with respect to the intensity of the same signal at the 8-s time point, when apoMb is completely folded. The decrease in peak intensity during the H/D exchange was curve-fitted, yielding the rate constant for the exchange; the protection factor was calculated by using intrinsic H/D exchange rate constants (18).
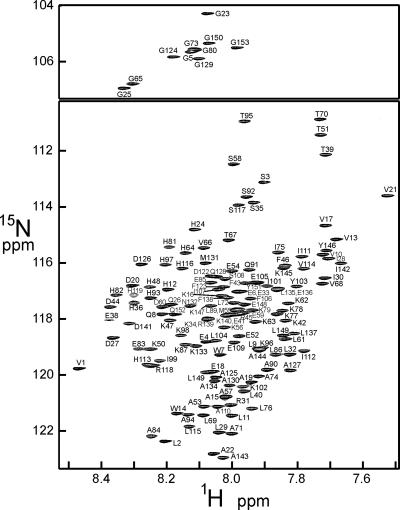
Backbone resonance assignments for unfolded apoMb in DMSO were made by using 3D HNCO (19), HNCA (19), HCA(CO)CANH (20), HNCACB (21), and CBCA(CO)NH (22) spectra collected at 30°C with a Bruker DRX800 spectrometer. A representative spectrum of the DMSO-unfolded protein is shown in Fig. 1. The HNCO and HCA(CO)CANH are particularly useful for the assignment of unfolded proteins (23). 13C,15N-double-labeled apoMb was dissolved in a similar amount of the buffer used in the pH-pulse-labeling experiment and lyophilized and redissolved to collect 3D NMR data under conditions as similar as possible to those of the pH-pulse-labeling and HD exchange experiments, because small variations in the concentration of salts and buffer components {3-(N-morpholino)propanesulfonic acid, 3-[cyclohexylamino]-1-propanesulfonic acid, etc.} were found to affect many of the chemical shifts under these conditions. In the 3D experiments, the 1H and 15N carrier was placed at 4.725 and 118 ppm, respectively. For HNCO and HCA(CO)CANH, the 13C carrier was placed at 175.7 ppm. Spectral widths of 12,802, 2,414.7, and 1,754.4 Hz were used for 1H, 13CCO, and 15N, respectively. For HNCACB and CBCA(CO)NH, the 13C carrier was placed at 45.691 ppm. Spectral widths of 12,802, 12,195.1, and 1,754.4 Hz were used for 1H, 13Cαβ, and 15N, respectively. For HNCA, the 13C carrier was placed at 57.69 ppm and a spectral width of 4,829.2 Hz was used for 13Cα. For all 3D experiments, 1,024, 64-75, and 43 points were acquired in the 1H, 13C, and 15N dimensions, respectively. The NMR data were collected in the indirect dimension with the States-time-proportional phase incrementation (TPPI) method (24) and were processed and analyzed by using nmrpipe (25) and nmrview (26), respectively.
Fig. 1.
1H,15N HSQC spectrum of apoMb in DMSO. The resonances were assigned on the basis of 3D experiments, and these assignments were used for the analysis of the H/D-exchange experiments for the pH 4 equilibrium intermediate and the kinetic experiments.
Results
The H/D exchange reaction is slowed in the presence of DMSO (27), which has no exchangeable protons. However, it is difficult to completely remove H2O from lyophilized protein samples, and thus additional artificial cross-exchange of amides can occur during the steps leading to the NMR measurements. Both rapid handling before data collection and maintaining the samples at low temperature and low pH (≈3) are important to prevent cross-exchange after pH-pulse labeling. Even with these precautions, amounts of H2O that were vanishingly small, but variable, remained in the lyophilized protein samples. We therefore used a solution composed of 99.6% d6-DMSO plus 0.4% D2O as a NMR solvent for the lyophilized protein. Under these conditions, most of the amide cross peaks showed a slow exponential decay of the peak intensity during data acquisition, due to exchange with the D2O added in the DMSO. For each sample, NMR data collection was started exactly 8 min after the addition of DMSO, and duplicate HSQC spectra were collected every 20 min. Extrapolation to zero time of the exponential curve fitted to the transition of signal intensity gives an estimate of the signal intensity in the absence of H/D exchange.
Interrupted H/D Exchange in Organic Solvent. H/D exchange experiments in organic solvents have been used to obtain information on the core structure of aggregated arrays such as amyloid fibrils (4) but are not generally used for such studies on stable globular proteins in solution: a simple procedure using a single NMR sample where the solvent has been exchanged from H2O to D2O can easily be used to follow the exchange in such proteins. For less stable proteins, such as the equilibrium intermediate of apoMb at pH 4, and even apoMb itself at pH 6, most amides in the molecule exchange too fast to be observed by conventional H/D-exchange methods. Information on such systems can be obtained by interrupted H/D exchange, where separate samples are prepared for different time points in the exchange reaction. Such a study has been described for the apoMb intermediate (7); the protein was stabilized at each time point in the H/D exchange series by converting it to fully folded myoglobin by increase of pH and the addition of heme. This method gave a significant amount of information on the stable secondary structure in the pH 4 state of apoMb, but many of the signals were lost through exchange in the fully folded state and during the manipulations. Here, we have applied a modified protocol employing organic solvents that can be used to obtain an enhanced picture of H/D exchange in unstable proteins.
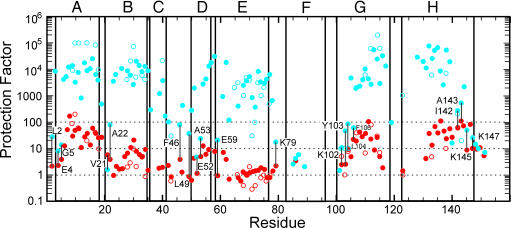
Samples were initially prepared by dilution of a stock solution in H2O into a D2O solution at a given pH* (4.0 or 6.0), thus initiating amide H/D exchange (7). After a certain time (the “exchange-out period”), the exchange reaction was quenched, by using a combination of low temperature, low pH, and lyophilization, and the sample was stored at -80°C before dissolving in DMSO for NMR analysis. The results of the interrupted H/D exchange in DMSO are shown with filled symbols in Fig. 2, and those from the experiment in D2O (7) are shown as open symbols. The results obtained in the two solutions are the same for both the intermediate state (red) and the native state (blue) H/D exchange, except for the greater number of data points in the DMSO experiments.
Fig. 2.
Plot of protection factor per residue obtained from the interrupted H/D exchange measurements made in DMSO solution (filled symbols) or in D2O solution [reproduced from the data of Hughson et al. (7)] (open symbols). Blue dots, pH 6.0; red dots, pH 4.0. Labels indicate residues for which the protection factor at the two pH values is comparable. The locations of helices in the folded structure of myoglobin are indicated with boxes. Dotted lines indicate protection factors of 1, 10, and 100. Letters A-H refer to helices.
Quench-Flow Experiments in Organic Solvent. The initial steps of the quench-flow experiment were conducted entirely in H2O-D2O solution, according to well established protocols. Unfolded protein in H2O was placed in one syringe of the quench-flow apparatus, and folding was initiated by mixing with folding buffer in D2O. Folding was allowed to proceed for a certain interval between 6 ms and 8 s, and the protein was pulse-labeled by mixing with a third solution at high pH in D2O. Amides that are not protected during the folding time were rapidly exchanged, leaving only the protected amides protonated and therefore visible in the NMR spectrum. In the original quench-flow experiments (1, 2) and subsequent refinements, the final step involves quenching of the exchange, followed by refolding of the protein and subsequent analysis of the refolded native protein by NMR or mass spectrometry (28). By contrast, in our method, the protein was not allowed to fold completely after the pulse-labeling step; instead the partly folded protein was rapidly frozen and lyophilized, then dissolved in DMSO for analysis of the NMR spectrum.
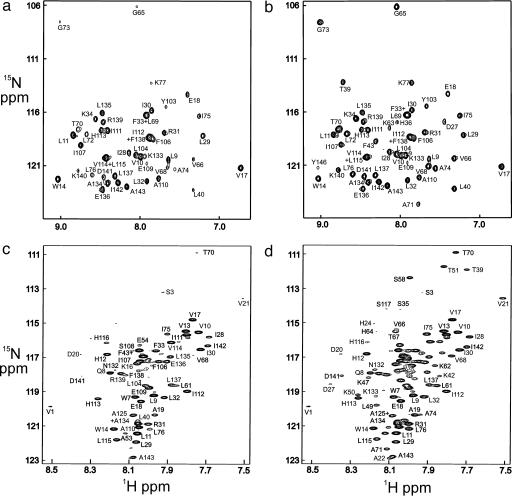
For apoMb, the quench-flow experiment in D2O is commonly performed with urea as denaturant (9-12, 29). However, the protein also may be unfolded at low pH, and the quench-flow results are very similar (12). Typical 1H,15N HSQC spectra obtained from a quench-flow experiment in D2O are shown in Fig. 3, for the shortest refolding time (6.4 ms) (Fig. 3a) and a long refolding time (Fig. 3b). The cross peaks form part of the HSQC spectrum of fully folded holomyoglobin under native conditions at pH* 5.6 and 35°C in D2O. Only a subset of the cross peaks is visible, representing amides that were both protected during the quench-flow experiment and persistent in the native form of the protein. A significant amount of information on the folding process was therefore lost because of exchange processes that occurred after folding was complete.
Fig. 3.
HSQC spectra of myoglobin refolded with various folding times in the quench-flow experiment. (a) Urea-jump experiment measured using the folded holoprotein in D2O solution with a folding time of 6.4 ms. (b) Urea-jump experiment using the folded holoprotein in D2O solution with a folding time of 3 s. (c) pH-jump experiment measured in DMSO solution using the unfolded apoprotein with a folding time of 6.4 ms. (d) pH-jump experiment measured in DMSO solution using the unfolded apoprotein with a folding time of 3 s. The upfield region of the spectrum, containing the glycine cross peaks, has been omitted from c and d, due to extremely low intensities.
Our method is, so far, able to be applied only for pH-jump experiments, because it is difficult to dissolve the lyophilized protein in DMSO in the presence of residual urea after urea-jump experiments. The results of a quench-flow experiment, identical in every way to those shown in Fig. 3 a and b but with the protein dissolved in DMSO without refolding, are shown in Fig. 3 c and d. These spectra correspond to the unfolded apoprotein rather than the folded holoprotein of Fig. 3 a and b. Qualitatively, the results are the same for the corresponding plots, Fig. 3 a and c and Fig. 3 b and d; that is, there are fewer amides protected at 6.4 ms than at 3 s. However, it is noticeable that there are significantly more cross peaks for the DMSO experiment in Fig. 3 c and d. Performing the quench-flow experiment in this way avoids the problems of amide proton exchange-out from the folded holoprotein that are encountered with the classical method.
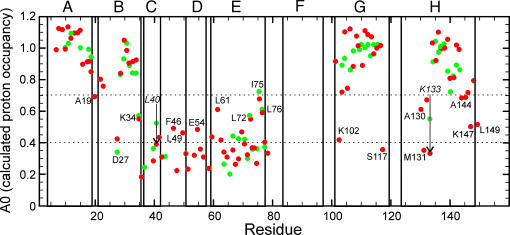
A direct comparison of the DMSO data with those obtained previously for apoMb in D2O-based quench-flow urea-jump experiments is shown in Fig. 4. In the previous experiments, data were obtained only for residues in the middle of the A, B, E, G, and H helices and for a few residues in the C helix and CD loop (9, 11-13). Additional data points are now available for the ends of all of the helices, as well as for the complete C and D helices and CD loop. Kinetic data for the F helix remain unreliable; the F helix is not folded even at the native state of apoMb (30), and, because the signal intensity at the longest refolding time is used as a standard to calculate the proton occupancy, we are not able to determine rates of folding for this helix. Over many repetitions of the quench-flow experiment (9, 11-13), the folding kinetics for wild-type apoMb remained consistent: the A, G, and H helices, together with a part of the B helix, folded rapidly, within the dead time of the quench-flow apparatus, whereas probes in the remainder of the molecule showed a slower, observable folding rate. The DMSO data (red dots in Fig. 4) closely resemble the previously determined data (green dots). Where information is available for a given residue from both experiments, the results are remarkably consistent. The information from the DMSO experiment shows that, as expected, neither the C and D helices nor the amides at the ends of helices are observed to be protected in the initial burst phase of folding, but instead fold on a slower time scale similar to that of the rest of the protein.
Fig. 4.
Plot of the calculated zero-time proton occupancy at a folding time of 6.4 ms for the burst phase intermediate in the pH-jump experiment in DMSO (red dots) and for the previously published urea-jump experiment in D2O (green dots) (13). Selected residues are labeled. The two italicized residues with arrows (L40 and K133) show significantly lower burst-phase proton occupancy in the DMSO experiment, changing from a classification of F+S to S. Dotted lines indicate the cut-off values for the classifications F, F+S, and S (see text). Letters A-H refer to helices.
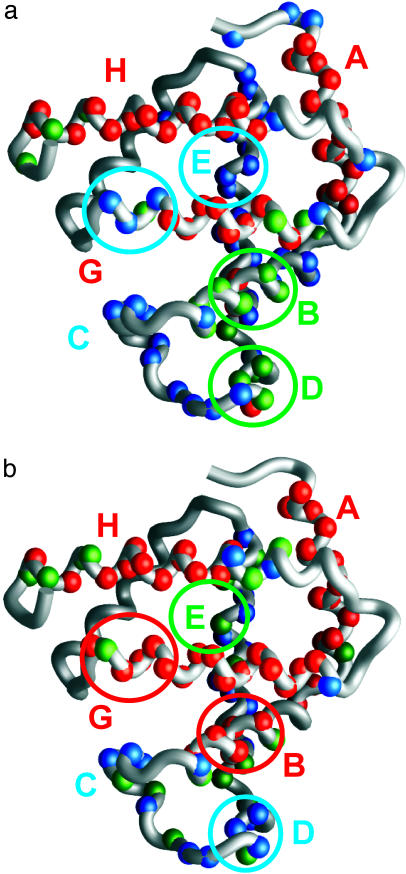
Comparison of the Equilibrium and Kinetic Intermediates. The interrupted H/D-exchange data shown in Fig. 2 for the pH 4 state of apoMb (red dots) provide a site-specific measurement of the amide proton protection factor in the equilibrium intermediate formed at pH 4 in low salt. These sites can be divided according to the protection factor, with the most protected amides having protection factors of >10, medium protection having factors between 7 and 10, and low protection with factors of <7. These divisions are illustrated on a structure of the fully folded myoglobin protein in Fig. 5a. Similarly, the calculated proton occupancy at the shortest folding time (6.4 ms) can be used to classify the kinetic data of Fig. 4. Previously, these classifications were defined as fast (F), biphasic (F+S), and slow (S) (13). The F amides belong to residues that are protected within the dead time of the quench-flow apparatus and show a proton occupancy of ≈1.0 throughout the time course of folding. The S amides are protected on a slower time scale, giving rise to an S-shaped curve from a proton occupancy value of ≈0.2 at short refolding times to a final value of 1.0 at long refolding times. The F+S amides show evidence of heterogeneity in their behavior: in some apoMb molecules in the conformational ensemble these amides are protected early, whereas in other molecules they are protected later. The points in Fig. 4 have been divided into the following three categories: F amides with A0 > 0.7, S amides with A0 < 0.4, and F+S amides with 0.4 < A0 < 0.7, and their locations are plotted onto a structure of the folded myoglobin in Fig. 5b. First, it is clear that the overall distribution of well protected amides (red spheres) is the same in the equilibrium and kinetic intermediates, but the degree of protection differs in some regions between the equilibrium and kinetic intermediates. This difference is greatest in the G helix, where the highly protected region is considerably shorter in the equilibrium intermediate (Fig. 5a) than in the kinetic intermediate (Fig. 5b). Similarly, the kinetic intermediate shows high protection in a number of residues in the B helix, whereas in the equilibrium intermediate only two residues, L29 and I30, are highly protected. Residues in the E helix that are in the F+S category in the kinetic intermediate (Fig. 4) appear to be uniformly lowered in protection in the equilibrium intermediate: the entire length of this helix is in the low-protection category (Fig. 2). By contrast, it appears that a small area of the D helix has been stabilized in the equilibrium intermediate; this finding is consistent with the observation of a propensity for nonnative helical structure in the D helix in apoMb unfolded at pH 2 (31).
Fig. 5.
Comparison of amide proton protection in the equilibrium intermediate at pH 4 and in the burst-phase intermediate, plotted onto the fully folded structure of myoglobin. (a) Amide proton protection measured in DMSO, using the interrupted H/D-exchange method. Red spheres show amides with high protection (protection factor of >10 in Fig. 2), green spheres indicate amides with medium protection (7 < protection factor < 10), and blue spheres show amides with low protection (protection factor of <7). (b) Amide proton protection in the burst-phase kinetic intermediate, measured in DMSO after the quench-flow pH-pulse-labeling experiment. Red spheres show high protection (proton occupancy >0.7 in Fig. 4), green spheres show medium protection (0.4 < proton occupancy <0.7), and blue spheres indicate low occupancy (proton occupancy <0.4).
Overall, the equilibrium intermediate appears to be less stable than the kinetic intermediate, particularly in the B, E, and G helices. We ascribe this difference primarily to the difference in the solution conditions of the two measurements. The kinetic folding intermediate is examined at a final pH of 6, whereas the measurements on the equilibrium intermediate are conducted at pH 4. This difference in pH probably accounts for the fraying of the structure at the ends of the helices and for the lowered stability of the B, E, and G helices. Fraying at the N terminus of the G helix and the C terminus of the H helix is observed even in the native apoprotein at pH 6, presumably related to the conformational exchange of the F helix in the absence of the heme prosthetic group (30, 32). However, we also observed destabilization in the pH 4 equilibrium intermediate of both the C terminus of the G helix (H113-H116, with no probes from S117 onward) and the N-terminal part of the B helix (A22-Q26). This area is precisely the region of the protein where a H-bond (between the imidazole side chain of H24 and the protonated imidazole side chain of H119) has been proposed as the triggering event for the folding of apoMb (33, 34). This hypothesis is quite consistent with our results for the pH 4 equilibrium intermediate. In the native state at pH 6, the H24-H119 H-bond stabilizes the two helical regions, A22-Q26 in the B-helix and H113-H116 in the G-helix. At pH 4, the H-bond is broken, due to protonation of H24; this result is reflected in the specific destabilization that we observe in the equilibrium intermediate (at pH 4) but not in the kinetic intermediate (at pH 6).
Conclusions
For apoMb, the use of the polar aprotic solvent DMSO gives an enhanced picture of the folding of the protein, especially in areas that have been invisible up to now as a result of rapid amide proton exchange rates in the fully folded holoprotein. Our picture of the folding pathway of wild-type sperm whale apoMb remains the same as that inferred from previous, more limited data sets. However, taking advantage of the greatly increased density of information now available using the DMSO technique, we have been able to make a detailed comparison of the equilibrium and kinetic intermediates for apoMb. The overall patterns of labeling and secondary structure formation remain similar between these two species, but local differences are observed, probably due to the difference in the pH. This method ought to be useful for studies of site-directed mutants, where the effects of the mutation may not be observable in areas that contain amide proton probes in the D2O-based experiment. We also anticipate wide application for comprehensive H/D exchange and folding kinetic studies on proteins that are intrinsically unstable and do not have many amide proton probes that can be used for classical quench-flow studies.
Acknowledgments
We thank John Chung and Gerard Kroon for help with NMR spectroscopy and Linda Tennant for technical assistance. This work was supported by National Institutes of Health Grant DK34909.
Author contributions: P.E.W. designed research; C.N. performed research; C.N. and H.J.D. analyzed data; and H.J.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: apoMb, apomyoglobin; HSQC, heteronuclear single quantum correlation.
References
- 1.Udgaonkar, J. B. & Baldwin, R. L. (1988) Nature 335, 694-699. [DOI] [PubMed] [Google Scholar]
- 2.Roder, H., Elöve, G. A. & Englander, S. W. (1988) Nature 335, 700-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavagnero, S., Thériault, Y., Narula, S. S., Dyson, H. J. & Wright, P. E. (2000) Protein Sci. 9, 186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332-336. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi, K., Katou, H., Hoshino, M., Hasegawa, K., Naiki, H. & Goto, Y. (2004) J. Mol. Biol. 338, 559-571. [DOI] [PubMed] [Google Scholar]
- 6.Griko, Y. V., Privalov, P. L., Venyaminov, S. Y. & Kutyshenko, V. P. (1988) J. Mol. Biol. 202, 127-138. [DOI] [PubMed] [Google Scholar]
- 7.Hughson, F. M., Wright, P. E. & Baldwin, R. L. (1990) Science 249, 1544-1548. [DOI] [PubMed] [Google Scholar]
- 8.Eliezer, D., Chung, J., Dyson, H. J. & Wright, P. E. (2000) Biochemistry 39, 2894-2901. [DOI] [PubMed] [Google Scholar]
- 9.Jennings, P. A. & Wright, P. E. (1993) Science 262, 892-896. [DOI] [PubMed] [Google Scholar]
- 10.Tsui, V., Garcia, C., Cavagnero, S., Siuzdak, G., Dyson, H. J. & Wright, P. E. (1999) Protein Sci. 8, 45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, C., Nishimura, C., Cavagnero, S., Dyson, H. J. & Wright, P. E. (2000) Biochemistry 39, 11227-11237. [DOI] [PubMed] [Google Scholar]
- 12.Cavagnero, S., Dyson, H. J. & Wright, P. E. (1999) J. Mol. Biol. 285, 269-282. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura, C., Dyson, H. J. & Wright, P. E. (2002) J. Mol. Biol. 322, 483-489. [DOI] [PubMed] [Google Scholar]
- 14.Jamin, M. & Baldwin, R. L. (1998) J. Mol. Biol. 276, 491-504. [DOI] [PubMed] [Google Scholar]
- 15.Jamin, M., Yeh, S. R., Rousseau, D. L. & Baldwin, R. L. (1999) J. Mol. Biol. 292, 731-740. [DOI] [PubMed] [Google Scholar]
- 16.Uzawa, T., Akiyama, S., Kimura, T., Takahashi, S., Ishimori, K., Morishima, I. & Fujisawa, T. (2004) Proc. Natl. Acad. Sci. USA 101, 1171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings, P. A., Stone, M. J. & Wright, P. E. (1995) J. Biomol. NMR 6, 271-276. [DOI] [PubMed] [Google Scholar]
- 18.Bai, Y., Milne, J. S., Mayne, L. & Englander, S. W. (1993) Proteins 17, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grzesiek, S. & Bax, A. (1992) J. Magn. Reson. 96, 432-440. [Google Scholar]
- 20.Löhr, F. & Rüterjans, H. (1995) J. Biomol. NMR 6, 189-197. [DOI] [PubMed] [Google Scholar]
- 21.Grzesiek, S. & Bax, A. (1992) J. Magn. Reson. 99, 201-207. [Google Scholar]
- 22.Grzesiek, S. & Bax, A. (1992) J. Am. Chem. Soc. 114, 6291-6293. [Google Scholar]
- 23.Yao, J., Dyson, H. J. & Wright, P. E. (1997) FEBS Lett. 419, 285-289. [DOI] [PubMed] [Google Scholar]
- 24.Marion, D., Ikura, M., Tschudin, R. & Bax, A. (1989) J. Magn. Reson. 85, 393-399. [Google Scholar]
- 25.Delaglio, F., Grzesiek, S., Vuister, G. W., Guang, Z., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 604-613. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y. Z., Paterson, Y. & Roder, H. (1995) Protein Sci. 4, 804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranker, A., Robinson, C. V., Radford, S. E., Aplin, R. T. & Dobson, C. M. (1993) Science 262, 896-900. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura, C., Prytulla, S., Dyson, H. J. & Wright, P. E. (2000) Nat. Struct. Biol. 7, 679-686. [DOI] [PubMed] [Google Scholar]
- 30.Eliezer, D. & Wright, P. E. (1996) J. Mol. Biol. 263, 531-538. [DOI] [PubMed] [Google Scholar]
- 31.Yao, J., Chung, J., Eliezer, D., Wright, P. E. & Dyson, H. J. (2001) Biochemistry 40, 3561-3571. [DOI] [PubMed] [Google Scholar]
- 32.Lecomte, J. T., Sukits, S. F., Bhattacharjya, S. & Falzone, C. J. (1999) Protein Sci. 8, 1484-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geierstanger, B., Jamin, M., Volkman, B. F. & Baldwin, R. L. (1998) Biochemistry 37, 4254-4265. [DOI] [PubMed] [Google Scholar]
- 34.Hennig, M. & Geierstanger, B. H. (1999) J. Am. Chem. Soc. 121, 5123-5126. [Google Scholar]