Abstract
Polymerase δ-interacting protein 2 (Poldip2) is a multi-functional protein originally described as a binding partner of the p50 subunit of DNA polymerase δ and proliferating cell nuclear antigen. In addition to its role in DNA replication and damage repair, Poldip2 has been implicated in mitochondrial function, extracellular matrix regulation, cell cycle progression, focal adhesion turnover and cell migration. However, Poldip2 functions are incompletely understood. In this review, we discuss recent literature on Poldip2 tissue distribution, subcellular localization, and function. We also address the putative function of Poldip2 in cardiovascular disease, neurodegenerative conditions and in renal pathophysiology.
Keywords: Poldip2, extracellular matrix, focal adhesions, DNA repair, redox signaling
Introduction
Polymerase δ-interacting protein 2 (Poldip2, also known as PDIP38 and Mitogenin-1) was discovered a little over ten years ago as a 368 amino acid binding partner of the p50 subunit of DNA polymerase δ and proliferating cell nuclear antigen (PCNA).1 Structurally, Poldip2 contains an N-terminal mitochondrial targeting sequence and two main functional domains: a DUF525 domain and a hemimethylated YccV-like domain (Figure 1A).2 DUF525 is found in some F-box proteins and in the bacterial protein ApaG, in which it may be involved in protein-protein interaction and cation efflux, respectively. YccV, identified in many taxa, may bind to DNA and regulate its expression. Poldip2 is highly conserved in metazoans (Figure 1B), but absent in prokaryotes, plants and fungi. Because several bacterial ApaG proteins have been crystallized, the tertiary structure of the DUF525 domain can be modeled accurately (Figure 1C). Poldip2 is translated as a 42-kDa protein, but can be post-translationally modified to a 37 kDa form by cleavage of the mitochondrial-targeting sequence. Recent work has demonstrated that in addition to polymerase δ and PCNA, Poldip2 in fact has a variety of binding partners and a plethora of functions, many of which appear to be a consequence of the specific associated protein (Table 1).
Figure 1. Bioinformatic analysis of Poldip2.
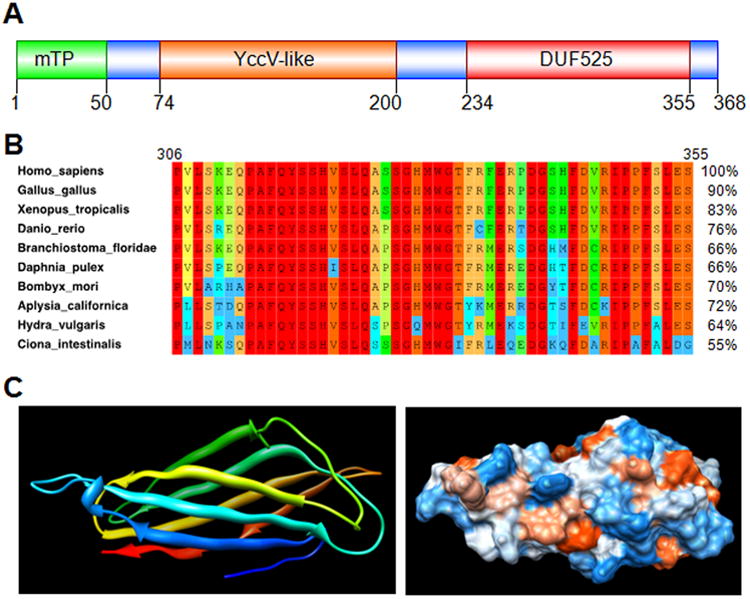
The entire human Poldip2 protein sequence (368 residues, NCBI accession number NP_056399.1) was used to search databases. A. Two domains were returned from the NCBI conserved domains database: the C-terminal DUF525domain, better conserved (E = 4.5e-32) than the upstream YccV-like domain (E = 1.3e-12). The N-terminus is predicted to be a cleavable mitochondrial targeting peptide (mTP) with near certainty, according to online software, such as TargetP,37, 38 Mitoprot39 and MitoFates.40 A BLAST search of the NCBI non-redundant protein sequences database returned about 600 homologous sequences from Eukaryotes (E < 1e-26). Each organism appeared to have a single gene homologous to Poldip2, suggesting that these hits are orthologs. Remarkably, BLAST detected no fungus, plant or prokaryote protein with homology to both conserved domains in Poldip2. These results suggest Poldip2 first appeared in Metazoans by fusion of two ancestral genes still found in bacteria, and separately coding for its conserved domains. Poldip2 is now present as a single gene in organisms as varied as sponges, sea anemones, molluscs, flat and round worms, arthropods and vertebrates. B. Ten Metazoan Poldip2 sequences were selected from BLAST results to represent a large variety of taxonomic groups. These proteins have a high percentage of identity to human Poldip2 in pairwise alignments, as shown on the right side of the panel. The left side shows a portion of a multiple sequence alignment including the last 50 residues of the DUF525 domain. Amino-acid conservation is indicated by color, from blue (10-19%) to red (100%). The rest of DUF525 and the YccV-like domain is also highly conserved across taxa (not shown). NCBI accession numbers for Homo sapiens: NP_056399.1; Gallus gallus: NP_001304285.1; Xenopus__tropicalis: NP_001017098.1; Danio__rerio: NP_997879.1; Branchiostoma__floridae: XP_002612268.1; Daphnia pulex: EFX67304.1; Bombyx_mori: XP_004926888.1; Aplysia_californica: XP_005102673.1; Hydra vulgaris: XP_002163974.2; Ciona_intestinalis: XP_002121208.2. C. Tertiary structure of the human Poldip2 DUF525 domain was predicted with similar results using two software packages: I-TASSER41-44 and Phyre2,45 based on homology to five solved ApaG structures available from the protein data bank. Homology to ApaG was sufficient to predict structure with high probability (estimated RMSD = 1.8±1.5Å in I-TASSER and 100% confidence in Phyre2). DUF525 is predicted to form a compact immunoglobulin-like beta-sandwich fold, which is represented using UCSF Chimera software,46 as ribbons colored blue to red from N to C-terminus (left) and as a space filling model with hydrophobicity increasing from blue to red (right). Hydrophobic residues at the surface of this domain could be involved in interactions with binding partners, or with other portions of Poldip2. One protein with homology to the human Poldip2YccV-like domain and of known structure is present in the protein data bank. However, relatively low homology does not allow structure prediction with high confidence (not shown).
Table 1.
Poldip2 binding partners Poldip2 binding partners reported in the literature. “Untested” indicates that the putative function has not been tested experimentally.
| Poldip2 binding protein | Putative function | Reference |
|---|---|---|
| Polymerase δ p50 subunit | DNA replication and damage repair | 1 |
| Proliferating cell nuclear antigen | DNA replication and damage repair | 1 |
| Polymerase η | DNA damage response | 22 |
| Rev1 polymerase | DNA damage response | 22 |
| Rev7 | DNA damage response | 22 |
| Polymerase λ | DNA damage response | 17 |
| PrimPol | DNA damage response | 21 |
| HPV 16 E7 | Viral DNA replication | 13 |
| p22phox | Reactive oxygen species production, cytoskeletal remodeling and cell migration; Modulation of transient receptor potential vanilloid 1 channel activity | 3,5 |
| CEACAM1 | Translocation from membrane to nucleus | 12 |
| Transcription factor A | mtDNA metabolism (untested) | 14 |
| Mitochondrial single stranded DNA binding protein 1 (mtSSB) | mtDNA metabolism (untested) | 14 |
| Leucine-rich protein (LPRC) | mtDNA metabolism (untested) | 14 |
| Lon protease homolog (LONM) | mtDNA metabolism (untested) | 14 |
| Heat shock protein 90-beta (HSP90β) | mtDNA metabolism (untested) | 14 |
| Heat shock protein 70 (HSP70) | mtDNA metabolism (untested) | 14 |
| ATP-dependent Clp protease (CLPX) | mtDNA metabolism (untested) | 14 |
| Heat shock protein 60 (HSP60) | mtDNA metabolism (untested) | 14 |
| ATP synthase beta chain (ATPB) | mtDNA metabolism (untested) | 14 |
| Squamous cell carcinoma antigen 1 (SCC1) | mtDNA metabolism (untested) | 14 |
| Squamous cell carcinoma antigen 2 (SCC2) | mtDNA metabolism (untested) | 14 |
| Aspartate aminotransferase (AATM) | mtDNA metabolism (untested) | 14 |
| Malate dehydrogenase (MDH) | mtDNA metabolism (untested) | 14 |
| 2,4-dienoyl-CoA reductase | mtDNA metabolism (untested) | 14 |
| Peroxiredoxin 3 (PDX3) | mtDNA metabolism (untested) | 14 |
| ATP synthase | mtDNA metabolism (untested) | 14 |
| Selenium binding protein 1 (SELENBP1) | Tumor suppressor (untested) | 47,48 |
| MAP3K7IP1 | TGFβ receptor and TAK1 signaling mediator (untested) | 47 |
| Histone deacetylase 6 (HDAC6) | Cancer metastasis (untested) | 47 |
| 3-Phosphoinositide Dependent Protein Kinase 1 (PDPK1) | Cancer metastasis (untested) | 47 |
| Activating signal cointegrator 1 complex subunit 2 (ASCC2) | Activation by multiple transcription factors (untested) | 47,48 |
| Carboxypeptidase E (CPE) | Biosynthesis of peptide hormones and neuropeptides (untested) | 48 |
| 5′-aminolevulinate synthase 1 (ALAS1) | Mitochondrial metabolism (untested) | 48 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 (ATP5A1) | ATP synthesis (untested) | 49 |
| chromosome 15 open reading frame 48 (C15orf48) | Carcinogenesis (untested) | 49 |
Tissue expression
Poldip2 is fairly ubiquitously expressed, having been reported in aorta,3, 4 lung,3 kidney,3, 5, 6 heart7 and thyroid,8 as well as many cell types including mouse aortic smooth muscle cells,9 astrocytes,10 vascular smooth muscle cells (VSMC),3, 11 brain endothelial cells,12 epithelial cells,12 HeLa,1, 12-15 fibroblasts,2, 13, 16, 17 kidney fibroblasts,18 myoblasts,19 human umbilical vein endothelial cells,20 human embryonic kidney cells 293 (HEK 293),13 MRC5 cells,21, 22 and cortical neurons.23 There is also evidence of Poldip2 expression in human breast cancer,24 human breast adenocarcinoma cells13 and human lung adenocarcinoma cells.15 Of note, Poldip2 expression in myeloid cells is very low.3
Subcellular localization
In vascular smooth muscle cells (VSMC), Poldip2 has a nuclear and cytoplasmic distribution, specifically in focal adhesions and in stress fibers.3 Poldip2 has also been identified in the mitochondria of myoblast C2C12 cells,19 and in the nucleus and mitochondria of HeLa cells.13, 14 In rat bladder carcinoma NBT-II cells, intestinal epithelial IEC18 cells, rat brain endothelial cells and HeLa cells, Poldip2 does not localize to the mitochondria.12 Instead, at least in NBT-II cells, its subcellular location seems to be regulated by the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in a cell cycle dependent manner. In non-proliferating, quiescent NBT-II cells, CEACAM1 antibody clustering induces Poldip2 translocation from the cytosol to the nucleus. In contrast, in subconfluent, proliferating NBT-II cells, CEACAM1 antibody clustering dramatically increases Poldip2 translocation from the cytoplasm to the plasma membrane.12 This raises the intriguing possibility that Poldip2 may regulate different cellular functions depending upon its subcellular localization as well as its binding partners.
Functions of Poldip2
It may seem peculiar for Poldip2 to be present in different subcellular compartments, where it appears to perform disparate functions. However, other pleiotropic proteins with similar localizations have been previously reported. For example, some families of LIM domain proteins are known to shuttle between the cytoplasm and the nucleus, where they regulate the cytoskeleton and gene transcription, respectively.25 These include the focal adhesion protein paxillin, which is important in cell migration and also interacts with nuclear androgen and glucocorticoid receptors. Similarly, CSRP1-3 bind to cytoskeletal proteins, such as α-actinin and nuclear transcription factors, including SRF, GATA and myogenin, thereby regulating actin dynamics and promoting expression of differentiation markers in smooth and striated muscle.25-29 Likewise, LIM kinases phosphorylate cofilin, an actin disassembly factor, and perform nuclear functions.25 Besides LIM domain family members, other pleiotropic proteins include the serine/threonine protein phosphatase PPP4C which regulates microtubule organization,30 as well as DNA repair.31 As a last example, the LPP protein shuttles between focal adhesions and the nucleus, thereby affecting both peripheral actin organization and nuclear transcription.32, 33 Thus, in addition to performing specific localized functions, Poldip2 may allow communication between subcellular compartments and enable coordinated regulation of architecture, signaling and metabolic pathways at the cellular level. However, in contrast to other pleiotropic proteins mentioned above, Poldip2 does not belong to a family, but appears to be a unique gene, performing functions that cannot be compensated by homologues, as suggested by the embryonic lethality of homozygous null mice.4 Nevertheless, the functions of Poldip2 are incompletely understood and current knowledge thus derives from indirect evidence and from a few published functional studies.
DNA repair
As noted, Poldip2 was originally described as a binding partner of p50, one of the polymerase δ subunits. Using a yeast two-hybrid screen with the p50 subunit of polymerase δ as a bait, Liu et al.1 identified Poldip2 as a binding partner and confirmed the interaction using coimmunoprecipitation and GST pull-down assays. They also found an interaction between Poldip2 and proliferating cell nuclear antigen (PCNA),1 suggesting that Poldip2 could potentially function as a nuclear protein involved in DNA replication and damage repair.
Subsequent research identified Poldip2 as a binding partner of the translesional DNA synthesis (TLS) polymerase η. Following DNA damage, a switch between replicative polymerases and TLS polymerases enables lesion bypass to occur. The interaction between Poldip2 and polymerase η was shown to occur at the ubiquitin-binding zinc finger domain of polymerase η and seems to modulate the focal localization of polymerase η. Depletion of Poldip2 increases the percentage of human fibroblast-derived MRC5 cells that form polymerase η nuclear foci at baseline and reduces cell survival following UV treatment. The authors proposed that Poldip2 is involved in the processing of the DNA damage response, contributing to the switch between replicative polymerases and TLS polymerases at damaged sites.22 Similarly, Poldip2 regulates the TLS polymerase and primer extension activities of PrimPol, a member of the archaeo-eukaryotic primase superfamily known to have both DNA primase and DNA polymerase activities. Poldip2 binds to the PrimPol catalytic domain and stimulates its activity by increasing PrimPol DNA binding. In addition, Poldip2 stimulates the efficiency and error-free 8-oxo-7,8-dihydroguanine (8-oxo-G)DNA lesion bypass by PrimPol.21 However, another study suggested that Poldip2 does not directly participate in the process of TLS, because it is not recruited to sites of DNA damage upon UV stimulation. Instead, Poldip2 translocates to spliceosomes and contributes to UV-induced MDM2 alternative splicing. Because Poldip2 lacks a canonical RNA binding domain, the authors of that study speculated that Poldip2 might influence mRNA processing by serving as a protein scaffold to allow protein–protein interactions, thus facilitating splicing events.15
In addition, Poldip2 has been shown to interact with the catalytic domain of TLS DNA polymerase λ. Poldip2 interaction with polymerase λ, as well as with polymerase η, increases the efficiency of error-free 8-oxo-G DNA lesion bypass.17 The same response was also observed with two other common DNA lesions, abasic site and cyclobutane thymine dimers. siRNA against Poldip2 increases mouse embryonic fibroblast sensitivity to oxidative stress induced by H2O2, similar to what was observed in polymerase λ−/− cells.17 As described below, Poldip2 can also increase H2O2 production by Nox4 and has been shown to associate with Nox4 in the nucleus.3 It is therefore intriguing to note that by promoting DNA repair, nuclear Poldip2 may prevent the accumulation of oxidative DNA lesions its own signaling functions might have caused.
Mitochondrial function
In 2005, a few years following the discovery that Poldip2 binds to p50 and PCNA, it was discovered that Poldip2 has a mitochondrial targeting sequence located within its first 35 N-terminal amino acid residues, suggesting that it might have functions other than associating with polymerase δ and PCNA.13 Indeed, almost simultaneously, two different research groups showed that Poldip2 is involved in mitochondrial function and morphology. In the first study, Cheng et al.14 found that Poldip2 localizes in the mitochondrial matrix and associates with components of the mitochondrial protein-DNA complex nucleoid. Almost immediately thereafter, in 2006, Poldip2 overexpression in central nervous system catecholaminergic neurons and in human umbilical endothelial cells was demonstrated to increase the number of elongated mitochondria, while silencing Poldip2 induced the fragmentation of mitochondria. Although the mechanism of how Poldip2 is involved in mitochondrial morphology is not well defined, the authors of this study suggested that Poldip2 plays a role in mitochondrial fusion.19
Even though significant evidence supports a role for Poldip2 in mitochondria, immunofluorescence studies using MitoTracker Red CMXRos indicate that Poldip2 does not colocalize with mitochondria in NBT-II, IEC18, rat brain endothelial cells or HeLa cells. Subcellular fractionation revealed a predominant cytoplasmic localization (68%), while nuclear fraction corresponded to 20% and mitochondrial fraction represented 11% of the total amount of Poldip2 in both NBT-II and HeLa cells.12 This finding is consistent with the growing number of proteins associated with Poldip2, and with the possibility that the functional roles of Poldip2 are compartment-specific. It seems likely that the subcellular localization of Poldip2 may differ depending on the cell type.
Proliferation
In parallel with the progress made toward understanding the involvement of Poldip2 in mitochondrial morphology, evidence suggested that the subcellular distribution of Poldip2 is altered during cell cycle progression. In myoblasts, Poldip2 was shown to be localized in the nucleus during G2/M to late G1 phase, which suggests a role in the cell cycle.19 Subsequent studies confirmed Poldip2 localization in the nucleus,3, 12 and, recently, our research group discovered that Poldip2 has multiple roles in cell division. Poldip2-/- mouse embryonic fibroblasts (MEFs) exhibit markedly reduced growth when compared to Poldip2+/+ cells. Using flow cytometry, we found that Poldip2-/-MEFs are arrested or delayed in both G1 and G2/M phases of the cell cycle, therefore decreasing the number of cells in S-phase. When assessing the protein levels of key cell cycle regulators, cdk1 and Cyclin A2 were found to be decreased in later passage Poldip2-/- MEFs compared to wild-type cells. Total p53and Rb, as well as phosphorylated p53 and Rb levels, were also investigated because these proteins have been shown to cause a delay in G1 and G2/M. Poldip2 depletion induced no change in Rb, but increased p53 phosphorylation at serine 20, as well as its transcriptional target, Sirt1, at early passages, and increased the expression of the cell cycle inhibitor p21Cip1 in late passages. Thus, Poldip2 increases the transcription of Cyclin A, Cdk1 and PCNA and decreases the expression of p21Cip1, promoting cell cycle progression.2
The exact mechanism by which Poldip2 regulates Cyclin A, Cdk1 and PCNA remains to be deciphered. However, because each of these genes is regulated by E2F, which remains inactive when bound to Rb, one possibility is that Poldip2 somehow disrupts E2F-Rb binding, releasing E2F transcriptional activity. In support of this possibility, SV40 immortalization of Poldip2-/- MEFs restored growth and cell cycle distribution to a wild type pattern,2 consistent with studies performed in SV40-transformed human fibroblasts in which siRNA against Poldip2 has no impact on cell cycle progression.22
Earlier studies in primary rat brain endothelial cells uncovered an additional mechanism by which Poldip2 can affect proliferation. Klaile et al.12 injected Poldip2 antibody or delivered siRNA against Poldip2 into these cells and observed disorganized spindles, disrupted chromosomal segregation and multinucleated cells. Such observations are consistent with a role for Poldip2 in cytoskeletal dynamics, as described in the next section.
Oxidative signaling and cell migration
Our laboratory became interested in Poldip2 and its regulation of cell migration in an indirect way. Using a yeast 2-hybrid screen of vascular smooth muscle cell (VSMC) proteins, we found that Poldip2 interacts with the C-terminal tail of p22phox, a NADPH oxidase (Nox) subunit.3 This interaction was verified in VSMC using GST pull-down and coimmunoprecipitation assays. Further characterization revealed that Poldip2 overexpression specifically increases the activity of the Nox4 enzyme, which produces reactive oxygen species (ROS), whereas Poldip2 depletion reduces basal ROS production. Interestingly, Poldip2, p22phox and Nox4 are colocalized at focal adhesions and stress fibers. Furthermore, manipulation of Poldip2 levels dramatically affects the cytoskeleton. Poldip2 overexpression impairs focal adhesion dissolution and markedly thickens the actin stress fiber network. Conversely, depletion of Poldip2 using siRNA, inhibits focal adhesion and stress fiber formation. These effects of Poldip2 are part of an oxidative signaling pathway mediated by p22phox, Nox4, the focal adhesion kinase FAK and the small GTPase RhoA, a master regulator of the actin cytoskeleton. Indeed, Poldip2 overexpression increases RhoA activity and cytoskeletal changes induced by Poldip2 overexpression are abrogated by depletion of p22phox, Nox4 or inhibition of RhoA. Conversely, alterations induced by Poldip2 depletion can be mimicked by depletion of p22phox or Nox4 and rescued by overexpression of FAK or constitutively active RhoA. Cell migration requires a dynamic rearrangement of actin stress fibers and continuous focal adhesion turnover, with new focal adhesions forming at the front of the cell and old ones dissolving at the rear. Both Poldip2 overexpression and depletion block PDGF-induced VSMC migration, due either to lack of focal adhesion dissolution or formation, respectively.3, 11 These results suggest that Poldip2 dynamically regulates Nox4 and its downstream effectors in different subcellular compartments to allow coordinated cytoskeletal changes required for cell migration.
Extracellular matrix regulation
Evidence for a role of Poldip2 in regulation of extracellular matrix comes from animal studies. Deletion of Poldip2 is perinatal lethal,2 but Poldip2 heterozygous mice exhibit altered arterial structure.4 Aortas isolated from Poldip2 heterozygous animals have increased extracellular matrix, with fragmented and disordered elastic laminae. Using Poldip2 siRNA in VSMC, we found that Poldip2 depletion increases both fibronectin and collagen secretion, which is abolished by treatment with glucose oxidase, suggesting that increased collagen secretion induced by lack of Poldip2 results from lower H2O2 levels.4
In a recent study, the mechanism responsible for collagen I accumulation was investigated in further detail in Poldip2+/− mouse aortic smooth muscle cells (MASMs). Corroborating previous work,4 collagen I secretion was found to be significantly increased under basal conditions in Poldip2+/− MASMs. Neither accumulation of ubiquitinated proteins nor autophagy is responsible for collagen accumulation stimulated by Poldip2 depletion, because collagen protein degradation is actually increased in these cells. Interestingly, the activity of the PI3K/Akt/mTOR signaling pathway, which is involved in regulation of protein synthesis,34 is upregulated following Poldip2 depletion in MASMs, suggesting that activation of this signaling pathway ultimately resulted in increased collagen synthesis and accumulation.9 This conclusion was confirmed by the ability of an Akt inhibitor and rapamycin to significantly inhibit collagen accumulation in Poldip2-depleted cells.
In addition to direct regulation of extracellular matrix protein synthesis, Poldip2 also regulates matrix degradation. The activity of both MMP-2 and MMP-9 is reduced in Poldip2+/− mice 21 days after hindlimb ischemia induction, which is associated with impaired neovascularization.20 Similar results were observed in ischemic tissue after stroke.10
Physiology and Pathophysiology of Poldip2
Role in the cardiovascular system
One of the earliest phenotypes described in mice heterozygous for deletion of Poldip2 was disorganization of aortic structure. Aortas from Poldip2+/- mice not only have increased collagen deposition as described above, but also exhibit disordered and broken elastic laminae. As a consequence, contraction to phenylephrine and potassium chloride is impaired, aortic compliance is reduced and vascular stiffness increases, with the latter considered to be a major cardiovascular risk factor.4
Extensive research has focused on identifying signaling pathways involved in the phenotypic diversity of VSMC. In these cells, the switch from a quiescent contractile phenotype to a synthetic, migratory phenotype is a physiological process that takes place during vascular development or tissue repair in response to vascular lesion.35 However, migration of VSMC is also involved in the pathophysiology of vascular diseases, including atherosclerosis and restenosis. As noted above, Poldip2 is involved not only in VSMC migration, but it also regulates cell proliferation and collagen synthesis, suggesting that it may contribute to proliferative vascular conditions such as restenosis and atherosclerosis. In fact, Poldip2+/- mice show less neointimal formation following wire injury of the femoral artery.11 However, the potential role of Poldip2 in atherosclerosis is likely more complex. Increased collagen deposition following Poldip2 heterozygous deletion might, for example, increase atherosclerotic plaque size. On the other hand, stabilizing of a vulnerable plaque by increasing the thickness of the collagen I-formed fibrous cap might be a promising strategy for preventing plaque rupture and therefore for preventing acute coronary syndrome and stroke. Further work is necessary to define the potential role of Poldip2 in atherosclerosis.
It is noteworthy that human aortic valve stenosis has been shown to be accompanied by a decrease in both Poldip2 and Nox4 protein expression, indicating that these proteins might be involved in the remodeling of the myocardium following human cardiac pressure overload secondary to aortic valve stenosis.7 Poldip2 has also been shown to promote ischemia-induced collateral vessel formation. Poldip2 heterozygous deletion impairs ischemia-induced revascularization following femoral artery ligation in mice.20 H2O2 production, macrophage infiltration and matrix MMP-2 and -9 activity are reduced in Poldip2+/- mice compared to Poldip2+/+ mice, suggesting that Poldip2 has multiple roles in the response to ischemia and is essential for the neovascularization process.20
Role in the central nervous system
The role of Poldip2 in the central nervous system has not been well studied, and is in fact limited to two investigations. The first involves the microtubule stabilizer protein Tau. Tau aggregation and consequent neurodegeneration is evident in a variety of neurodegenerative conditions, such as Alzheimer's disease, Parkinson's disease, Huntington's disease and amyotrophic lateral sclerosis.23 Interestingly, siRNA against Poldip2 alleviates H2O2-induced Tau aggregation in SH-SY5Y human neuroblastoma cells. Poldip2 does not influence Tau phosphorylation, but rather regulates Tau degradation via inhibition of autophagy. These results suggest that inhibiting Poldip2 may be an interesting strategy for novel treatments of neurodegenerative diseases, although much work remains to be done to determine the exact molecular mechanism by which Poldip2 inhibition exerts its beneficial effect.
The second study, published so far only in abstract form, addresses the role of Poldip2 in the permeability of the blood-brain-barrier (BBB) following brain ischemia. Our lab has demonstrated that cerebral ischemia induces the expression of Poldip2 in perivascular astrocytes. Poldip2+/- animals exhibit a significant decrease in Evans blue dye extravasation, a marker of increased permeability of the BBB, and have improved survival 24 hr following cerebral ischemia induction by transient middle cerebral artery occlusion. TNF-α, IL-6, MMP-2 and MMP-9 expression induced by cerebral ischemia is abrogated in Poldip2+/- mice compared to Poldip2+/+ animals.10 These results indicate that Poldip2 has a number of potential targets in the response to ischemia and represent a promising new area of investigation.
Role in the kidney
Investigation into the role of Poldip2 in the kidney has largely been based on the experimental evidence that Poldip2 is a Nox4 binding partner. In collaboration with our research group, Manickam et al.18 recently explored the role of Rac1 and RhoA/ROCK in Poldip2/Nox4 signaling in kidney myofibroblasts. Poldip2 significantly increases TGFβ-induced Nox4 activity and the expression of α-smooth muscle actin (α-SMA) and Fn-EIIIA, leading to myofibroblast differentiation through a redox-dependent signaling cascade that involves TGFβ receptor1 and RhoA/ROCK/Nox4/Poldip2, but not Rac1. Lin et al.5 investigated the role of the Nox4/p22phox/Poldip2 interaction on the activity of renal transient receptor potential vanilloid 1 channels. They found that Poldip2 co-immunoprecipitates with both Nox4 and p22phox in renal pelvic lysates and that the expression of Poldip2 is increased upon elevation of intrapelvic pressure (IPP). IPP increases Nox4/p22phox/Poldip2-derived H2O2 generation from sensory nerve terminals, which then activates TRPV1 channels and stimulates substance P release into the pelvic space. The role of Poldip2/Nox4 has also been investigated in the PKC-dependent ROS production induced by increased luminal flow in the renal thick ascending limb, where the function of these two proteins appears to be distinct. Poldip2 and Nox4 displayed a high degree of cytoplasmic colocalization in baseline conditions; however, Nox4 translocation to the nucleus in response to increased luminal flow was not associated with Poldip2 translocation.
Poldip2 and cancer
The role of Poldip2 in carcinogenesis remains largely unknown. Although there is no direct evidence that Poldip2 plays a pathophysiological role in cancer development, a study of the sense-antisense gene pair of TNFAIP1/POLDIP2 revealed that the expression of those genes is strongly correlated with breast cancer tumors.24 In contrast, Poldip2 mRNA expression in peripheral blood is inversely correlated with the risk of developing lung cancer.36 Given the functional data linking Poldip2 to cell proliferation, migration and matrix production, future studies of this protein in cancer are warranted.
Conclusions and future directions
Our understanding of the structure and function of Poldip2 remains at best rudimentary. To our knowledge, this review touches upon all of the publications focused on this fascinating protein to date, but it is evident that much more remains to be done. The biochemistry of Poldip2 is completely untouched, and there are likely many more binding partners yet to be uncovered. More work is also necessary to define the molecular mechanisms by which Poldip2 exerts its myriad effects. Very little is known about how intra- and extracellular environmental factors affect Poldip2 expression, localization and function. In fact, there have been no studies on the transcriptional/post-transcriptional regulation of Poldip2. It remains unclear if the two forms of Poldip2 (37-kDa and 42-kDa) have distinct functions. Finally, the physiology and pathophysiology of Poldip2 is also an area ripe for investigation. Given its expression profile and it role in basic cellular functions such as proliferation, migration and matrix production, it is likely to be involved in the physiology and diseases of the lung and liver as well as the kidney, cardiovascular system and nervous system (Figure 2). With the techniques now available in systems biology, and the availability of Poldip2 genetically modified mice, there is great opportunity for progress in understanding the many fundamental roles of Poldip2.
Figure 2. Poldip2 signal transduction pathways.
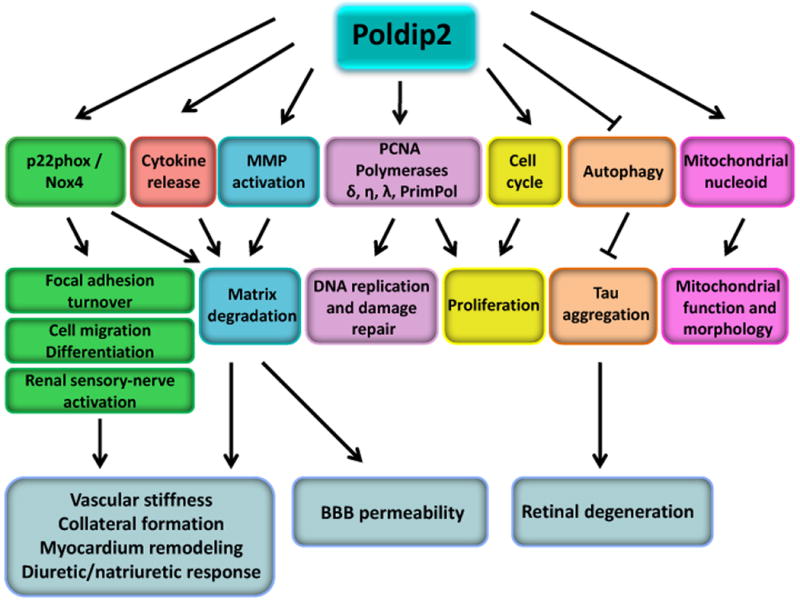
Poldip2 by itself or via interaction with its binding partners is involved in multiple signaling pathways, modulating focal adhesion turnover, ECM regulation, cell migration, proliferation and differentiation, renal sensory-nerve activation, DNA replication and damage repair, mitochondrial function and morphology and autophagy. Those processes contribute to vascular stiffness, collateral formation, myocardium remodeling, the diuretic/natriuretic response, BBB permeability and retinal degeneration. BBB, blood-brain-barrier; PCNA, proliferating cell nuclear antigen; MMPs, matrix metalloproteinases.
Acknowledgments
Disclosure of funding received for this work: This work was supported by NIH grants HL038206 and HL095070 to KKG and BL and FAPESP to MSH.
References
- 1.Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. The Journal of biological chemistry. 2003;278:10041–10047. doi: 10.1074/jbc.M208694200. [DOI] [PubMed] [Google Scholar]
- 2.Brown DI, Lassegue B, Lee M, Zafari R, Long JS, Saavedra HI, Griendling KK. Poldip2 knockout results in perinatal lethality, reduced cellular growth and increased autophagy of mouse embryonic fibroblasts. PLoS One. 2014;9:e96657. doi: 10.1371/journal.pone.0096657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyle AN, Deshpande NN, Taniyama Y, Seidel-Rogol B, Pounkova L, Du P, Papaharalambus C, Lassegue B, Griendling KK. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. 2009;105:249–259. doi: 10.1161/CIRCRESAHA.109.193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutliff RL, Hilenski LL, Amanso AM, Parastatidis I, Dikalova AE, Hansen L, Datla SR, Long JS, El-Ali AM, Joseph G, Gleason RL, Jr, Taylor WR, Hart CM, Griendling KK, Lassegue B. Polymerase delta interacting protein 2 sustains vascular structure and function. Arterioscler Thromb Vasc Biol. 2013;33:2154–2161. doi: 10.1161/ATVBAHA.113.301913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin CS, Lee SH, Huang HS, Chen YS, Ma MC. H2O2 generated by NADPH oxidase 4 contributes to transient receptor potential vanilloid 1 channel-mediated mechanosensation in the rat kidney. Am J Physiol Renal Physiol. 2015;309:F369–376. doi: 10.1152/ajprenal.00462.2014. [DOI] [PubMed] [Google Scholar]
- 6.Saez F, Hong NJ, Garvin JL. Luminal flow induces NADPH oxidase 4 translocation to the nuclei of thick ascending limbs. Physiol Rep. 2016;4 doi: 10.14814/phy2.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno MU, Gallego I, Lopez B, Gonzalez A, Fortuno A, San Jose G, Valencia F, Gomez-Doblas JJ, de Teresa E, Shah AM, Diez J, Zalba G. Decreased Nox4 levels in the myocardium of patients with aortic valve stenosis. Clin Sci (Lond) 2013;125:291–300. doi: 10.1042/CS20120612. [DOI] [PubMed] [Google Scholar]
- 8.Fortunato RS, Braga WM, Ortenzi VH, Rodrigues DC, Andrade BM, Miranda-Alves L, Rondinelli E, Dupuy C, Ferreira AC, Carvalho DP. Sexual dimorphism of thyroid reactive oxygen species production due to higher NADPH oxidase 4 expression in female thyroid glands. Thyroid. 2013;23:111–119. doi: 10.1089/thy.2012.0142. [DOI] [PubMed] [Google Scholar]
- 9.Fujii M, Amanso A, Abrahao TB, Lassegue B, Griendling KK. Polymerase delta-interacting protein 2 regulates collagen accumulation via activation of the Akt/mTOR pathway in vascular smooth muscle cells. J Mol Cell Cardiol. 2016;92:21–29. doi: 10.1016/j.yjmcc.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandes MSLB, Yepes M, Griendling K. Polymerase δ-Interacting Protein 2 Regulates Astrocyte Activation in Ischemic Stroke International Sroke Conference - American Heart Association. Los Angeles, CA: Stroke; 2016. [Google Scholar]
- 11.Datla SR, McGrail DJ, Vukelic S, Huff LP, Lyle AN, Pounkova L, Lee M, Seidel-Rogol B, Khalil MK, Hilenski LL, Terada LS, Dawson MR, Lassegue B, Griendling KK. Poldip2 controls vascular smooth muscle cell migration by regulating focal adhesion turnover and force polarization. Am J Physiol Heart Circ Physiol. 2014;307:H945–957. doi: 10.1152/ajpheart.00918.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klaile E, Muller MM, Kannicht C, Otto W, Singer BB, Reutter W, Obrink B, Lucka L. The cell adhesion receptor carcinoembryonic antigen-related cell adhesion molecule 1 regulates nucleocytoplasmic trafficking of DNA polymerase delta-interacting protein 38. The Journal of biological chemistry. 2007;282:26629–26640. doi: 10.1074/jbc.M701807200. [DOI] [PubMed] [Google Scholar]
- 13.Xie B, Li H, Wang Q, Xie S, Rahmeh A, Dai W, Lee MY. Further characterization of human DNA polymerase delta interacting protein 38. The Journal of biological chemistry. 2005;280:22375–22384. doi: 10.1074/jbc.M414597200. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Kanki T, Fukuoh A, Ohgaki K, Takeya R, Aoki Y, Hamasaki N, Kang D. PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid. J Biochem. 2005;138:673–678. doi: 10.1093/jb/mvi169. [DOI] [PubMed] [Google Scholar]
- 15.Wong A, Zhang S, Mordue D, Wu JM, Zhang Z, Darzynkiewicz Z, Lee EY, Lee MY. PDIP38 is translocated to the spliceosomes/nuclear speckles in response to UV-induced DNA damage and is required for UV-induced alternative splicing of MDM2. Cell Cycle. 2013;12:3184–3193. doi: 10.4161/cc.26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dosoki H, Stegemann A, Taha M, Schnittler H, Luger TA, Schroder K, Distler JH, Kerkhoff C, Bohm M. Targeting of NADPH oxidase in vitro and in vivo suppresses fibroblast activation and experimental skin fibrosis. Exp Dermatol. 2016 doi: 10.1111/exd.13180. [DOI] [PubMed] [Google Scholar]
- 17.Maga G, Crespan E, Markkanen E, Imhof R, Furrer A, Villani G, Hubscher U, van Loon B. DNA polymerase delta-interacting protein 2 is a processivity factor for DNA polymerase lambda during 8-oxo-7,8-dihydroguanine bypass. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18850–18855. doi: 10.1073/pnas.1308760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manickam N, Patel M, Griendling KK, Gorin Y, Barnes JL. RhoA/Rho kinase mediates TGF-beta1-induced kidney myofibroblast activation through Poldip2/Nox4-derived reactive oxygen species. Am J Physiol Renal Physiol. 2014;307:F159–171. doi: 10.1152/ajprenal.00546.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arakaki N, Nishihama T, Kohda A, Owaki H, Kuramoto Y, Abe R, Kita T, Suenaga M, Himeda T, Kuwajima M, Shibata H, Higuti T. Regulation of mitochondrial morphology and cell survival by Mitogenin I and mitochondrial single-stranded DNA binding protein. Biochim Biophys Acta. 2006;1760:1364–1372. doi: 10.1016/j.bbagen.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Amanso AM, Lassegue B, Joseph G, Landazuri N, Long JS, Weiss D, Taylor WR, Griendling KK. Polymerase delta-interacting protein 2 promotes postischemic neovascularization of the mouse hindlimb. Arterioscler Thromb Vasc Biol. 2014;34:1548–1555. doi: 10.1161/ATVBAHA.114.303873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilliam TA, Bailey LJ, Brissett NC, Doherty AJ. PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities. Nucleic Acids Res. 2016;44:3317–3329. doi: 10.1093/nar/gkw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tissier A, Janel-Bintz R, Coulon S, Klaile E, Kannouche P, Fuchs RP, Cordonnier AM. Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Poleta. DNA Repair (Amst) 2010;9:922–928. doi: 10.1016/j.dnarep.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Park H, Nah J, Moon S, Lee W, Hong SH, Kam TI, Jung YK. Essential role of POLDIP2 in Tau aggregation and neurotoxicity via autophagy/proteasome inhibition. Biochemical and biophysical research communications. 2015;462:112–118. doi: 10.1016/j.bbrc.2015.04.084. [DOI] [PubMed] [Google Scholar]
- 24.Grinchuk OV, Motakis E, Kuznetsov VA. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. 2010;11(1):S9. doi: 10.1186/1471-2164-11-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 26.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 27.Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Molecular and cellular biology. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louis HA, Pino JD, Schmeichel KL, Pomies P, Beckerle MC. Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. The Journal of biological chemistry. 1997;272:27484–27491. doi: 10.1074/jbc.272.43.27484. [DOI] [PubMed] [Google Scholar]
- 29.Papalouka V, Arvanitis DA, Vafiadaki E, Mavroidis M, Papadodima SA, Spiliopoulou CA, Kremastinos DT, Kranias EG, Sanoudou D. Muscle LIM protein interacts with cofilin 2 and regulates F-actin dynamics in cardiac and skeletal muscle. Molecular and cellular biology. 2009;29:6046–6058. doi: 10.1128/MCB.00654-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, Inagaki M, Hiraiwa N, Muramatsu M, Wynshaw-Boris A, Yoshiki A, Hirotsune S. Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. The Journal of cell biology. 2008;180:1133–1147. doi: 10.1083/jcb.200705148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Zhuang L, Reinhard M, Trueb B. The lipoma preferred partner LPP interacts with alpha-actinin. J Cell Sci. 2003;116:1359–1366. doi: 10.1242/jcs.00309. [DOI] [PubMed] [Google Scholar]
- 33.Petit MM, Fradelizi J, Golsteyn RM, Ayoubi TA, Menichi B, Louvard D, Van de Ven WJ, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis SF, Zahradka P. Vascular smooth muscle cell motility: From migration to invasion. Exp Clin Cardiol. 2010;15:e75–85. [PMC free article] [PubMed] [Google Scholar]
- 36.Chian CF, Hwang YT, Terng HJ, Lee SC, Chao TY, Chang H, Ho CL, Wu YY, Perng WC. Panels of tumor-derived RNA markers in peripheral blood of patients with non-small cell lung cancer: their dependence on age, gender and clinical stages. Oncotarget. 2016 doi: 10.18632/oncotarget.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European journal of biochemistry / FEBS. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 40.Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, Assmus HE, Andrade-Navarro MA, Wanker EE. A directed protein interaction network for investigating intracellular signal transduction. Sci Signal. 2011;4:rs8. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 48.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 49.Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, Gromek KA, Dolan BK, Ulbrich A, Stefely JA, Bohl SL, Werner KM, Jochem A, Westphall MS, Rensvold JW, Taylor RW, Prokisch H, Kim JJ, Coon JJ, Pagliarini DJ. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]


